Abstract
Background
Recurrent implantation failure (RIF) is a major limitation of assisted reproductive technology, which is associated with impaired endometrial receptivity. Although N6-methyladenosine (m6A) has been demonstrated to be involved in various biological processes, its potential role in the endometrium of women with RIF has been poorly studied.
Methods
Global m6A levels and major m6A methyltransferases/demethylases mRNA levels in mid-secretory endometrium from normal and RIF women were examined by colorimetric m6A quantification strategy and quantitative real-time PCR, respectively. The effects of METTL3-mediated m6A modification on embryo attachment were evaluated by an vitro model of a confluent monolayer of Ishikawa cells co-cultured with BeWo spheroids, and the expression levels of homeo box A10 (HOXA10, a well-characterized marker of endometrial receptivity) and its downstream targets were evaluated by quantitative real-time PCR and Western blotting in METTL3-overexpressing Ishikawa cells. The molecular mechanism for METTL3 regulating HOXA10 expression was determined by methylated RNA immunoprecipitation assay and transcription inhibition assay.
Results
Global m6A methylation and METTL3 expression were significantly increased in the endometrial tissues from women with RIF compared with the controls. Overexpression of METTL3 in Ishikawa cells significantly decreased the ration of BeWo spheroid attachment, and inhibited HOXA10 expression with downstream decreased β3-integrin and increased empty spiracles homeobox 2 expression. METTL3 catalyzed the m6A methylation of HOXA10 mRNA and contributed to its decay with shortened half-life. Enforced expression of HOXA10 in Ishikawa cells effectively rescued the impairment of METTL3 on the embryo attachment in vitro.
Conclusion
Increased METTL3-mediated m6A modification represents an adverse impact on embryo implantation by inhibiting HOXA10 expression, contributing to the pathogenesis of RIF.
Similar content being viewed by others
Background
Embryo implantation is an important and complex process in the establishment of pregnancy in mammals, which requires the simultaneous development of high-quality embryos and endometrial receptivity [1,2,3]. Endometrium is one of the most dynamic tissues in human body. Under the action of steroids during sexual cycle, endometrium undergoes cyclic developmental changes and highly ordered differentiation, leading to be receptive to blastocyst implantation ∼6 days after ovulation and remains receptive for 4 days (cycle days 20-24) [4, 5]. Endometrial receptivity deficiency may be the critical factor for women with recurrent implantation failure (RIF) who have high-quality embryos but undergo repeated implantation failure following in vitro fertilization-embryo transplantation (IVF-ET) treatment [6]. Several different signaling pathways and their associated genes have been demonstrated to be involved in the adjustment of endometrial receptivity [5, 7], in which homeo box A10 (HOXA10) has emerged as an important and well-characterized biomarker. The expression of HOXA10 is dynamic through the menstrual cycle and significantly increased at the time of implantation with increased progesterone levels [8, 9]. The gene HOXA10 has a highly conserved homeodomain that specifically recognizes the TTAT sequence in the promoter of downstream target genes, leading to expression changes of target genes, including β3-integrin (ITGB3) and empty spiracles homeobox 2 (EMX2) [10,11,12]. Studies have shown that ITGB3 expression is directly up-regulated by HOXA10 [10], whereas EMX2 expression is inhibited by HOXA10 [12]. ITGB3 is a transmembrane glycoprotein that presents on the surface of cells, which participates in cell adhesion and cell-surface-mediated signaling during embryo implantation [13,14,15]. EMX2 is a crucial transcription factor necessary for reproductive tract differentiation and development, but changes in endometrial EMX2 expression levels usually lead to abnormalities of the endometrium [16, 17]. Accumulating studies indicate that decreased HOXA10 expression contributes to the failure of embryo implantation [18,19,20,21]. However, the expression of HOXA10 and its underlying mechanisms for epi-transcriptomic regulation of HOXA10 in RIF remain to be characterized.
