Abstract
Background
Because severe acute respiratory syndrome coronarivus 2 (SARS-CoV-2) leads to severe conditions and thrombus formation, evaluation of the coagulation markers is important in determining the prognosis and phenotyping of patients with COVID-19.
Methods
In a prospective study that included 213 COVID-19 patients admitted to the intensive care unit (ICU) the levels of antithrombin, C-reactive protein (CRP); factors XI, XII, XIII; prothrombin and D-dimer were measured. Spearman’s correlation coefficient was used to assess the pairwise correlations between the biomarkers. Hierarchical and non-hierarchical cluster analysis was performed using the levels of biomarkers to identify patients´ phenotypes. Multivariate binary regression was used to determine the association of the patient´s outcome with clinical variables and biomarker levels.
Results
The levels of factors XI and XIII were significantly higher in patients with less severe COVID-19, while factor XIII and antithrombin levels were significantly associated with mortality. These coagulation biomarkers were associated with the in-hospital survival of COVID-19 patients over and above the core clinical factors on admission. Hierarchical cluster analysis showed a cluster between factor XIII and antithrombin, and this hierarchical cluster was extended to CRP in the next step. Furthermore, a non-hierarchical K-means cluster analysis was performed, and two phenotypes were identified based on the CRP and antithrombin levels independently of clinical variables and were associated with mortality.
Conclusion
Coagulation biomarkers were associated with in-hospital survival of COVID-19 patients. Lower levels of factors XI, XII and XIII and prothrombin were associated with disease severity, while higher levels of both CRP and antithrombin clustered with worse prognosis. These results suggest the role of coagulation abnormalities in the development of COVID-19 and open the perspective of identifying subgroups of patients who would benefit more from interventions focused on regulating coagulation.
Similar content being viewed by others
Background
According to disease severity, excessive production of thrombin, damage to the endothelial cells, inhibition of fibrinolysis, activation of the complement pathway, deposition of microthrombi and microvascular dysfunction, are characteristics of COVID-19 patients [1,2,3,4]. Patients admitted to the hospital due to severe COVID-19 are predisposed to endothelial cell activation and injury, platelet activation, and hypercoagulability. One of the consequences of COVID-19 is thrombotic and thromboembolic events, such as disseminated intravascular coagulation, deep vein thrombosis and pulmonary embolism [5, 6]. Arterial thrombosis, including ischemic stroke, acute coronary syndrome, limb ischemia and systemic arterial embolism can occur. The incidence varies according to the severity of the disease, with a higher prevalence in patients admitted to the intensive care unit (ICU) [7, 8].
Excessive inflammation caused by SARS-CoV-2 systemically activates blood clotting, probably due to the release of von Willebrand factor (vWF) and increased endothelial cell surface expression of adhesion molecules, favoring thrombus formation [9,10,11]. Consequently, the activation of the coagulation cascade promotes platelet aggregation, neutrophil activation and the release of neutrophil extracellular traps that propagate vascular and organ injury [9,10,11,12,13,14].
Elevated levels of clotting markers are important for determining the prognosis of patients with COVID-19, as infection with SARS-CoV-2 can lead to severe conditions and thrombus formation [6, 15]. Assessment of blood clotting factors, including pro-clotting factors such as fibrinogen, prothrombin, and factors XI, XII, and XIII, and natural anticoagulants such as antithrombin is critical in understanding the pathophysiological processes underlying the development of COVID-19 and its complications. Furthermore, analyzing these markers can help identify individuals at an increased risk of developing thrombosis and potentially guide thromboprophylaxis and treatment approaches [16, 17].
Patients with COVID-19 have high D-dimer levels, prolonged prothrombin time (PT) or activated partial thromboplastin time (aPTT), decreased factor V activity, and hypofibrinogenemia [18,19,20,21]. A recent meta-analysis on coagulation dysfunction found that the D-dimer levels, fibrinogen levels, aPTT and PT were significantly higher in severe COVID-19 patients [22]. However, most of the included studies were retrospective and did not consider the potential confounding factors affecting the association between coagulation markers and disease severity. In addition, data on the coagulation tests that are not routinely used in clinical practice are lacking. In this context, the prognostic value of coagulation biomarkers (CB) over and above the disease severity scores already in use has gained importance in prioritizing patient care.
The present prospective cohort study of hospitalized COVID-19 patients aimed to verify the following hypotheses: a) patient clustering of non-conventional coagulation parameters is predictive of in-hospital survival, and b) these biomarkers can be used in combination with conventional clinical parameters in the prognostication of in-hospital survival.
