Abstract
Background
Idiopathic pulmonary fibrosis is a chronic progressive, lethal disease in which ectopic lung fibroblast (LF) activation plays a vital part. We have previously shown that alamandine (ALA) exerts anti-fibrosis effects via the MAS-related G-protein coupled receptor D (MrgD). Here, we further investigate how it moderates transforming growth factor β1 (TGF-β1)-induced LF activation by regulating glucose metabolism and mitochondria autophagy (mitophagy).
Methods
In vitro, we examined glycolysis-related protein hexokinase 2 (HK2), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), and lactic acid in cells treated with TGF-β1. The oxygen consumption rate and the extracellular acidification rate were detected using Seahorse assays. Then, mitophagy was evaluated using transmission electron microscopy, mt-Keima, and the co-localization of Parkin and COX IV with LC3 and LAMP1, respectively. The autophagic degradation of HK2 and PFKFB3 was detected by 3MA and bafilomycin A1 and assessed by their co-localization with LC3 and LAMP1, respectively. The effects of ALA on LF activation markers collagen I and α-SMA were detected. The effects of ALA on glucose metabolism, mitophagy, and the activation of LF were also investigated in vivo.
Results
We found that the ALA/MrgD axis improved TGF-β1-mediated LF activation by repressing glycolysis by downregulating HK2 and PFKFB3 expression. Lactic acid sustained positive feedback between glycolysis and LF activation by maintaining the expression of HK2 and PFKFB3. We also showed that glycolysis enhancement resulted from blocking the autophagic degradation of HK2 and PFKFB3 while upregulated mRNA levels by TGF-β1, while all of those improved by ALA adding. Importantly, we determined that moderation of Parkin/LC3-mediated mitophagy by TGF-β1 also promotes glycolysis but is reversed by ALA. Furthermore, we proved that ALA counteracts the effects of bleomycin on HK2, PFKFB3, LC3, Parkin, and LF activation in vivo.
Conclusion
In this study, we show that the ALA/MrgD axis prevents TGF-β1-mediated fibroblast activation via regulation of aerobic glycolysis and mitophagy.
Similar content being viewed by others
Introduction
Aerobic glycolysis has a crucial role in the development of idiopathic pulmonary fibrosis (IPF) and transforming growth factor β1 (TGF-β1)-induced lung fibroblast (LF) activation [1, 2]. Targeting hexokinase 2 (HK2), the first key enzyme in the process of glycolysis, or 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), which produces fructose-2,6-bisphosphate, the most potent allosteric activator of the glycolytic rate-limiting enzyme phosphofructokinase-1, can alleviate TGF-β1-induced LF activation and lung fibrosis [3, 4]. However, the specific molecular mechanisms of the activation and degradation of HK2 and PFKFB3 by TGF-β1 remain obscure.
Mitophagy is a type of selective autophagy that clears damaged and potentially cytotoxic mitochondria through the autophagosome-lysosome pathway to maintain cellular homeostasis [5, 6]. As demonstrated by our previous research and that of others, mitophagy has an important but undefined role in multiple experimental animal models and human lung diseases and is associated with lung dysfunction and disease progression [7,8,9]. Mitophagy-deficient mice are more vulnerable to developing lung fibrosis [10]. Mitochondria represent the powerhouse and center of metabolism in eukaryotic cells, and evidence indicates that mitophagy is closely associated with metabolic disorders [11]. However, it is unclear whether mitophagy regulates key glycolysis enzymes HK2 and PFKFB3. The autophagosome–lysosome pathway is one of the main mechanisms of intracellular protein degradation [12]. There is evidence that HK2 and PFKFB3 may be degraded by this pathway, but whether they affect LF activation in IPF is unknown [13, 14].
Furthermore, it has been reported that simultaneous inhibition of autophagy and glycolysis can inhibit tumor growth, suggesting a close complementary relationship between glycolysis and autophagy [15]. Although it is tempting to postulate a single driver mediating LF activation and IPF, this is unlikely, given the multifactorial nature of pro-fibrotic events. Here, we focus on HK2 and PFKFB3 to better understand the mechanisms involved.
Alamandine (ALA), a novel member of the renin–angiotensin–aldosterone system (RAAS), exerts anti-inflammation and anti-fibrosis effects via the ALA/MrgD axis [16, 17]. Our previous studies have demonstrated the anti-fibrosis and autophagy-modulating effects of ALA in the lungs and liver [18, 19]. However, further studies are needed to investigate whether ALA protects against LF activation by regulating Parkin/LC3-mediated mitophagy. Moreover, different components of RAAS have been shown to be closely related to metabolic disturbance, but whether ALA plays a part in LF activation by improving glucose metabolism is unclear [20]. Building on previous findings, we performed experiments to explore the specific mechanism of how ALA regulates fibroblast activation and PF via mitophagy and aerobic glycolysis.
Materials and methods
Reagents
ALA was purchased from Biosyntan (Berlin, Germany); D-Pro7-Ang-(1–7) and β-alanine were synthesized by Sangon (Shanghai, China); TGF-β1, 3PO, 2DG, and bafilomycin A1 were purchased from MedChemExpress (HY-100558, USA); 3-methyladenine (3MA) was purchased from Selleck (S2767, USA); and Mdivi-1 was purchased from Abmole (M2830, USA). MrgD siRNA and Parkin siRNA were provided by Ribobio (Guangzhou, China). L-lactic acid was purchased from Solarbio (IL0540, Beijing, China). Other reagents were as described below.
Animal experiments
Male wild-type C57BL/6 mice (6–8 weeks old) were purchased from the Central Animal Care Facility of Southern Medical University, and the experiments were approved by the Committee on the Ethics of Animal Experiments of Southern Medical University and conducted following the regulations of the institution (Permit No. NFYY 2022-0153). Mice were housed in a specific-pathogen-free room with 12-h light/dark cycles and controlled temperature and humidity, with free access to water and food. Mice were randomly divided into three groups (10 mice per group): a control group, bleomycin (BLM) treatment group, and BLM + ALA treatment group. Initially, 100 μL of sterile saline containing BLM at a single dose of 5 mg/kg was instilled into the tracheas of mice in the two BLM groups after the mice were anesthetized using 1% pentobarbital (30 mg/kg); an equivalent dose of sterile saline was administered to the control group. ALA was continuously infused into the BLM + ALA mice using a subcutaneously implanted micro-osmotic pump at a rate of 2 μg/kg/h for 28 days. The other two groups simultaneously received the same dose of saline as a control. Animals were euthanized 28 days after establishing the PF model. The mice were anesthetized using pentobarbital before sample collection. Lung tissues were removed and fixed in 4% paraformaldehyde or stored at − 80 °C for further histological and biochemical analyses.
