Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is a serious public health issue globally, currently, the treatment of NAFLD lies still in the labyrinth. In the inchoate stage, the combinatorial application of food regimen and favorable gut microbiota (GM) are considered as an alternative therapeutic. Accordingly, we integrated secondary metabolites (SMs) from GM and Avena sativa (AS) known as potent dietary grain to identify the combinatorial efficacy through network pharmacology.
Methods
We browsed the SMs of AS via Natural Product Activity & Species Source (NPASS) database and SMs of GM were retrieved by gutMGene database. Then, specific intersecting targets were identified from targets related to SMs of AS and GM. The final targets were selected on NAFLD-related targets, which was considered as crucial targets. The protein–protein interaction (PPI) networks and bubble chart analysis to identify a hub target and a key signaling pathway were conducted, respectively. In parallel, we analyzed the relationship of GM or AS─a key signaling pathway─targets─SMs (GASTM) by merging the five components via RPackage. We identified key SMs on a key signaling pathway via molecular docking assay (MDA). Finally, the identified key SMs were verified the physicochemical properties and toxicity in silico platform.
Results
The final 16 targets were regarded as critical proteins against NAFLD, and Vascular Endothelial Growth Factor A (VEGFA) was a key target in PPI network analysis. The PI3K-Akt signaling pathway was the uppermost mechanism associated with VEGFA as an antagonistic mode. GASTM networks represented 122 nodes (60 GM, AS, PI3K-Akt signaling pathway, 4 targets, and 56 SMs) and 154 edges. The VEGFA-myricetin, or quercetin, GSK3B-myricetin, IL2-diosgenin complexes formed the most stable conformation, the three ligands were derived from GM. Conversely, NR4A1-vestitol formed stable conformation with the highest affinity, and the vestitol was obtained from AS. The given four SMs were no hurdles to develop into drugs devoid of its toxicity.
Conclusion
In conclusion, we show that combinatorial application of AS and GM might be exerted to the potent synergistic effects against NAFLD, dampening PI3K-Akt signaling pathway. This work provides the importance of dietary strategy and beneficial GM on NAFLD, a data mining basis for further explicating the SMs and pharmacological mechanisms of combinatorial application (AS and GM) against NAFLD.
Similar content being viewed by others
Background
Non-alcoholic fatty liver disease (NAFLD) is characterized by excessive fat accumulation in liver tissues, especially, in liver parenchyma [1]. Its pathophysiological spectrum is encompassed in simple fatty liver (SFL) to non-alcoholic steatohepatitis (NASH), eventually reaching at liver cirrhosis and hepatocellular carcinoma (HCC), via proceeding liver fibrosis (LF) [2]. Also, NAFLD is associated with diverse metabolic disorders: obesity, diabetes mellitus, hypertension, and cardiovascular diseases [3]. The surging incidence and complicated etiology cause clinical burden, searching for effective therapeutic strategies in epidemiological, and behavior approach of NAFLD patients with a primary option [4]. Recently, although several therapeutic options have been reported to ameliorate NAFLD, its noticeable therapeutic preferences are yet to be determined [5]. Formalized treatments for NAFLD are not documented and an available option is to be counselled concerning healthy lifestyle: regimen, abstinence of high fats and carbohydrates, and frequent physical exercise [6,7,8].
In the incomplete project, we pioneered the secondary metabolites (SMs) from Avena sativa (AS; known as oat) and gut microbiota (GM) to identify the key SMs in both AS and GM for the treatment of NAFLD. Furthermore, AS has a wide spectrum of pharmacological activities such as antioxidant, anti-inflammatory, antidiabetic and anticholesterolemic efficacy [9]. The AS is an ancient grain utilized as an important grain from primitive times, suggesting that AS can diminish cholesterol, control satiety, and even make positive effects on gastrointestinal (GI) health [10, 11]. Currently, several studies have demonstrated that natural products can regulate body metabolism including anti-obesity and anti-diabetes [12].
Specially, the flavonoids from AS ameliorated hyperlipidemia caused by high-fat-diet via modulating bile acid and GM in mice [13]. The AS supplement has alleviating effects to lower the blood pressure in hypertensive groups by increasing Bifidobacterium and Spirillum [14]. Furthermore, the bioactives of AS are key players to regulate the beneficial GM to relieve metabolic disorders: obesity, atherosclerosis, and even osteoporosis [15]. Thereby, it elicits that AS is a significant modulator to control GM community.
In parallel, GM in human intestine is significant community to control the physiological responses for host [16]. Some favorable GM (known as probiotics) can convert into key SMs (known as postbiotics) is implicated in many metabolic disorders including NAFLD [17]. Furthermore, probiotics and postbiotics are vital effectors to regulate PI3K/AKT pathway by interconnecting with AMP-activated kinase (AMPK) pathway [18, 19]. Some reports to approve the effects of postbiotics on inflammatory pathways have been shown different experimental results due to different postbiotic mixture in media and its derivative structures [20]. Collectively, our study is to manifest key SMs from AS and GM to keep consistent results for the treatment of NAFLD. With the help of network pharmacology concept, we performed integrative analysis to pinpoint crucial elements: key GM, signaling pathway(s), target(s), and SMs. Network pharmacology (NP) is a systemic methodology to decipher the complex biological pathways, which is a valuable tool to elucidate efficacy of complex natural products [21]. NP has been developed as a new methodology in drug discovery as it combines scattered valuable information with data science [22]. As a matter of fact, NP decodes the complicated interaction between compounds, targets, and diseases from holistic viewpoint on multiple components [23]. Most recently, a merged GM and NP study was contributed to decode the roles of GM against diarrhea-predominant irritable bowel syndrome (IBS-D), indicating that eighteen GM with treatment of Chinese traditional medicine were critical components to alleviate IBS-D [24]. Furthermore, key SMs of alcoholic liver diseases (ALD) and NAFLD were deciphered with the NP analysis [25, 26].