N6-methyladenosine (m6A) is the most abundant internal modification in messenger RNAs (mRNAs). The m6A modification is catalyzed by “writers” methyltransferases, including methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), RNA binding motif protein 15 (RBM15) and Wilms tumor 1-associated protein (WTAP). Meanwhile, m6A modification can be removed by “erasers” demethylase, such as fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5). In addition, m6A “readers” are responsible for recognition of the m6A modification [22]. Members of every classes of m6A regulators cooperatively participate in the regulation of mRNA stability and translation, affect gene expression output and thus play an important role in physiological and pathological conditions [22,23,24,25,26]. In the m6A methyltransferase complex, METTL3 functions as the the catalytic core while METTL14 serves as the RNA-binding platform [27]. METTL3 is a transferase that methylates mRNA, identifies methylated mRNA, and regulates mRNA translation. Recently, accumulating studies have identified multiple roles and molecular mechanisms associated with METTL3 in various biological processes [28,29,30]. However, whether METTL3-mediated m6A modification is involved in the regulation of endometrial receptivity and how this relates to RIF remains unclear.
Herein, we found that both the levels of global mRNA m6A methylation and METTL3 were significantly elevated in the endometrial tissues from RIF patients compared with the controls. Overexpression of METTL3 in Ishikawa cells significantly decreased the ration of BeWo spheroid attachment. METTL3 catalyzed the m6A methylation of HOXA10 mRNA and contributed to its decay with shortened half-life. Enforced expression of HOXA10 in Ishikawa cells effectively rescued the impairment of METTL3 on the BeWo spheroid attachment in vitro. Our study reveals that METTL3-mediated m6A modification could have an impact on embryo implantation and may contribute to the pathogenesis of RIF.
Materials and Methods
Patient samples and ethical approval
The patients enrolled in this study were recruited from in vitro fertilization unit of Reproductive Medicine Center of the Affiliated Changzhou Maternity and Child Health Care Hospital of Nanjing Medical University. All the endometrial samples were collected with written informed consent of the patients, and approval from the Scientific Research Ethics Committee was obtained for this study (2020103). The mid-secretory phase endometrial tissues were collected by endometrial biopsy from normal women and women with RIF according to the criteria described previously [19]. The normal control group was composed of women who were infertile due to male factors and proved to be fertile after the IVF-ET treatment. RIF was defined as the absence of implantation following two fresh or frozen embryo replacement cycles, during which at least four embryos with good quality were transferred to uterus. Women with endometriosis, adenomyosis, hydrosalpinx, uterine malformation, endometrial polyps or autoimmune disease were not included. The details of these patients are summarized in Table 1.
Cell culture
The human endometrial adenocarcinoma cell line Ishikawa was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and maintained in MEM medium (Thermo Fisher Scientific, Waltham, MA, USA). The human placental choriocarcinoma cell line BeWo was purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained in Ham's F-12K (Kaighn's) medium (Thermo Fisher Scientific). These mediums contain 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 100 U/mL penicillin, and 100 mg/mL streptomycin (HyClone, South Logan, UT, USA). Cells were incubated at 37°C with 5% CO2. Actinomycin D (S8946; Selleck Chemicals) was added for the indicated times at a final concentration of 10 μg/mL for the transcription inhibition assay.
Quantification of m6A RNA methylation
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), followed by the purification of polyadenylated mRNA using Dynabeads mRNA Purification Kit (Thermo Fisher Scientific) according to manufacturer’s protocol. An m6A RNA Methylation Assay Kit (Abcam, Cambridge, MA, USA) was used to evaluate the m6A content of total RNA according to the manufacturer’s instructions, as previously reported [31]. Equal amounts of total RNA (200 ng) were bound to strip wells using a RNA high binding solution. The m6A was captured and detected using the specific capture antibody and detection antibody. Then, the detected m6A signal was enhanced using enhancer solution, and quantified colorimetrically after adding color developing solutions by reading the absorbance at a wavelength of 450 nm in a microplate spectrophotometer.