Cell isolation and culture
Primary LFs were isolated from the lungs of C57BL/6 male mice as previously described [21]. The cells were cultured in Dulbecco’s modified Eagle medium (DMEM; C11995500BT-1, GIBCO, USA) containing 15% fetal bovine serum (FBS; FSP500, GIBCO, USA) plus 1% Pen-Strep (10,000 U/mL penicillin and 10,000 U/mL streptomycin; 15140122, GIBCO, USA) at 37 °C in a 5% CO2 incubator.
Isolation of mitochondria
Fibroblast mitochondria were isolated using a mitochondrial isolation kit (C3601, Beyotime, China) according to the manufacturer’s instructions. Briefly, cells were dispersed and centrifuged to obtain cell pellets. Then, they were resuspended with 1 mL lysis buffer and transferred into a glass grinder for homogenization. The homogenate was subsequently centrifuged at 800g for 5 min at 4 °C. The supernatant was collected and then centrifuged at 15,000g for 10 min at 4 °C, followed by giving up the medium buffer. Finally, 60 μL of lysis buffer containing protease inhibitor was used to resuspend the pellet to obtain the mitochondrial protein.
L-lactic acid and pH adjustment
L-lactic acid (IL0540, Solarbio) at 10 mM concentration was added to DMEM containing 15% FBS. Then, the pH of the medium was adjusted to 7.6 using 1 mol/L NaOH before it was used for the incubation of cell cultures.
Expression plasmid construction and transfection
An expression plasmid containing PFKFB3 (accession: NM_001177756) was generated by the following steps. Briefly, PFKFB3 cDNA reverse transcribed from mRNA (extracted from primary LFs) was amplified using PCR. The PCR amplicon appended with sites for restriction digest (BamHI and XhoI) was inserted into the pEnCMV plasmid (P27576, MiaoLing, Hubei, China). The resulting PFKFB3 plasmid was sequenced by Sangon Biotech (Shanghai, China). Transfection was performed using Lipofectamine 3000 (L3000015, ThermoFisher Scientific, USA) according to the manufacturer’s protocol. The efficiency of transfection was assessed by western blotting and quantitative reverse transcription PCR (qRT-PCR). The primary LFs were subjected to subsequent experiments after 24 h of transfection.
siRNA transfection
The siRNA transfection in fibroblasts was performed using Lipofectamine 3000 (L3000015, ThermoFisher Scientific) according to the manufacturer’s protocol.
Measurement of oxygen consumption rate and extracellular acidification rate
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined using a Seahorse XFe96 Extracellular Flux Analyzer (Agilent, CA, USA). Briefly, cells were seeded in XF96 Cell Culture Microplates and incubated at 37 °C for 24 h. The culture medium was changed to XF assay medium supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose for OCR assay or 1 mM glutamine for ECAR assay, and cells in the new medium were placed in a 37 °C incubator without CO2 for 1 h. OCR was measured by sequential injections of 1 µM oligomycin, 0.5 µM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone, and 1 µM rotenone plus antimycin A to perform a mitochondrial stress test using the XF Extracellular Flux Analyzer. ECAR was examined under basal conditions and during the sequential injection of 10 mM glucose, 1 µM oligomycin, and 50 mM 2-DG. After each assay, cells were lysed, and the protein concentration was measured to normalize the OCR and ECAR.
Extracellular lactate analysis
Primary mouse LFs were pretreated with 100 nM ALA for 1 h and then exposed to 4 ng/mL TGF-β1 (HY-P7117, MedChemExpress, USA) for 24 h. Then, the supernatant was collected to measure extracellular lactate production (A019-2-1, Nanjing Jiancheng, China) according to the manufacturer’s instructions. Lactate levels were measured at 530 nm using a microplate reader.
Mitotracker and lysotracker staining
Mitochondria were examined using Mitotracker Green (C1048, Beyotime, China), and lysosomes were examined using Lysotracker Red (C1046, Beyotime, China). Primary LFs were incubated for 20 min at 37 °C in DMEM supplemented with 20 nM Mitotracker Green and 50 nM Lysotracker together and protected from light. After washing with phosphate-buffered saline three times, cells were photographed under a confocal microscope (LSM880, Carl Zeiss, Germany).
Measurement of mitophagy using mt-Keima
Cells were transfected with the pH-dependent fluorescent protein mitochondria-targeted monomeric Keima-Red (mt-Keima, Genechem, Shanghai, China), a protein that emits green light (ex485 nm) at a neutral pH and emits red light in acidic lysosomes (ex561 nm). Then, the cells were pretreated with or without ALA for 1 h, followed by TGF-β1. Twenty-four hours later, live cells were observed with a confocal microscope (LSM 980, ZEISS, Germany). Then, using ImageJ software, ten cells per group were chosen randomly, and the mitophagy ratio was calculated as the ratio between the red area and the green area.
Mitochondrial superoxide detection
Mitochondrial superoxide levels were detected by MitoSOX™ Red (M36008, Invitrogen, USA) according to the manufacturer’s instructions. Briefly, live cells were incubated with MitoSOX Red (5 μM) for 10 min at 37 °C in the dark. Following washing with warm HBSS/Ca/Mg, cells were imaged by confocal microscopy.
Histopathological and immunohistochemical analyses
For histological analysis, lung tissue sections were subjected to hematoxylin and eosin (H&E) and Masson staining, and the severity of lung fibrosis was analyzed by Ashcroft scoring. For immunohistochemical staining, paraffin-embedded mouse lung sections were incubated with primary antibodies against PFKFB3 (1:200, A5593, Bimake, USA), HK2 (1:200, 66974-1-Ig, Proteintech, USA), and α-SMA (1:200, 19245T, Cell Signaling Technology, USA) according to the manufacturer’s instructions (GK500705, DAKO, Denmark). Images were captured by microscopy (IX73, Olympus or Imager D2, Carl Zeiss).