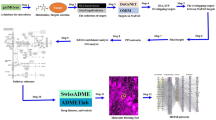
It is believed that NP might be a key to decrypt the therapeutic issue in dilemma, ending up with combinatorial application. As aforementioned, our study has established that the combinatorial application of AS and GM is to be expected as an alternative therapeutic strategy for NAFLD. Thus, this approach might be given critical hints to further clinical trials and advancement of the combined applications with AS and GM. The process of this study is displayed in Fig. 1. Given the limited microbiome data, the pharmacological pathway of combining AS and GM in the alleviation of NAFLD is only dependent upon mining data. The integrative methodology to reveal the combinatorial effects on AS and GM provides significant clues for favorable diet and well-designed microbiota compositions. The purpose of this study was to uncover combinatorial effects on both AS and GM for the treatment on NAFLD.
Materials and methods
The employment of the web-based public bioinformatics
At present, the development of data science provides a great deal of information related to biological pathways between compounds and targets, which can be a platform by merging the important databases. Based on it, we gathered valuable data to utilize NP as a drug discovery methodology. We profiled the available bioinformatics resources in Table 1.
The identification of SMs and its targets from AS
Natural Product Activity & Species Source (NPASS) database (http://bidd.group/NPASS/) (accessed on 28 September 2022) was utilized to select the significant SMs from AS [27], indicating that targets related to the SMs were retrieved by Similarity Ensemble Approach (SEA) (https://sea.bkslab.org/) (accessed on 28 September 2022) [28] and SwissTargetPrediction (STP) (http://www.swisstargetprediction.ch/) (accessed on 28 September 2022) [29]. With the exactness and rigor, the intersecting targets between SEA and STP were considered as important targets associated with SMs from AS. It was defined as AS-related targets. Crucially, SEA database is a mining platform to select some major targets linked to targets, developed by Dr Shoichet’s group. It is to be specified that the number of 23 in 30 targets extracted by SEA was confirmed by experimentation [30]. Apparently, STP has been used to identify the putative targets for ligands, for instance, the attained targets for cudraflavone C hit the mark experimentally [31].
The selection of SMs and its targets from GM
The gutMGene database was used to obtain the SMs converted by GM (http://bio-annotation.cn/gutmgene/) (accessed on 29 September 2022) [32]. The obtained SMs were input into both SEA and STP platform to select the targets. The overlapping targets identified by SEA and STP were identified as critical targets associated with SMs from GM. It is defined as GM-related targets.
The determination of core targets from AS and GM against NAFLD
The intersecting targets were identified between AS-related targets and GM-related targets, which were considered as significant targets for combinatorial therapeutics. The NAFLD- related targets extracted by DisGeNET (https://www.disgenet.org/) (accessed on 30 September 2022) [33] and OMIM (https://www.omim.org/) (accessed on 30 September 2022) [34]. Finally, we selected the core targets against NAFLD, by comparing the targets between combinatorial therapeutics’ targets and NAFLD-related targets.
The protein–protein interaction networks
We utilized String database (https://string-db.org/) (accessed on 01 October 2022) [35] to identify protein–protein interaction (PPI) networks, which was described by R Package. On the PPI networks, we found a target with the highest degree value, thus it was to be defined as a key target to ameliorate NAFLD.
The construction of bubble plot
The construction of bubble plot was established by Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway enrichment analysis. The signaling pathways on the bubble plot were depicted, according to Rich factor value. We discerned a key signaling pathway for the treatment of NAFLD, suggesting that the mechanism might be inhibitive effect on NAFLD. The bubble plot was constructed by R package.
The construction of GM or AS—a key signaling pathway-targets-SMs (GASTM) networks
We described GASTM network to know the relationships of each component: GM or AS, a key signaling pathway, targets, and secondary metabolites. The GASTM network was constructed by utilizing R Package. Taken together with GM or AS, a key signaling pathway, targets, and SMs as nodes, matching associations above components were assembled with Microsoft Excel, then input into R package to identify the interaction network of GASTM against NAFLD.
Molecular docking assay (MDA)
The Molecular docking assay (MDA) was implemented with AutodockTools-1.5.6 to understand what the most significant SMs in both GM and AS are. Commonly, the threshold of AutodockTools-1.5.6. was fitted as -6.0 kcal/mol [36] or SM with lowest Gibbs energy (the greatest negative value) was regarded as the uppermost SM to have therapeutic value in the treatment of NAFLD. The SMs were selected as.sdf format from PubChem (https://pubchem.ncbi.nlm.nih.gov/) (accessed on 02 October 2022), changing into.pdb format via Pymol tool. The.pdb format was transformed into.pdbqt format to prepare for the MDA on targets. The targets were selected by the Protein Data Bank (PDB) (https://www.rcsb.org/) (accessed on 01 October 2022) for.pdb format, which were switched into.pdbqt format by setting parameter in AutodockTools-1.5.6. The MDA was conducted on.pdbqt format by preparing for conformer between SMs and targets. The docking site was set in cubic box (x = 40 Å, y = 40 Å, and z = 40 Å) in a central point of each target.