Dot blotting assay
A dot blotting assay was performed essentially as previously reported [32]. Total RNA or poly (A) + mRNA was isolated as described above. Equal amounts of total poly (A) + mRNA samples (2 μg) were denatured at 65°C for 5 min. Then the samples were loaded onto nylon membranes (GE Healthcare, USA) with ice-cold 20× saline sodium citrate solution (Sigma Aldrich) in a dot blot apparatus (Bio-Rad, USA). The membranes were then UV-crosslinked for 5 min, blocked with 5% non-fat milk for 1 hour, incubated with an m6A antibody (1:400; ab151230, Abcam) overnight at 4 °C and horseradish peroxidase-conjugated anti-rabbit IgG for 1 hour at room temperature, and finally detected with a 3,3’-diaminobenzidine peroxidase substrate kit. At the same time, the same poly (A) + mRNA (2 μg) samples were spotted onto membranes, UV-crosslinked twice, stained with 0.02% methylene blue in 0.3 M sodium acetate for 2 hours, and washed with ribonuclease-free water for 5 hours, followed by the scanning to indicate the total content of input RNA.
Quantitative real-time PCR (qRT-PCR)
Total RNA was lysed using TRIzol reagent and used for the synthesis of cDNA with a One-Step RT-PCR Kit (Thermo Fisher Scientific). Reactions of qRT-PCR were performed using the ABI Vii7 system (Applied Biosystems, USA). GAPDH was used as a housekeeping gene. Relative gene expression was calculated by the 2-△△CT cycle threshold method [33]. The primers used for qRT-PCR analysis are listed in Table 2.
Cell transfection and stable cell line construction
Recombinant lentiviruses expressing wild-type METTL3 (OE-METTL3-WT) or control (OE-con), catalytic domain mutant METTL3 (D395A and W398A; OE-METTL3-Mut) and HOXA10 (OE-HOXA10) were purchased from Biosmedi (Shanghai, China). The cell line Ishikawa was transfected with concentrated lentiviruses (OE-METTL3-WT, 50 MOI; OE-METTL3-Mut, 50 MOI; OE-HOXA10 50 MOI), and stable cell lines were selected by treatment with puromycin for 2 weeks.
Western blotting analysis
Western blotting analysis was performed as previously described [34] using antibodies against METTL3 (1:1,500; ab221795, Abcam), HOXA10 (1:2,000; A8550, Abclonal), ITGB3 (1:1,000; ab119992, Abcam), EMX2 (1:1,500; ab171818, Abcam) and GAPDH (1:6,000; KC-5G5, Aksomics). GAPDH was used as an endogenous control to normalize protein loading. The relative band intensities were measured using a quantitative scanning densitometer and image analysis software, ImageJ.
In vitro embryo implantation assay
We used multicellular spheroids of human placental choriocarcinoma BeWo cells co-cultured with a confluent monolayer of endometrial adenocarcinoma Ishikawa cells as an in vitro model of embryo attachment [19]. First, a single-cell suspension of BeWo cells was placed in a 35 mm2 culture dish pre-coated with an anti-adhesive polymer poly-2-hydroxyethyl methacrylate (Sigma Aldrich). Multicellular spheroids of BeWo cells were induced after 48 hours of culture and 150-200 μm in diameter. Meanwhile, E2 (10-8 M) and P4 (10-6 M) were added into the medium of the monolayer stable METTL3- and/or HOXA10-overexpressing Ishikawa cells after reaching 70%–80% confluence in a 24-well culture plate. Simultaneously, BeWo spheroids were transferred onto the confluent monolayer of Ishikawa cells. After incubation at 37°C for 2 hours, cells were washed with phosphate buffer saline containing 0.1 mg /L Ca2+ and Mg2+ to remove the unattached spheroids. The attached spheroids were then counted under a light microscope, and the adhesion rate was expressed as a percentage of the total number of BeWo spheroids added onto the Ishikawa monolayer (% adhesion).
Methylated RNA immunoprecipitation (Me-RIP) assay
A previously described procedure was used for Me-RIP [32]. The Dynabeads mRNA Purification Kit (Thermo Fisher Scientific) was used to purify mRNA from total RNA and the RNA quality was analyzed by NanoDrop 2000. Then, a Magna MeRIP™ m6A Kit (17-10499, Merck Millipore) was used to measure the changes in the m6A levels of the mRNA according to the manufacturer’s protocol. We saved 0.5 μg of the mRNA as input and used the remaining mRNA for m6A immunoprecipitation. After being immunoprecipitated with Magna ChIP protein A/G Magnetic Beads and eluted twice with elution buffer, the m6A-precipitated RNA was recovered by ethanol precipitation. The RNA concentration was measured with NanoDrop 2000 and the immunoprecipitated m6A RNA was used as templates for qRT-PCR.