Immunofluorescence staining
Paraffin sections (4 µm) and cells were prepared for immunofluorescence and incubated with primary antibody overnight at 4 °C. Then, lung sections and cells were stained with the appropriate secondary antibody and mounted with DAPI (F6057, Sigma Aldrich, USA). Images were captured with a confocal microscope (LSM880, Carl Zeiss) or fluorescence microscope (BX63, OLYMPUS). The primary antibodies used included anti-PFKFB3 (1:200, A5593, Bimake), anti-HK2 (1:100, 66974-1-Ig, Proteintech), anti-Parkin (1:100, 66674-1-Ig, Proteintech), anti-cytochrome c oxidase IV (COX IV; 1:100, 66110-1-Ig, Proteintech), anti-LC3B (1:100, A5202, Bimake/1:200, 83506S, Cell Signaling Technology), anti-LAMP1 (1:100, 32731, Signalway Antibody, USA), and anti-Collagen I (1:100, ab260043, Abcam, USA/1:100, 67288-1-Ig, Proteintech).
Western blot analysis
Equal amounts of proteins from cell lysate or tissues, as determined by BCA protein estimation, were boiled with 5× loading buffer and separated by 10% or 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The proteins were transferred to 0.22 μm polyvinylidene fluoride membranes and then blocked with 5% bovine serum albumin for 1 h at room temperature. Next, the membranes were incubated with primary antibodies, including antibodies against collagen I (1:1000, 14695-1-AP, Proteintech), α-SMA (1:1000, 19245 T, Cell Signaling Technology), PFKFB3 (1:1000, A5593, Bimake), HK2 (1:1000, 66974-1-Ig, Proteintech), β-actin (1:1000, AM1021B, Abcepta, China), LC3B (1:1000, A5202, Proteintech), VDAC1 (1:1000, A5224, Bimake), and Parkin (1:1000, 66674-1-Ig, Proteintech) overnight at 4 °C. The membranes were washed three times with TBS-T buffer and then incubated with secondary antibodies (FDR007, FDM007, Fdbio Science, China) at room temperature for another 1 h. After further washing of the membranes three times with TBS-T buffer, protein signals were detected using a BLT GelView 6000 Pro system.
qRT-PCR
RNA was extracted using TRIzol (9109, Takara, Japan), and cDNA was synthesized using PrimeScript™ RT Master Mix (RR036A, Takara, Japan). qPCR was performed using TB Green™ Premix Ex Taq™ (RR420B, Takara, Japan) with a Roche LightCycler® 480. The primers used in this study are presented in Table 1. The relative fold change was calculated by the comparative CT method.
RNA-seq data analysis
The RNA-seq data of lung tissue and fibroblasts isolated from IPF patients were downloaded from GEO DataSets of NCBI (GEO accession: GSE185492, GSE52463). Heatmap analysis was performed using the pheatmap (v1.0.12) package via R scripts.
Statistical analysis
All the results are expressed as the mean ± SD. Data analysis was performed using GraphPad Prism 8. Intergroup comparisons of mean values were performed by one-way analysis of variance. Statistical significance was defined as P < 0.05.
Results
ALA/MrgD axis attenuated bleomycin-induced pulmonary fibrosis and TGF-β1-induced fibroblast activation
To explore the precise mechanism of pulmonary fibrosis, BLM-induced lung fibrosis was established. Sections of lung tissue from the BLM group displayed severe lung fibrosis compared with the control group as determined by H&E and Masson staining and Ashcroft score (Fig. 1A, B). The expression of α-SMA in lung tissue also increased in the BLM group, as detected by immunohistochemistry (Fig. 1C). Furthermore, the protein expression of collagen I and α-SMA was enhanced in BLM-induced fibrotic lung (Fig. 1D). By contrast, the BLM + ALA group showed an apparent reduction in lung fibrosis (Fig. 1A, B). Moreover, treatment with ALA inhibited the expression of collagen I and α-SMA in the BLM + ALA group (Fig. 1C, D). In vitro, the protein and mRNA levels of fibroblast markers collagen I and α-SMA in primary LFs were significantly decreased in the TGF-β1 + ALA group compared with the TGF-β1 group, and the MrgD receptor agonist β-alanine enhanced the effect of ALA whereas the MrgD receptor antagonist D-pro7-Ang-(1-7) or siRNA knockdown of MrgD had the opposite effect (Fig. 1E–G). These data suggest that the ALA/MrgD axis has an attenuating effect on TGF-β1-induced fibroblast activation and BLM-induced PF.
ALA attenuates bleomycin-induced lung fibrosis and TGF-β1-induced lung fibroblast activation. A Representative images of lung sections stained with H&E and Masson’s trichrome. Bar: 100 μm. B Ashcroft scores for each group. C Immunohistochemical staining was performed to determine the localization and expression of α-SMA. Bar: 50 μm. D Protein levels of collagen I and α-SMA in lung tissues were analyzed by western blotting. E, F Protein levels of collagen I and α-SMA. G mRNA levels of Col1a1 and Acta2 in fibroblasts. Original magnification: 200×. Data are presented as mean ± SEM. *P < 0.05 versus vehicle group; #P < 0.05 versus BLM group or TGFβ1 group; △P < 0.05 versus the TGFβ1 + ALA group (n = 3)
ALA/MrgD axis inhibited fibroblast metabolic reprogramming induced by TGF-β1
Next, we investigated whether the ALA/MrgD axis inhibited TGF-β1-induced metabolic reprogramming, and some glycolysis indicators were tested. We found that ALA treatment increased OCR levels compared to the TGF-β1 group (Fig. 2A–C). In addition, ECAR analysis showed that TGF-β1 treatment increased glycolysis and glycolytic activity, both of which were reduced by ALA (Fig. 2D–F). Furthermore, ALA significantly decreased extracellular lactic acid levels but increased ATP production compared with the TGF-β1 group (Fig. 2F, G), indicating that metabolic reprogramming induced by TGF-β1 was mitigated by the ALA/MrgD axis. Next, to determine the precise mechanism by which ALA regulates glucose metabolism, we detected the expression of PFKFB3 and HK2, two key enzymes of the glycolytic pathway. TGF-β1 upregulated their expression at both the protein and mRNA levels, whereas these effects were reversed by the ALA/MrgD axis (Fig. 2I–K). As lactic acid could induce LF activation [22] and ALA could downregulate lactic acid (Fig. 2G), we next explored whether lactic acid could induce glycolysis, with ALA addition having the opposite effect. We found that L-lactic acid alone upregulated the expression of HK2 and PFKFB3, whereas ALA reversed this effect (Fig. 2L). These data suggest that the induction of glycolysis by TGF-β1 could be counteracted by the ALA/MrgD axis through multiple targets.