Methods
Study design
A prospective cohort study was conducted on patients admitted to two tertiary hospitals in southern Brazil between June 2020 and November 2020. The study was approved by the Institutional Review Board of each institution. In addition, all patients or their surrogates provided written informed consent before their inclusion in the study.
Setting
The study sample consisted of all consecutive patients admitted to the intensive care unit (ICU) of participating hospitals from June 2020 to November 2020.
Participants
Patients aged > 18 years who were diagnosed with COVID-19 through reverse transcriptase reaction or rapid antigen test and required supplementary oxygen (World Health Organization (WHO) class 4), noninvasive ventilation (WHO class 5), or invasive mechanical ventilation (WHO class 6) due to COVID-19 pneumonia were included in the study. By contrast, patients with severe chronic diseases (chronic kidney disease undergoing dialysis, Child–Pugh class C cirrhosis, severe chronic obstructive pulmonary disease, severe heart failure) or diseases that alter the inflammatory response, such as those with long-term use of immunosuppressants, with cancer without disease control, with human immunodeficiency virus infection without disease control, and who received palliative care, or with a life expectancy of less than 24 h as judged by the attending physician were excluded.
Procedures
After patient inclusion, venous blood samples were collected within 24 h after ICU admission; meanwhile the sociodemographic and clinical information was collected directly from the patient, their surrogate, or the electronic medical records. The levels of CB (antithrombin; C-reactive protein (CRP); factors XI, XII, and XIII; and prothrombin) were measured using the Coagulation 6-Plex Human ProcartaPlex Panel 1 (Cat. #EPX060-10824–901), from Thermo Fisher Scientific, (Waltham, MA, USA) in a Luminex MAGPIX® system (Luminex Corporation -Austin, TX, USA). Final protein concentrations were calculated using the online Procarta Plex Analysis Application (Thermo Fisher Scientific) and expressed as arbitrary units from the reference plasma. Additionally, as a conventional coagulation biomarker D-dimer was measured using an ELISA kit according to manufacturer instructions (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
The independent groups were compared using a Mann–Whitney U test or a Kruskal–Wallis test and chi-square test for continuous and categorical variables in the univariate analyses. Spearman’s correlation coefficient was used to assess the pairwise correlations between the numerical variables. A hierarchical cluster of the corresponding correlation matrix was performed to identify the clusters of co-expressed biomarkers. Additionally, a non-hierarchical K-means clustering analysis was performed to assess for CB clustering, which was subsequently related to the outcome. Multivariate binary (logistic) regression was used to determine the association of the patient´s outcome with other independent variables, conceptually divided into core demographic (age and sex) and clinical prognostic factors (SAPS III, Charlson score, chest computed tomography (CT) score, and body mass index (BMI)), hitherto denominated “core predictors”, and the aforementioned CB. The latter were categorized into quintiles owing to their highly non-normal distributions, with missing values fitted as an additional category to avoid the impact of selective dropout. The receiver operating curves (ROC) for core alone and core plus the CB predictors were compared in terms of accuracy, measured by the area under the ROC (AUROC), as well as the sensitivity, specificity, positive and negative predictive values, and likelihood ratios of positive and negative tests. The AUROC was cross-validated on five independent samples to avoid model fitting and evaluation of the same sample. Bootstrap bias corrected 95% confidence intervals (CI) were used to express the AUROC uncertainty.
Data were analyzed using IBM SPSS Statistics version 22.0 (IBM Corp., Armonk, N.Y., USA) and Stata version 13.1 (StataCorp, College Station, TX, USA) software. The type I error level was set to 0.05 in all statistical analyses.
Results
Clinical characteristics of the sample
The demographic data, comorbidities, disease characteristics at hospital admission and clinical outcomes of the 213 patients in the analytical sample are listed in Table 1. The in-hospital mortality in the analytical sample was 25% (53 of 213 patients). Results of the univariate analysis showed that age, need for mechanical ventilation, comorbidities (measured based on the Charlson score), BMI, disease severity at ICU admission (measured based on the SAPS III score), and degree of organ dysfunction (measured based on the total SOFA score at admission), were all associated with mortality.