The validation of drug-likeness and toxic parameters on the uppermost SMs
The properties of drug-likeness on the uppermost SMs were performed by SwissADME (http://www.swissadme.ch/) (accessed on 02 October 2022) [37]. The filtering standard was based on Lipinski’s rule: Molecular weight (< 500 g/mol) or Topological Polar Surface Area (TPSA) (< 140 Å2) or Moriguchi octanol–water partition coefficient (MLogP) (≤ 4.15) or Hydrogen Bonding Acceptor (HBA) (< 10) or Hydrogen Bonding Donor (HBD) (≤ 5). To accept the rule, the molecules should not be violated more than 2 parameters out of 5 parameters. The toxicity of the uppermost SMs was confirmed by ADMETlab 2.0 [38] and ProTox-II [39], its parameters are as follows: Human ether-a-go-go-related gene (hERG) [40]; Human Hepatotoxicity (H-HT) [41]; Carcinogens [42]; Cytotoxicity [43]; and Eye corrosion [44].
Results
The secondary metabolites (SMs) of AS and its targets
The number of 12 SMs from AS was documented from NPASS database, all of which were accepted by Lipinski’s rule (Table 2). The targets related to the 12 SMs were retrieved by SEA (260) and STP (260), thus, the number of 25 overlapping targets was identified between the cheminformatics databases (Fig. 2A). The 25 overlapping targets were considered as significant protein-coding gene associated with AS.
A The number of overlapping 25 targets from Avena sativa between SEA and STP. B The number of overlapping 668 targets from gut microbiota between SEA and STP. C The number of overlapping 23 targets between Avena sativa and gut microbiota. D The number of overlapping 16 targets via intersecting targets of Avena sativa and gut microbiota against NAFLD
The secondary metabolites (SMs) of GM
We identified the number of 208 SMs (Additional file 1: Table S1) in gutMGene database, the targets connected to the 208 SMs were confirmed by SEA (1256) and STP (947) (Additional file 1: Table S1). The overlapping 668 targets were regarded as significant protein-coding genes related to SMs from GM (Additional file 1: Table S1) (Fig. 2B).
The overlapping targets between AS-related targets and GM-related targets
The number of 23 targets was identified between the number of 25 overlapping targets from AS and 668 targets from GM, suggesting that the 23 targets are significant targets to exert the combinatorial efficacy on both AS and GM (Fig. 2C).
The identification of core targets against NAFLD
The number of 23 targets obtained from the combined AS-related targets and GM-related targets was compared with NAFLD-associated targets (1836) (Additional file 1: Table S1), the final 16 targets were identified as bona fide targets to be expected to exert combinatorial efficacy on AS-based and GM-based application (Fig. 2D).
A key target on PPI network and MDA
The final 16 targets PPI network consisted of 11 nodes and 17 edges, and 5 (ALDH2, PON1, ERN1, NR4A1, and GPR35) out of 16 targets were not interacted with one another (Fig. 3A). The Vascular Endothelial Growth Factor A (VEGFA) in the networks was the highest degree of value, followed by ESR1 (7), ESR2 (3), IL2 (3), and TERT (3) (Table 3). We considered the VEGFA as the uppermost target against NAFLD. In addition, results of the MDA showed that the binding energy of myricetin, quercetin was -8.2 kcal/mol as the lowest score in the number of 20 SMs (Fig. 3B), indicating that these SMs (myricetin, quercetin) could exert a potent binding affinity with VEGFA (Fig. 3C, D). The GM is enabled to convert myricitrin into myricetin, the applicable GM is Escherichia sp. 12, Escherichia sp. 33, and Enterococcus sp. 45 [45]. Moreover, quercitrin by Bacteroides sp. 45 [46], rutin by Bifidobacterium dentium, Bacteroides uniformis, and Bacteroides ovatus [47, 48], avicularin Bacillus sp. 46 [49], myricitrin by Enterococcus sp. 45, and Escherichia sp. 33 [45], isoquercitrin by Enterococcus casseliflavus [50], can convert into quercetin. It implies that the GM is beneficial probiotics to produce favorable postbiotics, in parallel, myricetin and quercetin are good effectors to bind stably on VEGFA.
Bubble plot and GASTM network
The bubble plot provided by PPI shows three signaling pathways: Prolactin signaling pathway, T cell receptor signaling pathway, and PI3K-Akt signaling pathway (Fig. 3E). Among the three signaling pathways, PI3K-Akt signaling pathway linked directly to VEGFA was considered as a key signaling pathway (Table 4). In parallel, VEGFA, IL2, GSK3B, and NR4A1 related to PI3K-Akt signaling pathway are regarded as promising targets significantly. Noticeably, PI3K-Akt signaling pathway had the lowest rich factor, suggesting that the signaling pathway might function as antagonistic mode. It means that lower rich factor can be defined as less number of expressed genes in annotated signaling pathways [51].
The GASTM network shows the relationships between GM (60 nodes, green circle) or AS (1 node, green circle), PI3K-Akt signaling pathway (1 node, green circle), targets (4 nodes, orange circle), and metabolites (56 nodes, blue sky circle), consisting of 122 nodes and 155 edges (Fig. 4). On a holistic viewpoint, the integrated four components can exert therapeutic effects, orchestrate with each other against NAFLD.
Molecular docking assay
The molecular docking assay (MDA) shows what the promising SM(s) are on the PI3K- Akt signaling pathway, and the key SM(s) are derived from either GM or AS. As mentioned previously, the key SMs (myricetin, and quercetin) of VEGFA were derived from GM (Escherichia sp. 12, Escherichia sp. 33, Enterococcus sp. 45, Bacteroides sp. 45, Bifidobacterium dentium, Bacteroides uniformis, Bacteroides ovatus, Bacillus sp. 46, and Enterococcus casseliflavus). Likewise, myricetin on GSK3B had the highest affinity with -10.6 kcal/mol (Fig. 5A), originated from Escherichia sp. 12, Escherichia sp. 33, and Enterococcus sp. 45. Diosgenin bound most stably to IL2 had the greatest affinity with -9.1 kcal/mol (Fig. 5B), which can be converted from SCHEMBL20481776 (PubChem ID: 135312912) [52]. However, GM can convert diosgenin are yet to be revealed. Vestitol on NR4A1 formed the most stable conformer with -9.0 kcal/mol (Fig. 5C), the vestitol was derived from AS. The information of the MDA was profiled in Additional file 2: Table S2. Collectively, the combinatorial application of AS and beneficial GM can involve in the treatment on NAFLD via the PI3K-Akt signaling pathway by multiple-compounds, and multiple-targets.