ALA/MrgD axis inhibited glycolysis in TGF-β1-induced activated fibroblasts. A OCR was measured using the Seahorse XF. Basal respiration (B) and maximal respiration (C) are quantified and shown as histograms. D ECAR was measured using Seahorse XF. Glycolysis (E) and glycolysis capacity (F) are quantified and shown as histograms. Extracellular lactate concentration (G) and intracellular ATP levels (H). Protein (I, J, L) and mRNA (K) levels of PFKFB3 and HK2 in fibroblasts. Data are presented as mean ± SEM. *P < 0.05 versus control group; #P < 0.05 versus TGF-β1 group or L-lactic acid group; △P < 0.05 versus TGFβ1 + ALA group (n = 3)
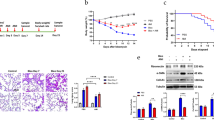
ALA inhibited HK2 and PFKFB3 expression in bleomycin-induced pulmonary fibrosis
Next, we tested whether ALA could inhibit the expression of HK2 and PFKFB3 in vivo. The expression of PFKFB3 and HK2 was increased by BLM but decreased by ALA infusion (Fig. 3A). The protein levels in lung tissue displayed the same tendency (Fig. 3B). Then, using collagen I as a marker of fibroblasts, we found that the co-localization of collagen I and PFKFB3 or HK2 was elevated in the BLM group but decreased by ALA (Fig. 3C, D). These findings reveal that HK2/PFKFB3-mediated glycolysis in fibroblasts has a non-negligible role in BLM-induced lung fibrosis.
ALA inhibited activation of glycolysis in bleomycin-induced pulmonary fibrosis. A Immunohistochemical staining to determine the localization and expression of HK2 and PFKFB3. Bar: 50 μm. B Protein levels of PFKFB3 and HK2 in lung tissues. C, D Co-localization of PFKFB3 and HK2 with collagen I in lung tissues. Original magnification: 200× or 100 pixels. Data are presented as mean ± SEM. *P < 0.05 versus vehicle group; #P < 0.05 versus BLM group
ALA inhibited fibroblast activation by repressing glycolysis
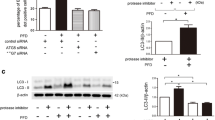
Then, we explored whether ALA exerted an unfavorable effect on fibroblast activation by repressing glycolysis. 3PO and 2DG, inhibitors repressing PFKFB3 and HK2, respectively, could counteract the pro-fibrotic effect of TGF-β1 (Fig. 4A), which is in line with previous research [4, 23]. Next, we constructed a PFKFB3 overexpression plasmid and transferred it to fibroblasts (Fig. 4B, C). PFKFB3 overexpression reversed the effect of ALA on TGF-β1-induced fibroblast activation (Fig. 4D). Overexpression of PFKFB3 alone promoted the expression of collagen I and α-SMA in fibroblasts, and this phenomenon was repressed by ALA (Fig. 4E). L-lactic acid alone also induced fibroblast activation, but this was repressed by ALA (Fig. 4F). Taken together, the above data demonstrate that ALA inhibits fibroblast activation by repressing TGF-β1-induced PFKFB3/HK2/lactate-mediated glycolysis.
ALA inhibited lung fibroblast activation by repressing glycolysis. A Protein levels of collagen I and α-SMA in fibroblasts treated with TGF-β1 with or without 3PO and 2DG, the inhibitor of PFKFB3 and HK2, respectively. B, C mRNA and protein expression of PFKFB3 in fibroblasts processed with PFKFB3 overexpression plasmid or mock plasmid. D–F Collagen I and α-SMA expression. Data are presented as mean ± SEM. *P < 0.05 versus control group; #P < 0.05 versus TGF-β1 group or PFKFB3 overexpression group or L-lactic acid group; △P < 0.05 versus TGFβ1 + ALA group (n = 3)
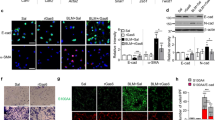
ALA increased Parkin/LC3B-driven mitophagy in fibroblasts and fibrotic lungs
Next, to determine whether metabolic reprogramming is related to mitochondrial homeostasis, we detected mitophagy levels and investigated how ALA regulated mitophagy in fibroblasts. After TGF-β1 treatment, mitochondria degradation by lysosomes declined in LF, while ALA addition visibly improved mitochondria degradation (Fig. 5A). Similarly, ALA increased the co-localization of Mitotracker and Lysotracker, which had been decreased by TGF-β1 (Fig. 5B). To further confirm the effect of TGF-β1 and ALA on mitophagic flux, cells were transfected with mt-Keima. Twenty-four hours after TGF-β1 treatment, the ratio of red to green areas had decreased, while ALA increased mitophagic flux (Fig. 5C, D). The MitoSOX assay also showed that levels of mitochondria reactive oxygen species (mitoROS) were reduced by ALA (Fig. 5E). These results explain how ALA could induce mitophagy to clear damaged mitochondria. Next, the co-localization of LC3B and COX IV (a mitochondrial marker) or Parkin was shown to be decreased by TGF-β1, but both co-localizations were further reversed by adding ALA, revealing that Parkin/LC3B-mediated insufficient mitophagy participated in the process of TGF-β1-induced LF activation and that ALA could upregulate mitophagy (Fig. 5F, G). The proteins expressed in cell lysate and mitochondria showed the same tendency (Fig. 5H, I). In vivo, ALA also upregulated the co-localization of Parkin and LC3B and fibroblast marker collagen I in lung tissue compared with that in the BLM group (Fig. 5J). Collectively, these data indicate that ALA could upregulate Parkin/LC3B-mediated mitophagy to repress TGF-β1-induced LF activation and BLM-induced PF.