Statistical analysis
Data are presented as mean ± SD of at least three independent experiments. Statistical analyses were performed using GraphPad Prism 9 software (La Jolla, CA, USA). Differences between group means were evaluated with the Student’s t-test or one-way analysis of variance. P < 0.05 shows a statistical significance.
Results
Upregulation of m6A modification and METTL3 in the endometrial tissues of women with RIF
To explore the potential role of m6A modification in RIF, we first examined global m6A levels in total RNA from mid-secretory phase endometrial tissues of normal and RIF women by colorimetric m6A quantification strategy. We found that endometrial m6A levels were significantly increased in the RIF patients than in the controls (Fig. 1a). This increase was further confirmed by dot blotting assay (Fig. 1b).
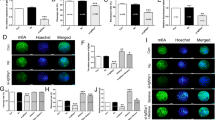
Increased m6A RNA methylation and METTL3 expression in RIF endometrial tissues. a The levels of m6A RNA methylation in the endometrial tissues from RIF patients (n=9) and normal control women (n=8) were evaluated by the m6A RNA Methylation Assay Kit. b The m6A levels in the endometrial tissues from RIF patients (n=6) and health control women (n=6) were evaluated by dot blotting assay. c The mRNA levels of major m6A methyltransferases (METTL3, METTL14, RBM15, WTAP and VIRMA) and demethylases (FTO and ALKBH5) in the endometrial tissues from RIF patients (n=13) and normal control women (n=13) were detected by qRT-PCR. d The protein levels of METTL3 in the endometrial tissues from RIF patients (n=8) and normal control women (n=4) were detected by Western blotting. *P<0.05, **P<0.01, ***P<0.001compared with the controls
Then, we detected the mRNA levels of major m6A methyltransferases (METTL3, METTL14, RBM15, WTAP and VIRMA) and demethylases (FTO and ALKBH5) in the normal and RIF endometrial tissues. The m6A methyltransferases (METTL3, METTL14, and RBM15) and demethylase FTO were significantly increased in the RIF endometrial tissues compared with normal controls, while the m6A methyltransferase WTAP was significantly decreased in the RIF endometrial tissues (Fig. 1c). However, there were no significant differences in the mRNA levels of VIRMA or ALKBH5 between normal and RIF patients.
Considering the increased m6A modification in RIF endometrium and the catalytic abilities of these m6A regulators, we selected METTL3 as the candidate molecule for further studies of aberrant m6A modification in RIF. Protein levels of METTL3 in the RIF endometrial tissues were significantly increased in comparison to normal controls (Fig. 1d), similar to the result obtained from qRT-PCR.
METTL3 overexpression impairs embryo attachment in vitro
To further investigate the effects of METTL3-mediated m6A modification on embryo attachment, we established METTL3 overexpression cell models in Ishikawa cells by lentivirus. The efficiency of overexpressing METTL3 at the mRNA and protein levels were verified by qRT-PCR (Fig. 2a) and Western blotting (Fig. 2b), respectively. We found that METTL3 overexpression significantly enhanced total m6A levels in Ishikawa cells, as indicated in the colorimetric m6A quantification assay (Fig. 2c) and dot blotting assay (Fig. 2d). In an vitro model of a confluent monolayer of Ishikawa cells co-cultured with BeWo spheroids, METTL3 overexpression significantly decreased the ration of BeWo spheroids attachment (Fig. 2e). As HOXA10 is a well-characterized marker of endometrial receptivity and a critical upstream regulator of ITGB3 and EMX2, we then evaluated the expressions of HOXA10 and its downstream targets in METTL3-overexpressing Ishikawa cells. Notably, significant decreases in both HOXA10 mRNA (Fig. 2a) and protein (Fig. 2b) levels were observed in METTL3-overexpressing Ishikawa cells. Moreover, the METTL3-overexpressing Ishikawa cells exhibited a lower expression of ITGB3 and a higher expression of EMX2 compared with the control cells (Fig. 2a and b). Collectively, METTL3 may impair embryo attachment in vitro by inhibiting HOXA10 expression.