The verification of drug-likeness and toxicity on key SMs
The number of four SMs (myricetin, quercetin, diosgenin, and vestitol) was accepted by Lipinski’s rule, thus, which could be important agents to develop therapeutics. Accordingly, the parameters of toxicity were all confirmed: hERG, Human Hepatotoxicity, Carcinogens, Cytotoxicity, and Eye corrosion. Thus, the identified four SMs are promising candidates against NAFLD (Table 5). The four SMs had no physicochemical hindrances to be therapeutic agents. The chemical structures of the key SMs were exhibited in Fig. 6.
Discussion
The PPI network shows that VEGFA is the uppermost target to regulate other 10 targets. The key SMs (myricetin, quercetin) have been revealed by MDA, which could form the most stable conformers on VEGFA. The myricetin diminishes the lipid synthesis in liver cell and inflammatory response by tuning GM [53]. Additionally, an animal test demonstrated that the quercetin enhances NAFLD by alleviating inflammation, free-radicals, and lipid degradation in type 2 diabetic mice [54]. Commonly, expression level of the VEGFA increases during obesity, and neutralization of VEGFA relieves the metabolic disorders occurred by diet [55]. It elicits that VEGFA inhibitors can be candidates to improve lipid metabolic dysfunction including NAFLD.
The three signaling pathways confirmed by PPI are related to occurrence and development of NAFLD, which concisely discussed in Table 6.
The most stable SM on IL2 was diosgenin, suggesting that the diosgenin is a representative compound in saponin derivatives [56]. A report shows that diosgenin reduces triglyceride content in the liver, and stimulates the excretion of cholesterol [57]. Another report to support the therapeutic efficacy of diosgenin demonstrated that diosgenin interrupts the lipid absorption in intestine, triggers cholesterol transformation into bile acid and its elimination as well as interferes with lipid biosynthesis [58]. Also, IL2 inhibitor is a potent therapeutic agent to treat diverse inflammatory responses including NAFLD [59, 60]. It elicits that diosgenin is not only an effector to control lipid content but can also be used as NAFLD alleviator. A typical SM conformed to Glycogen Synthase Kinase 3 Beta (GSK3B) was myricerin with highly therapeutic values such as antioxidant, antidiabetic, antiinflammation, and even anticancer [61]. Noticeably, GSK3 antagonist alleviates hepatic steatosis which is accompanied by mitochondrial abnormality [62]. A report demonstrated that GSK3B inhibitor can be a promising therapeutic effector to control NAFLD [63]. Thus, it has been supported that antagonists of GSK3B might be significant agents for the treatment of NAFLD. A representative SM bound to Nuclear receptor subfamily 4 group A member 1 (NR4A1) was vestitol derived from AS, which is a species of isoflavonoid [64]. The vestitol is known as a potent anti-inflammatory agent by reducing leukocyte rolling [64]. At present, few studies of vestitol have been reported. The NR4A1 involved in chronic inflammatory state and dysfunction of lipid metabolism in type2 diabetic (T2D) patients [65]. It implies that the inhibition of NR4A1 can help regulate lipid biosynthesis against NAFLD.
Overall, this study shows that dampening of PI3K-Akt signaling pathway might be a potential mechanism to relieve NAFLD. In detail, the key effectors that we suggested are myricetin, quercetin from Escherichia sp. 12, Escherichia sp. 33, Enterococcus sp. 45, Bacteroides sp. 45, Bifidobacterium dentium, Bacteroides uniformis, Bacteroides ovatus, Bacillus sp. 46, and Enterococcus casseliflavus. However, the GM that converts diosgenin is still veiled. Although there were no evident relationships either positive or negative feedback between AS and nine GM, we could expect the high possibility as relievers on NAFLD, obtaining the four SMs: (1) myricetin, (2) quercetin, (3) diosgenin from GM, and (4) vestitol from AS (known as oatmeal).
All in all, these findings shed light on the importance of GM as therapeutics, and AS with auxiliary role in the context NAFLD. Despite that, our study is required to do clinical tests and extensive investigation with rigorousness. The key findings of this study are represented in Fig. 7.
The pros and cons of this study
From the incorporation of NP, this study exploratory organizes on the key GM, signaling pathways, targets, and SMs with the application of AS in NAFLD treatment, suggesting theoretical evidence for further clinical verification. As limited borderline of NP, the pharmacological pathway of integrating analysis on AS or GM against NAFLD is only dependent on data-driven analysis, and its combinatorial effects, the interaction between AS and GM in vivo were ignored, which is integral to validate the authenticity through preclinical and clinical tests. Hence, the performance of our analysis needs to be advanced more, for instance, by integrating new dataset continually. At this point, our approach platform is easy to merge new data to improve the performance and might be a hallmark to elucidate the relationships between diet and GM. In addition, our study provides a rationale for how to improve accuracy prior to clinical trials.
Conclusion
In conclusion, our study highlights the therapeutic effects and mechanisms of the treatment on NAFLD via combinatorial application: gut microbiota (GM), and Avena sativa (AS), indicating antagonists (myricetin, quercetin, diosgenin, and vestitol) to inhibit PI3K-Akt signaling pathway. These findings provide a new insight to utilize the endogenous species (gut microbiota) and exogenous species (Avena sativa) on microbiome-based therapeutics. However, this study should be taken in vitro or in vivo experimentation into consideration to uncover bona fide pharmacological efficacy.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its Additional files).