ALA upregulated Parkin/LC3B-driven mitophagy in lung fibroblasts. A Transmission electron microscopy analysis showing autophagosome/autolysosome (arrows) after treatment of lung fibroblasts. Bar: 500 μm. B Immunofluorescence analysis of the co-localization of Mitotracker and Lysotracker. Original magnification: 20 μm. C Mitophagy as determined by mt-Keima transfection. Green indicates mitochondria located in the cytoplasm, and red indicates mitochondria located in lysosomes. Original magnification: 50 pixels. D Mitophagy ratios are calculated as the ratio of red area to green area. E mtROS levels as examined using the mitoSOX indicator. Original magnification: 100 μm. F, G Co-localization of LC3B and COX IV or Parkin was assessed by confocal microscopy. Original magnification: 50 pixels and 20 μm. H, I Parkin and LC3B in cell lysate and mitochondrial components. J Co-localization of LC3B, Parkin, and collagen I in lung tissues. Original magnification: 20 μm. Data are presented as mean ± SEM. *P < 0.05 versus control group; #P < 0.05 versus TGF-β1 group (n = 3)
ALA upregulated mitophagy to repress fibroblast activation
To determine why mitophagy was reduced by TGF-β1 and increased by ALA, we focused on LC3B. To clarify whether the increase in LC3B caused by ALA was a result of activated autophagy or blocked autophagic flux, cells were pretreated with bafilomycin A1, which blocks the fusion of autophagosomes and lysosomes. As shown in Fig. 6A, bafilomycin A1 further increased the expression of LC3BII, suggesting that the elevation of LC3BII levels by ALA was due to autophagy induction. We also found that inhibiting autophagy with 3MA could reverse the upregulation of LC3BII by ALA in both mitochondria components and cell lysate (Fig. 6B, C). 3MA could also mediate the expression of collagen I and α-SMA in fibroblast, suggesting that upregulation of LC3B by ALA may make up for the mitochondrial flux and therefore repress fibroblast activation (Fig. 6D). Next, we treated fibroblasts with Parkin siRNA or Mdivi-1 to uncover the relationship between mitophagy and fibroblast activation. Both reversed the inhibitory effect of ALA on collagen I and α-SMA (Fig. 6E, F). Therefore, we conclude that ALA may inhibit LF activation via the upregulation of Parkin/LC3B-mediated mitophagy.
ALA upregulated Parkin/LC3B-mediated mitophagy to repress lung fibroblast activation. A–C Protein levels of LC3BII in cell lysate and mitochondrial components. D–F Collagen I and α-SMA were assessed after treatment with 3MA (10 μmol/L), parkin siRNA, or Mdivi-1 (10 μmol/L). Data are presented as mean ± SEM. *P < 0.05 versus control group; #P < 0.05 versus TGF-β1 group; △P < 0.05 versus TGFβ1 + ALA group; &P < 0.05 versus TGFβ1 + ALA + bafilomycin A1 group (n = 3)
ALA prevented glycolysis via promoting mitophagy/autophagy of fibroblasts
Finally, to confirm the extent to which glycolysis was influenced by mitophagy, we evaluated the expression of PFKFB3 and HK2 in fibroblasts. The results showed that repressing mitophagy using a Parkin inhibitor or siRNA could reverse the suppression of PFKFB3 and HK2 by ALA (Fig. 7A, B). Moreover, 3MA and bafilomycin A1 upregulated the expression of HK2 and PFKFB3, but activation of autophagy with Rapa had the opposite effect (Fig. 7C–F). The co-localization of lysosome marker LAMP1 or autophagosome marker LC3 and HK2 or PFKFB3 is also upregulated by ALA, respectively (Fig. 7H, I). These results show that ALA could induce the autophagic degradation of HK2 and PFKFB3, which is repressed by TGF-β1. As HK2 and PFKFB3 could also be degraded by proteasomes, we investigated whether ALA represses their expression by inducing the proteasomes degradation pathway (Fig. 7G). Collectively, these results suggest that ALA inhibits TGF-β1-induced LF activation by repressing glycolysis via the promotion of mitophagy in fibroblasts.
ALA prevented glycolysis by promoting mitophagy/autophagy in lung fibroblasts. A–G PFKFB3 and HK2 in cell lysate were measured after treatment with Parkin siRNA, Mdivi-1, 3MA, bafilomycin A1, Rapa, and MG132. H, I Co-localization of LC3 or LAMP1 and HK2 or PFKFB3, measured by immunofluorescence. Bar: 20 μm. Data are presented as mean ± SEM. *P < 0.05 versus control group; #P < 0.05 versus TGF-β1 group; △P < 0.05 versus TGFβ1 + ALA group; &P < 0.05 versus TGFβ1 + ALA + bafilomycin A1 (n = 3)
Bioinformatic analysis of glycolysis and mitophagy in LF and lung tissue isolated from IPF patients
Finally, we assessed the gene expression profiles of glycolysis and mitophagy in LF and lung tissue isolated from IPF patients by using the transcriptome data from GEO datasets (Additional file 1: Figs. S1, S2). In LF, the mRNA expression of COL1A1, a marker of activated fibroblasts, is clearly upregulated, while LC3B and PRKN, two mitophagy molecules, are obviously downregulated. In lung tissues isolated from IPF patients, the mRNA expression levels of COL1A1 and ACTA2, markers of activated fibroblasts, are upregulated, while PRKN is mildly downregulated. Two key glycolysis enzymes, HK2 and PFKFB3, show a small increase in fibroblasts and lung tissue, which is consistent with our conclusion.
Discussion
In the present study, we unraveled the mechanism of how the ALA/MrgD axis regulates LF activation in lung fibrosis. Our results showed that TGF-β1-induced LF activation was due to the regulation of metabolic reprogramming. TGF-β1 upregulated glycolysis by dysregulation of transcription levels of HK2 and PFKFB3 while inhibiting autophagic degradation of the corresponding proteins; both these effects were accompanied by a decrease in mitophagy, leading to a block in oxidative phosphorylation. All these effects were ameliorated by ALA (Fig. 8).