Coagulation parameters and disease severity
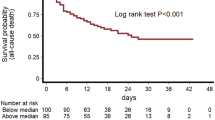
First, the differences in the levels of measured CB between the three crescent WHO ordinal scale severities (from 4 to 6, Fig. 1 A-F). Only the concentrations of factors XI (Fig. 1B) and XIII (Fig. 1F) were significantly different between the groups (p = 0.01 and p = 0.013, respectively), being higher in those with less severe types of COVID-19. The levels of prothrombin (Fig. 1C) and factor XII (Fig. 1E) were higher in the WHO category 4 patients. D-dimer levels did not differ between groups (p = 0.55). Additionally, the levels of antithrombin (Fig. 2D) and factor XIII (Fig. 2F) were significantly associated with mortality. Furthermore, D-dimer levels were also associated with mortality (453 ± 308 vs 704 ± 470, p = 0.0001, in survivors and non-survivors respectively).
Spearman’s correlation coefficient was used to assess the pairwise correlation of these biomarkers (Fig. 3). Differently from non-conventional CB, D-dimer did not significantly correlate to any of another measured parameter (Fig. 3). Using hierarchical clusters it was observed a cluster between factor XIII and antithrombin reinforced the observed association between factor XIII and antithrombin levels and in-hospital mortality. Interestingly, this hierarchical cluster was extended to CRP. To further explore a potential relationship, a non-hierarchical K-means cluster analysis was performed. Two phenotypes were identified based on the CRP and antithrombin levels, but they did not include factor XIII (Table 1, Supplementary Material). Despite the higher mortality rate among phenotype 2 patients compared with phenotype 1 patients, their clinical characteristics were similar, except for the higher total organ dysfunction score (Table 2). D-dimer did not significatively add to non-hierarchical K-means cluster. Adding D-dimer to the other biomarkers phenotype 1 included only 2 patients.
We further investigated the diagnostic performance of the CB in the multivariate logistic regression analysis of in-hospital death (Table 2, Supplementary Material). Adding the CB to the core clinical predictors (Table 3) significantly improved the AUROC (p = 0.036), from 79% (95% CI 73%—84%) to 86% (95% CI 81%—90%). Significant improvements were also observed in the sensitivity value (from 38 to 57% (p = 0.02), positive predictive value (PPV) (from 65 to 81% (p = 0.054), and for likelihood ratios of a positive/negative test (p < 0.001). A two-fold increase in the likelihood ratio of a positive test following the addition of the biomarkers (Table 3) suggests a two-fold increase in the odds of correctly predicting in-hospital mortality. Adding D-dimer to core clinical predictors also improved the AUROC (from 79%—95% CI 73% to 84% vs 80%—95% CI 72% to 88).
If the false positive test result ranges from 10–20%, which is considered acceptable in several screening applications, the addition of CB increases the chance of correctly predicting the in-hospital death among COVID-19 patients by approximately 20%-30% compared with the ROC without the CB (Fig. 1, Supplementary Material).
Discussion
Here, a significant improvement was observed in predicting COVID-19 in-hospital mortality when non-conventional CB were added to the core clinical indicators of disease severity assessed on ICU admission. The significant improvements in various diagnostic parameters, such as sensitivity, PPV, accuracy, and diagnostic odds, indicate the relevant gains in various aspects of predicting in-hospital death. Additionally, two different biomarkers were associated with mortality, and were clustered based on CRP and antithrombin levels. Conventional coagulation parameters, such as D-dimer, were also associated with mortality, but surprisingly did not help to clustering patients in phenotypes.
From the beginning of the pandemic, blood coagulation, assessed based on the D-dimer levels, has been associated with higher mortality rates [23]. Additional data suggest that several other routinely assessed coagulation parameters, such as platelet count, PT, and fibrinogen, were associated with disease severity and mortality [24]. From a mechanistic point of view, hypercoagulability is associated with elevated vWF, endothelial dysfunction, elevated fibrinogen and factor VIII levels, and reduced thrombomodulin levels [25]. Using a similar approach, Ceballos et al. 2021 [16] found a significant reduction in factor XI, factor XII, and factor XIII levels in patients with severe COVID-19. Herein, the levels of factors XI and XIII were statistically higher in the WHO category 4 group than in the more severe forms group. The levels of factor XII (p = 0.068) and prothrombin (p = 0.067) showed a trend to be higher, reinforcing the possible role of a procoagulant state in the progression of COVID-19. However, Ceballos et al. 2021 [16] study has several limitations. For example, noninvasive ventilated patients were not mentioned; blood samples were collected within a median time of 2 days (IQR 4 days). Additionally, Ceballos et al. [16] divided the patients into three quantiles (low, medium and high protein levels) and reported lower levels of coagulant factors in non-survivors; however, adding coagulation proteins to the survival models that included sex and age did not improve the survival prediction. In our study, categorizing CB into quintiles led to an improvement in prognostic assessment compared with the use of clinical variables alone. Furthermore, the sensitivity increased in the region with a low (< 20%) false-positive rate, which may be useful for prioritizing ICU resources.