Abbreviations
- AMPK:
-
AMP-activated Kinase
- AS:
-
Avena sativa
- GASTM:
-
GM or AS- a key Signaling pathway-Targets-SMs
- GM:
-
Gut Microbiota
- HBA:
-
Hydrogen Bonding Acceptor
- HBD:
-
Hydrogen Bonding Donor
- hERG:
-
Human Ether-a-go-go-Related Gene
- H-HT:
-
Human HepatoToxicity
- KEGG:
-
Kyoto Encyclopedia of Gene and Genomes
- LF:
-
Liver Fibrosis
- MDA:
-
Molecular Docking Assay
- MLogP:
-
Moriguchi octanol–water partition coefficient
- NAFLD:
-
Non-Alcoholic Fatty Liver Disease
- NASH:
-
Non-Alcoholic SteatoHepatitis
- NP:
-
Network Pharmacology
- NPASS:
-
Natural Product Activity & Species Source
- PDB:
-
Protein Data Bank
- PPI:
-
Protein–Protein Interaction
- SEA:
-
Similarity Ensemble Approach
- SM:
-
Secondary Metabolite
- SMs:
-
Secondary Metabolites
- STP:
-
SwissTargetPrediction
- T2D:
-
Type2 Diabetic
- TPSA:
-
Topological Polar Surface Area
- VEGFA:
-
Vascular Endothelial Growth Factor A
References
Abenavoli L, Larussa T, Corea A, Procopio AC, Boccuto L, Dallio M, Federico A, Luzza F. Dietary polyphenols and non-alcoholic fatty liver disease. Nutrients. 2021;13:494–503. https://doi.org/10.3390/NU13020494.
Sherif ZA, Sherif ZA. The rise in the prevalence of nonalcoholic fatty liver disease and hepatocellular carcinoma. Nonalcoholic Fatty Liver Dis An Update. 2019. https://doi.org/10.5772/INTECHOPEN.85780.
Godoy-Matos AF, Silva Júnior WS, Valerio CM. NAFLD as a continuum: from obesity to metabolic syndrome and diabetes. Diabetol Metab Syndr. 2020;12:1–20. https://doi.org/10.1186/S13098-020-00570-Y/TABLES/5.
Targher G, Corey KE, Byrne CD. NAFLD, and cardiovascular and cardiac diseases: factors influencing risk, prediction and treatment. Diabetes Metab. 2021;47(101215):1–14. https://doi.org/10.1016/J.DIABET.2020.101215.
Peng C, Stewart AG, Woodman OL, Ritchie RH, Qin CX. Non-Alcoholic steatohepatitis: a review of its mechanism, models and medical treatments. Front Pharmacol. 2020;11:1–22. https://doi.org/10.3389/FPHAR.2020.603926/BIBTEX.
Abenavoli L, Boccuto L, Federico A, Dallio M, Loguercio C, Di Renzo L, De Lorenzo A. Diet and non-alcoholic fatty liver disease: the mediterranean way. Int J Environ Res Public Health. 2019;16:3011–9. https://doi.org/10.3390/IJERPH16173011.
Carneros D, López-Lluch G, Bustos M. Physiopathology of lifestyle interventions in non-alcoholic fatty liver disease (NAFLD). Nutrients. 2020;12:3472–94. https://doi.org/10.3390/NU12113472.
Romero-Gómez M, Zelber-Sagi S, Trenell M. Treatment of NAFLD with diet, physical activity and exercise. J Hepatol. 2017;67:829–46. https://doi.org/10.1016/J.JHEP.2017.05.016.
Singh R, De S, Belkheir A. Avena sativa (Oat), a potential neutraceutical and therapeutic agent: an overview. Critical Rev Food Sci Nutr. 2012;53:126–44. https://doi.org/10.1080/10408398.2010.526725.
Zamaratskaia G, Gerhardt K, Wendin K. Biochemical characteristics and potential applications of ancient cereals—an underexploited opportunity for sustainable production and consumption. Trends Food Sci Technol. 2021;107:114–23. https://doi.org/10.1016/J.TIFS.2020.12.006.
Korczak R, Kocher M, Swanson KS. Effects of oats on gastrointestinal health as assessed by in vitro, animal, and human studies. Nutr Rev. 2020;78(5):343–63. https://doi.org/10.1093/nutrit/nuz064.
Pan C, Guo Q, Lu N, Dai C. Role of gut microbiota in the pharmacological effects of natural products. Evidence-Based Compl Alter Med eCAM. 2019;2019:1–8. https://doi.org/10.1155/2019/2682748.
Duan R, Guan X, Huang K, Zhang Y, Li S, Xia J, Shen M. Flavonoids from whole-grain oat alleviated high-fat diet-induced hyperlipidemia via regulating bile acid metabolism and gut microbiota in mice. J Agric Food Chem. 2021;69:7629–40. https://doi.org/10.1021/ACS.JAFC.1C01813/ASSET/IMAGES/LARGE/JF1C01813_0007.JPEG.
Qin J, Liu L, Su XD, Wang BB, Fu BS, Cui JZ, Liu XY. The effect of PCSK9 inhibitors on brain stroke prevention: a systematic review and meta-analysis. Nutr Metab Cardiovasc Dis. 2021;31:2234–43. https://doi.org/10.1016/J.NUMECD.2021.03.026.
Yu Y, Zhou L, Li X, Liu J, Li H, Gong L, Zhang J, Wang J, Sun B. The progress of nomenclature, structure, metabolism, and bioactivities of oat novel phytochemical: avenanthramides. J Agric Food Chem. 2022;70:446–57. https://doi.org/10.1021/ACS.JAFC.1C05704.