Studies have demonstrated that ALA plasmatic concentrations are decreased in IPF patients [24] and that ALA alleviates BLM-induced mice lung fibrosis and angiotensin II (Ang II)-induced fibroblast activation by binding to the MrgD receptor [16, 19]. Other studies found that myocardial, vascular, and liver fibrosis could be inhibited by ALA [17, 18, 25, 26]. However, the role of the ALA/MrgD axis in LF activation remained incompletely understood. Furthermore, interference with glycolysis switch PFKFB3 was found to rescue a mouse abdominal aortic aneurysm disease model induced by Ang II [27]. High glucose is closely related to aberrant glycolysis induction, and Ang II can increase glucose uptake, whereas angiotensin 1–7 (Ang 1–7) decreases it [28, 29]. Given this complex interplay and close associations with glucose metabolism of various components of RAAS, we explored how ALA regulated glycometabolism in LF by binding to the MrgD receptor.
Aerobic glycolysis, also called the Warburg effect, occurs when cells rely mainly on glycolysis to metabolize even in the presence of oxygen [30]. Glycolysis promotes fibroblast activation and proliferation and participates in the development of fibrosis in various organs, including the lung [3, 4, 31, 32]. Our findings presented here are consistent with those of previous studies. To explore whether ALA regulated the development of lung fibrosis through glycolysis, we detected the two main glycolytic switches and found that ALA could modulate the expression of PFKFB3 and HK2 at both the protein and transcript levels and induce them to be degraded by the autophagy-lysosome pathway. In line with Kottmann et al., we further found that lactate, the primary by-product of glycolysis, alone could induce fibroblast activation [22] while maintaining high expression of HK2 and PFKFB3. This indicates the existence of positive feedback involving glycolysis that maintains fibroblasts in their activated state, further supporting the continuous development of LF activation under inhibition by the ALA/MrgD receptor axis. Based on the finding that TGF-β1 activation is often accompanied by extremes of pH in vitro, we speculate that ALA represses lactate-induced fibroblast activation by inhibiting the activation of TGF-β1 [22, 33]; however, further study is needed to explore this in the future. Overexpression of PFKFB3 alone also induced fibroblast activation, suggesting that PFKFB3/lactate-driven glycolysis may have a pivotal role in maintaining fibroblast activation as well as being an indicator of activated fibroblasts. Furthermore, ALA increased the production of ATP repressed by TGF-β1, although the specific mechanism by which this occurred was not explored in the current work. As ALA has many similarities in terms of structure and function with Ang 1–7, we speculate that ALA may increase ATP production by upregulating the expression of citrate synthase, a key enzyme in the tricarboxylic acid cycle, further accelerating the oxidative phosphorylation process to promote ATP production in the same way as Ang 1–7 [34]. The evidence suggests that ALA may be a promising therapeutic target for the repression of LF activation through targeted glycolysis pathways.
Mitophagy is a crucial mean of mitochondrial quality control [10, 35,36,37]. The co-localization of LC3 and mitochondria indicates that mitophagy is in progress [38]. Reduced expression of Parkin and LC3B, two of the crucial factors in this process, indicates insufficient mitophagy in activated fibroblasts [39, 40]. The expression of Parkin in fibroblasts has been shown to decline after treatment with TGF-β1 [41]. Here, we observed reduced expression of Parkin and LC3B both in cell lysate and mitochondria after treatment with TGF-β1. ALA upregulated the production of LC3B to induce autophagy and repressing autophagy with 3MA abolished the anti-fibroblast-activation effect of ALA. We speculated that the upregulation of LC3B by ALA compensated for the mitochondrial flux, thereby maintaining the nonactivated state of fibroblasts. MitoROS, an indicator of mitochondrial homeostasis, was upregulated by TGF-β1 but downregulated by ALA, similar to the effects of Ang 1–7 reported in our previous work [42]; this indicates that insufficient mitophagy flux may cause large numbers of abnormal mitochondria to accumulate in cells such that mitoROS is not removed effectively. Moreover, Li Xiao et al. found that mitoQ, a mitochondria-targeted antioxidant that removes superfluous mitoROS, could also upregulate mitophagy by promoting Parkin expression. All these results indicate a close relationship between mitoQ and mitophagy. How mitoROS affect glycolysis and fibroblast activation through improving mitophagy will be studied in more detail in future research.
As mitochondria are the pivotal organelles for glucose metabolism, our results indicate that abnormal mitophagy may be the main reason for the enhanced glycolysis observed in lung fibrosis. The role of mitophagy in metabolic reprogramming has been controversial [43, 44]. Mitophagy-dependent loss of mitochondria has been reported to induce glycolysis during tumor cells' mitotic arrest [39]. Mitophagy deficiency was also found to lead to aerobic glycolysis, whereas restoration of Parkin could reverse this effect [45]. However, the specific relationship between mitophagy and glycolysis has not been elucidated, nor has the mechanism by which they regulate fibroblast activation and lung fibrosis. In the present study, downregulating Parkin or blocking autophagy flux with bafilomycin A1 increased the expression of glycolysis key molecules; therefore, we concluded that Parkin/LC3B-driven mitophagy/autophagy influences the autophagic degradation of HK2 and PFKFB3 in LF, with further repression of fibroblast activation by ALA through activating mitophagy/autophagy. Zhang et al. previously showed that mitophagy and glycolysis are not regulated in one direction and that glycolysis could also regulate mitophagy through different targets [46]; however, further studies are needed to determine whether the latter leads to LF activation. We also analyzed the public databases from NCBI and found that compared to the normal control group, the tendency of glycolysis and mitophagy in LF and lung tissue isolated from IPF patients are similar to our conclusions. Other researches have also found that the expressions of glycolysis increased, while mitophagy-related molecules LC3B and Parkin decreased in the lung tissue of IPF patients [3, 4, 39, 47]. Despite the promising implications of the finding that ALA alleviated fibroblast pro-fibrotic activity, the present study was limited in that it did not further evaluate the expression of ALA or the relationship between glycolysis and mitophagy in patients with IPF. These aspects will be examined using further experiments in future studies.