Another relevant aspect of coagulation disorders and COVID-19 is whether a specific treatment aimed at coagulation would benefit the patients. Early in the pandemic, some authors recommended stratifying doses according to the D-dimer levels or extended post-discharge thromboprophylaxis [26]. Existing evidence shows that therapeutic-dose heparin benefits non-critically ill patients hospitalized due to COVID-19 [27,28,29], but it could induce harm in the critically ill [27]. Thus, the characteristics of patients who would benefit more from anticoagulation therapy should be indicated. Both hierarchical and k-cluster analyses could be used to stratify patients based on CB. In the k-cluster analyses, one phenotype was associated with the most severe organ dysfunction and higher mortality rates. It was not expected that D-dimer did not improve patients´ phenotyping. This finding opens the perspective that different phenotypes would respond differently to anticoagulation treatments, and further studies should address this issue. Using creatinine, albumin, CRP, white blood cell count and clinical characteristics five phenotypes of hospitalized COVID-19 patients were identified [30]. Patients with one of these phenotypes had renal failure, hypoalbuminemia, anemia, lymphopenia, and elevated CRP level (median, 9.0 mg/dL), and had the highest likelihood of ICU transfer or in-hospital mortality (59%). The use of 22 candidate variables for clustering analysis that included demographic information, disease history, major clinical symptoms, and medications on the day of positive diagnosis could also help determine the phenotype [31]. In these patients, three sub-phenotypes, determined mainly by a history of chronic hypertension, the presence of fever, development of respiratory and non-respiratory symptoms, and age, were associated with clinical deterioration. Recently, using four distinct clinical phenotypes described in non-COVID-19 septic patients it was determined that some of these phenotypes were more common in bacterial, when compared to viral sepsis [32]. Additionally, some phenotypes were associated with better outcomes after the introduction of dexamethasone therapy in COVID-19, reinforcing the idea that phenotyping patients would impact on prognostication and treatment stratification. To the best of our knowledge the present study was the first to provide discriminative phenotypes based only on the levels of CB.
This study has some limitations. First, the data were collected only from two hospitals in South Brazil, thus multicenter studies are needed to verify the generalizability of the study findings. Nevertheless, many patients with sufficient outcomes (severity and mortality) were included to allow robust regression and cluster analyses. Second, only a restricted number of coagulation proteins were analyzed at a single time point after hospital admission, leading to the lack of information on coagulation modifications over time and on more conventional coagulation parameters. However, several studies were conducted in the latter [24], hence our findings significantly add to the literature. Third, all included patients were not vaccinated, thus it is not possible to ascertain how the post-vaccination status impacts these results. Some data support that clinically these patients are similar regarding clinical characteristics, at least when hospitalization is needed [33]. Not only the vaccination status, but reinfection could also interfere in the immune response, and consequently in biomarker profile. For example, increased risks of haematological and vascular events that led to hospital admission or death were observed for short time intervals after first doses of vaccines. The risks of most of these events were substantially higher and more prolonged after SARS-CoV-2 infection than after vaccination in the same population [34]. Thus, coagulation biomarkers or coagulation-related events could be potentiated in vaccinated patients that develop severe COVID-19. Fourth, we did not measure coagulation biomarkers in healthy individuals, and this could add some valuable information to our data interpretation.
Conclusion
CB were associated with the in-hospital survival of COVID-19 patients over and above the core clinical factors on admission. Adding these biomarkers significantly increased the sensitivity, PPV, accuracy, and diagnostic odds. Lower levels of some blood clotting factors were associated with disease severity, while higher levels of both CRP and antithrombin clustered with worse prognosis. These results suggest the role of coagulation abnormalities in COVID-19 development and help to identify the subgroups of patients who would benefit more from interventions focused on coagulation.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- aPTT:
-
Activated partial thromboplastin time
- AUROC:
-
Area under receiver operating curve
- BMI:
-
Body mass index
- CB:
-
Coagulation biomarkers
- CT:
-
Computed tomography
- COVID-19:
-
Coronavirus disease 2019
- CRP:
-
C-reactive protein
- ICU:
-
Intensive care unit
- PPV:
-
Positive predictive value
- PT:
-
Prothrombin time
- SOFA:
-
Sequential Organ Failure Assessment
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronarivus 2
- SAPS:
-
Simplified Acute Physiology Score
- vWF:
-
Von Willebrand factor
- WHO:
-
World Health Organization
References
Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924.
Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–8.
Chen TL, Dai Z, Mo P, Li X, Ma Z, Song S, et al. Clinical Characteristics and Outcomes of Older Patients with Coronavirus Disease 2019 (COVID-19) in Wuhan, China: a single-centered, retrospective study. J Gerontol A Biol Sci Med Sci. 2020;75:1788–95.
Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708–20.
Dolhnikoff M, Duarte-Neto AN, de Almeida Monteiro RA, da Silva LFF, de Oliveira EP, Saldiva PHN, et al. Pathological evidence of pulmonary thrombotic phenomena in severe COVID-19. J Thromb Haemost. 2020;18:1517–9.
Perico L, Benigni A, Casiraghi F, Ng LFP, Renia L, Remuzzi G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol. 2021;17:46–64.
Burn E, Duarte-Salles T, Fernandez-Bertolin S, Reyes C, Kostka K, Delmestri A, et al. Venous or arterial thrombosis and deaths among COVID-19 cases: a European network cohort study. Lancet Infect Dis. 2022. https://doi.org/10.1016/s1473-3099(22)00223-7.
Gorog DA, Storey RF, Gurbel PA, Tantry US, Berger JS, Chan MY, et al. Current and novel biomarkers of thrombotic risk in COVID-19: a Consensus Statement from the International COVID-19 Thrombosis Biomarkers Colloquium. Nat Rev Cardiol. 2022;19(7):475–95.
Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–62.
Savla SR, Prabhavalkar KS, Bhatt LK. Cytokine storm associated coagulation complications in COVID-19 patients: pathogenesis and management. Expert Rev Anti Infect Ther. 2021;19:1397–413.
Vinayagam S, Sattu K. SARS-CoV-2 and coagulation disorders in different organs. Life Sci. 2020;260:118431.
Hidalgo A. A NET-thrombosis axis in COVID-19. Blood. 2020;136:1118–9.
Schurink B, Roos E, Radonic T, Barbe E, Bouman CSC, de Boer HH, et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. The Lancet Microbe. 2020;1:e290–9.
Wichmann D, Sperhake JP, Lütgehetmann M, Steurer S, Edler C, Heinemann A, et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med. 2020;173:268–77.
Polimeni A, Leo I, Spaccarotella C, Mongiardo A, Sorrentino S, Sabatino J, et al. Differences in coagulopathy indices in patients with severe versus non-severe COVID-19: a meta-analysis of 35 studies and 6427 patients. Sci Rep. 2021;11:10464.
Ceballos FC, Ryan P, Blancas R, Martin-Vicente M, Vidal-Alcántara EJ, Peréz-García F, et al. Are reduced levels of coagulation proteins upon admission linked to COVID-19 severity and mortality? Front Med. 2021;8:718053.
Zhou Y, Qian X, Liu Z, Yang H, Liu T, Chen K, et al. Coagulation factors and the incidence of COVID-19 severity: Mendelian randomization analyses and supporting evidence. Signal Transduct Target Ther. 2021;6(1):222.
Miesbach W, Makris M. COVID-19: Coagulopathy, risk of thrombosis, and the rationale for anticoagulation. Clin Appl Thromb Hemost. 2020;26:1076029620938149.
Salamanna F, Maglio M, Landini MP, Fini M. Platelet functions and activities as potential hematologic parameters related to Coronavirus Disease 2019 (Covid-19). Platelets. 2020;31:627–32.
Townsend L, Fogarty H, Dyer A, Martin-Loeches I, Bannan C, Nadarajan P, et al. Prolonged elevation of D-dimer levels in convalescent COVID-19 patients is independent of the acute phase response. J Thromb Haemost. 2021;19:1064–70.
Ribes A, Vardon-Bounes F, Mémier V, Poette M, Au-Duong J, Garcia C, et al. Thromboembolic events and Covid-19. Adv Biol Regul. 2020;77:100735.