Martin AM, Sun EW, Rogers GB, Keating DJ. The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front Physiol. 2019;10:428–38. https://doi.org/10.3389/FPHYS.2019.00428/BIBTEX.
Arellano-García L, Portillo MP, Martínez JA, Milton-Laskibar I. Usefulness of probiotics in the management of NAFLD: evidence and involved mechanisms of action from preclinical and human models. Int J Mol Sci. 2022;23:1–25. https://doi.org/10.3390/IJMS23063167.
Zhang M, Wang C, Wang C, Zhao H, Zhao C, Chen Y, Wang Y, McClain C, Feng W. Enhanced AMPK phosphorylation contributes to the beneficial effects of Lactobacillus rhamnosus GG supernatant on chronic-alcohol-induced fatty liver disease. J Nutr Biochem. 2015;26:337–44. https://doi.org/10.1016/J.JNUTBIO.2014.10.016.
Lew LC, Hor YY, Jaafar MH, Lau ASY, Ong JS, Chuah LO, Yap KP, Azzam G, Azlan A, Liong MT. Lactobacilli modulated AMPK activity and prevented telomere shortening in ageing rats. Beneficial Microbes. 2019;10:883–92. https://doi.org/10.3920/BM2019.0058.
Jastrząb R, Graczyk D, Siedlecki P. Molecular and Cellular Mechanisms Influenced by Postbiotics. Int J Mol Sci. 2021;22:1–34. https://doi.org/10.3390/IJMS222413475.
Kibble M, Saarinen N, Tang J, Wennerberg K, Mäkelä S, Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32:1249–66. https://doi.org/10.1039/C5NP00005J.
Dong Y, Hao L, Fang K, Han XX, Yu H, Zhang JJ, Cai LJ, Fan T, Zhang WD, Pang K, Ma WM, et al. A network pharmacology perspective for deciphering potential mechanisms of action of Solanum nigrum L. in bladder cancer. BMC Compl Med Ther. 2021;21:1–14. https://doi.org/10.1186/S12906-021-03215-3/FIGURES/5.
Wu SS, Hao LJ, Shi YY, Lu ZJ, Yu JL, Jiang SQ, Liu QL, Wang T, Guo SY, Li P, et al. Network pharmacology-based analysis on the effects and mechanism of the wang-bi capsule for rheumatoid arthritis and osteoarthritis. ACS Omega. 2022;7:7825–36. https://doi.org/10.1021/ACSOMEGA.1C06729/ASSET/IMAGES/LARGE/AO1C06729_0006.JPEG.
Zhen Z, Xia L, You H, Jingwei Z, Shasha Y, Xinyi W, Wenjing L, Xin Z, Chaomei F. An integrated gut microbiota and network pharmacology study on fuzi-lizhong pill for treating diarrhea-predominant irritable bowel syndrome. Front Pharmacol. 2021;12(3330):1–14. https://doi.org/10.3389/FPHAR.2021.746923/BIBTEX.
Oh KK, Choi YR, Gupta H, Ganesan R, Sharma SP, Won SM, Jeong JJ, Lee SB, Cha MG, Kwon GH, et al. Identification of gut microbiome metabolites via network pharmacology analysis in treating alcoholic liver disease. Curr Issues Mol Biol. 2022;44:3253–66. https://doi.org/10.3390/CIMB44070224.
Oh KK, Gupta H, Min BH, Ganesan R, Sharma SP, Won SM, Jeong JJ, Lee SB, Cha MG, Kwon GH, et al. Elucidation of prebiotics, probiotics, postbiotics, and target from gut microbiota to alleviate obesity via network pharmacology study. Cells. 2022;11:1–14. https://doi.org/10.3390/CELLS11182903.
Zeng X, Zhang P, He W, Qin C, Chen S, Tao L, Wang Y, Tan Y, Gao D, Wang B, et al. NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. 2018;46:D1217–22. https://doi.org/10.1093/NAR/GKX1026.
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. https://doi.org/10.1038/NBT1284.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–3664. https://doi.org/10.1093/NAR/GKZ382.
Singh N, Chaput L, Villoutreix BO. Virtual screening web servers: designing chemical probes and drug candidates in the cyberspace. Brief Bioinform. 2021;22:1790–818. https://doi.org/10.1093/BIB/BBAA034.
Soo HC, Chung FFL, Lim KH, Yap VA, Bradshaw TD, Hii LW, Tan SH, See SJ, Tan YF, Leong CO, et al. Cudraflavone C induces tumor-specific apoptosis in colorectal cancer cells through inhibition of the phosphoinositide 3-kinase (PI3K)-AKT pathway. PLoS ONE. 2017;12:1–20. https://doi.org/10.1371/JOURNAL.PONE.0170551/PONE_0170551_PDF.PDF.
Cheng L, Qi C, Yang H, Lu M, Cai Y, Fu T, Ren J, Jin Q, Zhang X. gutMGene: a comprehensive database for target genes of gut microbes and microbial metabolites. Nucleic Acids Res. 2022;50:D795–800. https://doi.org/10.1093/NAR/GKAB786.
Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960–7. https://doi.org/10.1016/J.CSBJ.2021.05.015.
Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–98. https://doi.org/10.1093/NAR/GKU1205.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–12. https://doi.org/10.1093/NAR/GKAA1074.
Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinform Chem AABC. 2014;7:23–36. https://doi.org/10.2147/AABC.S63749.
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:1–13. https://doi.org/10.1038/srep42717.
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, et al. ADMETlab 2 0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–14. https://doi.org/10.1093/NAR/GKAB255.
Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–63. https://doi.org/10.1093/NAR/GKY318.