Conclusion
In conclusion, we present a previously unrecognized mechanism by which the ALA/MrgD axis could inhibit TGF-β1-induced LF activation by counteracting glycolysis via activating Parkin/LC3-mediated mitophagy. This indicates a potential new strategy for targeted therapy of lung fibrosis with potential clinical applications.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- IPF:
-
Idiopathic pulmonary fibrosis
- PF:
-
Pulmonary fibrosis
- LF:
-
Lung fibroblast
- ALA:
-
Alamandine
- MrgD:
-
MAS-related G-protein coupled receptor D
- ECM:
-
Excessive extracellular matrix
- BLM:
-
Bleomycin
- TGF-β1:
-
Transforming growth factor-beta1
- α-SMA:
-
α-Smooth muscle actin
- RAAS:
-
Renin–angiotensin–aldosterone system
- HK2:
-
Hexokinase 2
- PFKFB3:
-
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- COX IV:
-
Cytochrome c oxidase IV
- LC3B:
-
Light chain 3B
- 3MA:
-
3-Methyladenine
- H&E:
-
Hematoxylin and eosin
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
References
Kang YP, Lee SB, Lee JM, Kim HM, Hong JY, Lee WJ, Choi CW, Shin HK, Kim DJ, Koh ES, et al. Metabolic profiling regarding pathogenesis of idiopathic pulmonary fibrosis. J Proteome Res. 2016;15:1717–24.
Ung CY, Onoufriadis A, Parsons M, McGrath JA, Shaw TJ. Metabolic perturbations in fibrosis disease. Int J Biochem Cell Biol. 2021;139: 106073.
Yin X, Choudhury M, Kang JH, Schaefbauer KJ, Jung MY, Andrianifahanana M, Hernandez DM, Leof EB. Hexokinase 2 couples glycolysis with the profibrotic actions of TGF-β. Sci Signal. 2019. https://doi.org/10.1126/scisignal.aax4067.
Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu RM, Bernard K, Thannickal VJ, Liu G. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am J Respir Crit Care Med. 2015;192:1462–74.
Goodall EA, Kraus F, Harper JW. Mechanisms underlying ubiquitin-driven selective mitochondrial and bacterial autophagy. Mol Cell. 2022;82:1501–13.
Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120:1812–24.
Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31.
Katzen J, Beers MF. Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J Clin Invest. 2020;130:5088–99.
Zhang Y, Huang W, Zheng Z, Wang W, Yuan Y, Hong Q, Lin J, Li X, Meng Y. Cigarette smoke-inactivated SIRT1 promotes autophagy-dependent senescence of alveolar epithelial type 2 cells to induce pulmonary fibrosis. Free Radic Biol Med. 2021;166:116–27.
Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, Corey C, Herazo-Maya JD, Sembrat J, Lee JS, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest. 2015;125:521–38.
Ferro F, Servais S, Besson P, Roger S, Dumas JF, Brisson L. Autophagy and mitophagy in cancer metabolic remodelling. Semin Cell Dev Biol. 2020;98:129–38.
Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–22.
Jiao L, Zhang HL, Li DD, Yang KL, Tang J, Li X, Ji J, Yu Y, Wu RY, Ravichandran S, et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy. 2018;14:671–84.
La Belle FA, Calhoun BC, Sharma A, Chang JC, Almasan A, Schiemann WP. Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat Commun. 2019;10:3668.
Doménech E, Maestre C, Esteban-Martínez L, Partida D, Pascual R, Fernández-Miranda G, Seco E, Campos-Olivas R, Pérez M, Megias D, et al. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nat Cell Biol. 2015;17:1304–16.
Lautner RQ, Villela DC, Fraga-Silva RA, Silva N, Verano-Braga T, Costa-Fraga F, Jankowski J, Jankowski V, Sousa F, Alzamora A, et al. Discovery and characterization of alamandine: a novel component of the renin-angiotensin system. Circ Res. 2013;112:1104–11.
de Souza-Neto FP, Carvalho Santuchi M, de Morais ESM, Campagnole-Santos MJ, da Silva RF. Angiotensin-(1–7) and alamandine on experimental models of hypertension and atherosclerosis. Curr Hypertens Rep. 2018;20:17.
Huang Y, Li Y, Lou A, Wang GZ, Hu Y, Zhang Y, Huang W, Wang J, Li Y, Zhu X, et al. Alamandine attenuates hepatic fibrosis by regulating autophagy induced by NOX4-dependent ROS. Clin Sci. 2020;134:853–69.
Liu Q, Zheng B, Zhang Y, Huang W, Hong Q, Meng Y. Alamandine via MrgD receptor attenuates pulmonary fibrosis via NOX4 and autophagy pathway. Can J Physiol Pharmacol. 2021;99:885–93.
Roscioni SS, Heerspink HJ, de Zeeuw D. The effect of RAAS blockade on the progression of diabetic nephropathy. Nat Rev Nephrol. 2014;10:77–87.
Meng Y, Li T, Zhou GS, Chen Y, Yu CH, Pang MX, Li W, Li Y, Zhang WY, Li X. The angiotensin-converting enzyme 2/angiotensin (1–7)/Mas axis protects against lung fibroblast migration and lung fibrosis by inhibiting the NOX4-derived ROS-mediated RhoA/Rho kinase pathway. Antioxid Redox Signal. 2015;22:241–58.
Kottmann RM, Kulkarni AA, Smolnycki KA, Lyda E, Dahanayake T, Salibi R, Honnons S, Jones C, Isern NG, Hu JZ, et al. Lactic acid is elevated in idiopathic pulmonary fibrosis and induces myofibroblast differentiation via pH-dependent activation of transforming growth factor-β. Am J Respir Crit Care Med. 2012;186:740–51.
Hu X, Xu Q, Wan H, Hu Y, Xing S, Yang H, Gao Y, He Z. PI3K-Akt-mTOR/PFKFB3 pathway mediated lung fibroblast aerobic glycolysis and collagen synthesis in lipopolysaccharide-induced pulmonary fibrosis. Lab Invest. 2020;100:801–11.
Sipriani TS, dos Santos RAS, Rigatto K. The renin-angiotensin system: alamandine is reduced in patients with idiopathic pulmonary fibrosis. J Cardiol Cardiovasc Med. 2019;4:210–5.
Iwata M, Cowling RT, Gurantz D, Moore C, Zhang S, Yuan JX, Greenberg BH. Angiotensin-(1–7) binds to specific receptors on cardiac fibroblasts to initiate antifibrotic and antitrophic effects. Am J Physiol Heart Circ Physiol. 2005;289:H2356-2363.
Yang C, Wu X, Shen Y, Liu C, Kong X, Li P. Alamandine attenuates angiotensin II-induced vascular fibrosis via inhibiting p38 MAPK pathway. Eur J Pharmacol. 2020;883: 173384.