Len P, Iskakova G, Sautbayeva Z, Kussanova A, Tauekelova AT, Sugralimova MM, et al. Meta-analysis and systematic review of coagulation disbalances in COVID-19: 41 studies and 17,601 patients. Front Cardiovasc Med. 2022;9:794092.
Tang N, Bai H, Chen X, Gong J, Li D, Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost. 2020;18:1094–9.
Mitra S, Ling RR, Yang IX, Poon WH, Tan CS, Monagle P, et al. Severe COVID-19 and coagulopathy: a systematic review and meta-analysis. Ann Acad Med Singap. 2021;50:325–35.
Goshua G, Pine AB, Meizlish ML, Chang CH, Zhang H, Bahel P, et al. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. The Lancet Haematology. 2020;7:e575–82.
Gomez K, Laffan M, Bradbury C. Debate: Should the dose or duration of anticoagulants for the prevention of venous thrombosis be increased in patients with COVID-19 while we are awaiting the results of clinical trials? Br J Haematol. 2021;192:459–66.
Pr L, Ec G, Js B, Md N, Bj M, Jc N, et al. Therapeutic anticoagulation with heparin in noncritically ill patients with Covid-19. N Engl J Med. 2021;385:790–802.
Sholzberg M, Tang GH, Rahhal H, Alhamzah M, Kreuziger LB, Áinle FN, et al. Effectiveness of therapeutic heparin versus prophylactic heparin on death, mechanical ventilation, or intensive care unit admission in moderately ill patients with covid-19 admitted to hospital: RAPID randomised clinical trial. BMJ. 2021;375:n2400.
Spyropoulos AC, Goldin M, Giannis D, Diab W, Wang J, Khanijo S, et al. Efficacy and safety of therapeutic-dose heparin vs standard prophylactic or intermediate-dose heparins for thromboprophylaxis in high-risk hospitalized patients with COVID-19: The HEP-COVID randomized clinical trial. JAMA Intern Med. 2021;181:1612–20.
Wang X, Jehi L, Ji X, Mazzone PJ. Phenotypes and subphenotypes of patients With COVID-19: a latent class modeling analysis. Chest. 2021;159:2191–204.
Differences in clinical deterioration among three sub-phenotypes of COVID-19 patients at the time of first positive test: results from a clustering analysis. Intensive care medicine. 2021;47:113–5.
Bruse N, Kooistra EJ, Jansen A, van Amstel RBE, de Keizer NF, Kennedy JN, et al. Clinical sepsis phenotypes in critically ill COVID-19 patients. Crit Care. 2022;26:244.
Rahman S, Rahman MM, Miah M, Begum MN, Sarmin M, Mahfuz M, et al. COVID-19 reinfections among naturally infected and vaccinated individuals. Sci Rep. 2022;12:1438.
Hippisley-Cox J, Patone M, Mei XW, Saatci D, Dixon S, Khunti K, et al. Risk of thrombocytopenia and thromboembolism after covid-19 vaccination and SARS-CoV-2 positive testing: self-controlled case series study. BMJ. 2021;374: n1931.
Acknowledgements
Not applicable
Funding
MCTIC/CNPq/FNDCT/MS/SCTIE/DECIT, 07/2020, grant number 401263/2020–7.
BRF S.A. Hub unrestricted donation.
FAPERGS-PPSUS #21/2551–0000073-2
The funding sources did not have any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
E.C. contributed to the study design, data collection and interpretation, and writing of manuscript. R. G. contributed to data collection and interpretation and literature search. C.S.G. contributed to data collection and interpretation. L.S. contributed to data collection and interpretation. J.C.F.M. contributed to the study design and to writing of the manuscript. D.P.G contributed to the study design and to writing of the manuscript. G.A.W. contributed to the study design and to writing of the manuscript. E.K. contributed to the study design, data interpretation and to writing of the manuscript. R.W. contributed to the study design, data interpretation and to writing of the manuscript. C. R. contributed to the study design and to writing of the manuscript. F.D-P, contributed to the study design, data interpretation, and writing of manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki and the Brazilian National Health Council resolution 466. The Ethics Committee of the São José Hospital (31384620.6.1001.5364) and UNIMED Hospital (31384620.6.2002.5362) approved the protocol. All patients or their surrogates gave written consent before inclusion in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Corneo, E., Garbelotto, R., Prestes, G. et al. Coagulation biomarkers and coronavirus disease 2019 phenotyping: a prospective cohort study. Thrombosis J 21, 80 (2023). https://doi.org/10.1186/s12959-023-00524-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12959-023-00524-0