Lamothe SM, Guo J, Li W, Yang T, Zhang S. The human Ether-a-go-go-related Gene (hERG) potassium channel represents an unusual target for protease-mediated damage. J Biol Chem. 2016;291:20387–401. https://doi.org/10.1074/JBC.M116.743138.
Mulliner D, Schmidt F, Stolte M, Spirkl HP, Czich A, Amberg A. Computational models for human and animal hepatotoxicity with a global application scope. Chem Res Toxicol. 2016;29:757–67. https://doi.org/10.1021/ACS.CHEMRESTOX.5B00465/SUPPL_FILE/TX5B00465_SI_003.ZIP.
Gaylor DW. Are tumor incidence rates from chronic bioassays telling us what we need to know about carcinogens? Regul Toxicol Pharmacol. 2005;41:128–33. https://doi.org/10.1016/j.yrtph.2004.11.001.
Niles AL, Moravec RA, Riss TL. Update on in vitro cytotoxicity assays for drug development. Expert Opin Drug Discov. 2008;3:655–69. https://doi.org/10.1517/17460441.3.6.655.
Clippinger AJ, Raabe HA, Allen DG, Choksi NY, Van Der Zalm AJ, Kleinstreuer NC, Barroso J, Lowit AB, Ao Barroso J. Human-relevant approaches to assess eye corrosion/irritation potential of agrochemical formulations Human-relevant approaches to assess eye corrosion/irritation potential of agrochemical formulations. Cutaneous Ocular Toxicol. 2021;40(2):145–67. https://doi.org/10.1080/15569527.2021.1910291.
Du LY, Zhao M, Xu J, Qian DW, Jiang S, Shang EX, Guo JM, Liu P, Su SL, Duan JA, et al. Identification of the metabolites of myricitrin produced by human intestinal bacteria in vitro using ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Expert Opin Drug Metab Toxicol. 2014;10:921–31. https://doi.org/10.1517/17425255.2014.918954.
Jiang S, Yang J, Qian D, Guo J, Shang EX, Duan JA, Xu J. Rapid screening and identification of metabolites of quercitrin produced by the human intestinal bacteria using ultra performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. Arch Pharmacal Res. 2014;37:204–13. https://doi.org/10.1007/S12272-013-0172-9.
Bang SH, Hyun YJ, Shim J, Hong SW, Kim DH. Metabolism of rutin and poncirin by human intestinal microbiota and cloning of their metabolizing α-L-rhamnosidase from Bifidobacterium dentium. J Microbiol Biotechnol. 2015;25:18–25. https://doi.org/10.4014/JMB.1404.04060.
Selma MV, Espín JC, Tomás-Barberán FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem. 2009;57:6485–501. https://doi.org/10.1021/JF902107D.
Zhao M, Jun X, Qian D, Guo J, Jiang S, Shang E-x, Duan J-a, Yang J, Le-yue D. Ultra performance liquid chromatography/quadrupole-time-of-flight mass spectrometry for determination of avicularin metabolites produced by a human intestinal bacterium. J Chromatography B. 2014. https://doi.org/10.1016/J.JCHROMB.2014.01.005.
Blaut M, Schoefer L, Braune A. Transformation of flavonoids by intestinal microorganisms. Int J Vitamin Nut Res. 2003;73:79–87. https://doi.org/10.1024/0300-9831.73.2.79.
Zhang YJ, Sun YZ, Gao XH, Qi RQ. Integrated bioinformatic analysis of differentially expressed genes and signaling pathways in plaque psoriasis. Mol Med Rep. 2019;20:225–35. https://doi.org/10.3892/MMR.2019.10241/HTML.
Feng JF, Tang YN, Ji H, Xiao ZG, Zhu L, Yi T. Biotransformation of dioscorea nipponica by rat intestinal microflora and cardioprotective effects of diosgenin. Oxid Med Cell Longev. 2017;2017:1–9. https://doi.org/10.1155/2017/4176518.
Sun WL, Li XY, Dou HY, Wang XD, Li J, Da; Shen, L., Ji, H.F. Myricetin supplementation decreases hepatic lipid synthesis and inflammation by modulating gut microbiota. Cell Rep. 2021;36(109641):1–19. https://doi.org/10.1016/J.CELREP.2021.109641.
Yang H, Yang T, Heng C, Zhou Y, Jiang Z, Qian X, Du L, Mao S, Yin X, Lu Q. Quercetin improves nonalcoholic fatty liver by ameliorating inflammation, oxidative stress, and lipid metabolism in db/db mice. Phytotherapy Res PTR. 2019;33:3140–52. https://doi.org/10.1002/PTR.6486.
Wu LE, Meoli CC, Mangiafico SP, Fazakerley DJ, Cogger VC, Mohamad M, Pant H, Kang MJ, Powter E, Burchfield JG, et al. Systemic VEGF-A neutralization ameliorates diet-induced metabolic dysfunction. Diabetes. 2014;63:2656–67. https://doi.org/10.2337/DB13-1665.
Wang D, Wang X. Diosgenin and its analogs: potential protective agents against atherosclerosis. Drug Des Dev Ther. 2022;16:2305–23. https://doi.org/10.2147/DDDT.S368836.
Kusano Y, Tsujihara N, Masui H, Shibata T, Uchida K, Takeuchi W. Diosgenin supplementation prevents lipid accumulation and induces skeletal muscle-fiber hypertrophy in rats. J Nutr Sci Vitaminol. 2019;65:421–9. https://doi.org/10.3177/JNSV.65.421.
Sun F, Yang X, Ma C, Zhang S, Yu L, Lu H, Yin G, Liang P, Feng Y, Zhang F. <p>The effects of diosgenin on hypolipidemia and its underlying mechanism: a review</p>. Diabetes Metab Syn Obesity Targets Ther. 2021;14:4015–30. https://doi.org/10.2147/DMSO.S326054.