Gäbel G, Northoff BH, Balboa A, Becirovic-Agic M, Petri M, Busch A, Maegdefessel L, Mahlmann A, Ludwig S, Teupser D, et al. Parallel murine and human aortic wall genomics reveals metabolic reprogramming as key driver of abdominal aortic aneurysm progression. J Am Heart Assoc. 2021;10: e020231.
Jiang F, Yang J, Zhang Y, Dong M, Wang S, Zhang Q, Liu FF, Zhang K, Zhang C. Angiotensin-converting enzyme 2 and angiotensin 1–7: novel therapeutic targets. Nat Rev Cardiol. 2014;11:413–26.
Li J, Liu H, Takagi S, Nitta K, Kitada M, Srivastava SP, Takagaki Y, Kanasaki K, Koya D. Renal protective effects of empagliflozin via inhibition of EMT and aberrant glycolysis in proximal tubules. JCI Insight. 2020. https://doi.org/10.1172/jci.insight.129034.
Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–64.
Li Z, Sun C, Qin Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics. 2021;11:8322–36.
Eming SA, Murray PJ, Pearce EJ. Metabolic orchestration of the wound healing response. Cell Metab. 2021;33:1726–43.
Kottmann RM, Trawick E, Judge JL, Wahl LA, Epa AP, Owens KM, Thatcher TH, Phipps RP, Sime PJ. Pharmacologic inhibition of lactate production prevents myofibroblast differentiation. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1305-1312.
Song LN, Liu JY, Shi TT, Zhang YC, Xin Z, Cao X, Yang JK. Angiotensin-(1–7), the product of ACE2 ameliorates NAFLD by acting through its receptor Mas to regulate hepatic mitochondrial function and glycolipid metabolism. Faseb j. 2020;34:16291–306.
Song M, Zhou Y, Fan X. Mitochondrial quality and quantity control: mitophagy is a potential therapeutic target for ischemic stroke. Mol Neurobiol. 2022. https://doi.org/10.1007/s12035-022-02795-6.
Wang S, Tan J, Miao Y, Zhang Q. Mitochondrial dynamics, mitophagy, and mitochondria-endoplasmic reticulum contact sites crosstalk under hypoxia. Front Cell Dev Biol. 2022;10:848214.
Wu NN, Zhang Y, Ren J. Mitophagy, mitochondrial dynamics, and homeostasis in cardiovascular aging. Oxid Med Cell Longev. 2019;2019:9825061.
Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14.
Kobayashi K, Araya J, Minagawa S, Hara H, Saito N, Kadota T, Sato N, Yoshida M, Tsubouchi K, Kurita Y, et al. Involvement of PARK2-mediated mitophagy in idiopathic pulmonary fibrosis pathogenesis. J Immunol. 2016;197:504–16.
Kurita Y, Araya J, Minagawa S, Hara H, Ichikawa A, Saito N, Kadota T, Tsubouchi K, Sato N, Yoshida M, et al. Pirfenidone inhibits myofibroblast differentiation and lung fibrosis development during insufficient mitophagy. Respir Res. 2017;18:114.
Sosulski ML, Gongora R, Danchuk S, Dong C, Luo F, Sanchez CG. Deregulation of selective autophagy during aging and pulmonary fibrosis: the role of TGFβ1. Aging Cell. 2015;14:774–83.
Pan M, Zheng Z, Chen Y, Sun N, Zheng B, Yang Q, Zhang Y, Li X, Meng Y. Angiotensin-(1–7) attenuated cigarette smoking-related pulmonary fibrosis via improving the impaired autophagy caused by nicotinamide adenine dinucleotide phosphate reduced oxidase 4-dependent reactive oxygen species. Am J Respir Cell Mol Biol. 2018;59:306–19.
Saxena S, Mathur A, Kakkar P. Critical role of mitochondrial dysfunction and impaired mitophagy in diabetic nephropathy. J Cell Physiol. 2019;234:19223–36.
Panigrahi DP, Praharaj PP, Bhol CS, Mahapatra KK, Patra S, Behera BP, Mishra SR, Bhutia SK. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin Cancer Biol. 2020;66:45–58.
Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci USA. 2011;108:16259–64.
Zhang T, Liu Q, Gao W, Sehgal SA, Wu H. The multifaceted regulation of mitophagy by endogenous metabolites. Autophagy. 2022;18:1216–39.
Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, Rosas IO, Morse D. Autophagy in idiopathic pulmonary fibrosis. PLoS ONE. 2012;7: e41394.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81570064, No. 81870068, No. 82070063) and Key Laboratory of Emergency and Trauma (Hainan Medical University), Ministry of Education (Grant. KLET-202102).
Author information
Authors and Affiliations
Contributions
YM: conceptualization, validation, resources, data curation, writing—review and editing, supervision, project administration, funding acquisition; XL: conceptualization, validation, resources, data curation, writing—review and editing, supervision, project administration, funding acquisition; WW: methodology, validation, formal analysis, investigation, writing—original draft preparation; YZ: methodology, software, formal analysis, investigation; WHH: software, formal analysis, investigation; YFY: formal analysis; HQH: investigation; ZZX: investigation; LJL: investigation; YXC: investigation. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All experimental procedures on mice were approved by Committee on the Ethics of Animal Experiments of Southern Medical University (Permit No. SYXK 2015-0056).
Consent for publication
Yes.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1
. Gene expression of glycolysis and mitophagy in fibroblast isolated from IPF patients. A This heatmap shows the expression level of glycolysis-related and mitophagy-related genes. B Raw counts were converted into transcripts per million (TPM) for comparison in different samples. Boxplot of transcripts per million (TPM) showing glycolysis-related, mitophagy-related, and marker genes in fibroblast that with slight variation and corresponding trends. Figure S2. Gene expression of glycolysis and mitophagy in lung tissue isolated from IPF patients. A This heatmap shows the expression level of glycolysis-related and mitophagy-related genes. B Raw counts were converted into transcripts per million (TPM) for comparison in different samples. Boxplot of transcripts per million (TPM) showing glycolysis-related, mitophagy-related, and marker genes in lung tissue with slight variation and corresponding trends.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, W., Zhang, Y., Huang, W. et al. Alamandine/MrgD axis prevents TGF-β1-mediated fibroblast activation via regulation of aerobic glycolysis and mitophagy. J Transl Med 21, 24 (2023). https://doi.org/10.1186/s12967-022-03837-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-022-03837-2