Kaur M, Bahia MS, Silakari O. Inhibitors of interleukin-2 inducible T-cell kinase as potential therapeutic candidates for the treatment of various inflammatory disease conditions. Eur J Pharm Sci. 2012;47:574–88. https://doi.org/10.1016/J.EJPS.2012.07.013.
Duan Y, Pan X, Luo J, Xiao X, Li J, Bestman PL, Luo M. Association of inflammatory cytokines with non-alcoholic fatty liver disease. Front Immunol. 2022;13:1–15. https://doi.org/10.3389/FIMMU.2022.880298/FULL.
Semwal DK, Semwal RB, Combrinck S, Viljoen A. Myricetin: a dietary molecule with diverse biological activities. Nutrients. 2016;8:1–31. https://doi.org/10.3390/NU8020090.
Li Y, Lin Y, Han X, Li W, Yan W, Ma Y, Lu X, Huang X, Bai R, Zhang H. GSK3 inhibitor ameliorates steatosis through the modulation of mitochondrial dysfunction in hepatocytes of obese patients. iScience. 2021;24(102149):1–50. https://doi.org/10.1016/J.ISCI.2021.102149.
Cao J, Feng XX, Yao L, Ning B, Yang ZX, Fang DL, Shen W. Saturated free fatty acid sodium palmitate-induced lipoapoptosis by targeting glycogen synthase kinase-3β activation in human liver cells. Dig Dis Sci. 2014;59:346–57. https://doi.org/10.1007/S10620-013-2896-2.
Franchin M, Cólon DF, Castanheira FVS, Da Cunha MG, Bueno-Silva B, Alencar SM, Cunha TM, Rosalen PL. Vestitol isolated from brazilian red propolis inhibits neutrophils migration in the inflammatory process: elucidation of the mechanism of action. J Nat Prod. 2016;79:954–60. https://doi.org/10.1021/ACS.JNATPROD.5B00938/ASSET/IMAGES/MEDIUM/NP-2015-00938K_0008.GIF.
Huang Q, Xue J, Zou R, Cai L, Chen J, Sun L, Dai Z, Yang F, Xu Y. NR4A1 is associated with chronic Low-Grade inflammation in patients with type 2 diabetes. Exp Ther Med. 2014;8:1648–54. https://doi.org/10.3892/ETM.2014.1958/HTML.
Zhang P, Ge Z, Wang H, Feng W, Sun X, Chu X, Jiang C, Wang Y, Zhu D, Bi Y. Prolactin improves hepatic steatosis via CD36 pathway. J Hepatol. 2018;68:1247–55. https://doi.org/10.1016/j.jhep.2018.01.035.
Breuer DA, Pacheco MC, Washington MK, Montgomery SA, Hasty AH, Kennedy AJ. CD8+ T cells regulate liver injury in obesity-related nonalcoholic fatty liver disease. Am J Physiol Gastroin Liver Physiol. 2020;318:G211–24. https://doi.org/10.1152/AJPGI.00040.2019.
Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P, Stroncek DF, Terabe M, Kapoor V, ElGindi M, et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–7. https://doi.org/10.1038/NATURE16969.
Coia H, Ma N, Hou Y, Permaul E, Berry DL, Cruz MI, Pannkuk E, Girgis M, Zhu Z, Lee Y, et al. Theaphenon E prevents fatty liver disease and increases CD4+ T cell survival in mice fed a high-fat diet. Clin Nutrition. 2021;40:110–9. https://doi.org/10.1016/J.CLNU.2020.04.033.
Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P, Stroncek DF, Terabe M, Kapoor V, ElGindi M, et al. NAFLD causes selective CD4+ T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–7. https://doi.org/10.1038/NATURE16969.
Huang X, Liu G, Guo J, Su ZQ. The PI3K/AKT pathway in obesity and type 2 diabetes. Int J Biol Sci. 2018;14:1483–96. https://doi.org/10.7150/IJBS.27173.
Matsuda S, Kobayashi M, Kitagishi Y. Roles for PI3K/AKT/PTEN pathway in cell signaling of nonalcoholic fatty liver disease. ISRN Endocrinol. 2013;2013:1–7. https://doi.org/10.1155/2013/472432.
Acknowledgements
This study has been implemented with the support of the Hallym University Research Fund, the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology.
Funding
This research was supported by the Hallym University Research Fund, the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology (NRF-2020R1I1A3073530 and NRF-2020R1A6A1A03043026).
Author information
Authors and Affiliations
Contributions
Supervision, Project administration, and Investigation: KTS, DJK, Conceptualization, Methodology: KTS, KKO, Formal analysis: KTS, KKO, SJY, Visualization, Data Curation: RG, SMW, JJJ, SPS, SBL, SYL, Writing—Original Draft: KTS, KKO, Software, Investigation, and Data Curation: KKO, HG, Validation and Writing: KTS, DJK, KKO, HG, Review and Editing: KTS, DJK. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
There is no conflict of interest declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
: The number of 208 SMs of gut microbiota from gutMGene; The number of 1256 targets from SEA; The number of 947 targets from STP; The number of 668 targets between SEA (1256) and STP (947); The number 1836 targets related to NAFLD.
Additional file 2: Table S2
: The binding energy and coordinated amino acid bonds of SMs on each key target.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Oh, KK., Yoon, SJ., Lee, SB. et al. The convergent application of metabolites from Avena sativa and gut microbiota to ameliorate non-alcoholic fatty liver disease: a network pharmacology study. J Transl Med 21, 263 (2023). https://doi.org/10.1186/s12967-023-04122-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-023-04122-6