METTL3 overexpression impairs embryo attachment in vitro. a-b The expressions of METTL3, HOXA10, ITGB3 and EMX2 at the mRNA (a) and protein (b) levels in the METTL3-overexpressing Ishikawa cells were analyzed by qRT-PCR (a) and Western blotting(b), respectively. c The levels of m6A RNA methylation in the METTL3-overexpressing Ishikawa cells were evaluated by m6A RNA Methylation Assay Kit. d The levels of m6A in the METTL3-overexpressing Ishikawa cells were evaluated by dot blotting assay. e A vitro model of a confluent monolayer of Ishikawa cells co-cultured with BeWo spheroids was used to evaluate the embryo attachment. Bar 100 μm. The data are the average of three independent experiments (n=3). *P<0.05, **P<0.01 versus the indicated group
METTL3 epigenetically decreases HOXA10 expression
We next investigated the mechanism by which METTL3 participates in the regulation of HOXA10 and its downstream targets expressions, contributing to the impairment of embryo implantation. We first assessed the mRNA levels of HOXA10 in endometrial tissues from RIF and normal subjects. The mRNA levels of HOAX10 were significantly decreased in the endometrial tissues from RIF patients (Fig. 3a). Then, we analyzed transcriptome m6A mapping data by an online m6A modification site predictor (http://www.cuilab.cn/sramp/) and found that at least 11 m6A residues were located across the HOXA10 sequence. Consistently, METTL3 overexpression significantly enhanced m6A methylation of HOXA10 mRNA in Ishikawa cells (Fig. 3b). When we generated a mutated METTL3 (D395A and W398A) construct with disordered enzymatic activity, as described previously [35], we found that the mutated METTL3 (D395A and W398A) failed to elevate the m6A methylation of HOXA10 mRNA in Ishikawa cells (Fig. 3b). The levels of HOXA10 mRNA and protein were both decreased by wild-type METTL3, but not mutated METTL3 (D395A and W398A) in Ishikawa cells (Fig. 3c and d). Furthermore, the decay rate of HOXA10 mRNA was accelerated rapidly by wild-type METTL3, but not mutated METTL3 (D395A and W398A), in the transcription inhibition assay (Fig. 3e). These results demonstrated that METTL3 epigenetically decreases HOXA10 expression.
METTL3 epigenetically decreases HOXA10 expression. a The mRNA levels of HOXA10 in the endometrial tissues from RIF patients (n=13) and normal control women (n=13) were evaluated by qRT-PCR. b The methylation of HOXA10 mRNA in the wild-type/mutant (D395A and W398A) METTL3-overexpressing Ishikawa cells were analyzed by Me-RIP-qRT-PCR. c-d The mRNA and protein levels of HOXA10 in the wild-type/mutant (D395A and W398A) METTL3-overexpressing Ishikawa cells were analyzed by qRT-PCR (c) and Western blotting (d), respectively. e The curve and statistical analysis of HOXA10 mRNA decay slope in the wild-type/mutant (D395A and W398A) METTL3-overexpressing Ishikawa cells after transcriptional inhibition. *P<0.05, ***P<0.001 versus the indicated group. N.S. means P >0.05
HOXA10 overexpression rescues METTL3-impaired embryo attachment in vitro
To determine whether decreased HOXA10 expression was responsible for the reduced ration of BeWo spheroids attachment upon METTL3 overexpression in Ishikawa cells, we attempted to rescue this phenotype by overexpressing HOXA10. The efficiency of METTL3 and HOXA10 overexpression were verified by qRT-PCR (Fig. 4a) and Western blotting (Fig. 4b). As above, METTL3 overexpression induced corresponding changes in the expressions of HOXA10, ITGB3 and EMX2, whereas HOXA10 overexpression dramatically enhanced the expression of ITGB3 and decreased the expression of EMX2 despite in the METTL3-overexpressing Ishikawa cells (Fig. 4a and b). We further found that overexpression of METTL3 dramatically increased total m6A levels with or without HOXA10 overexpression in Ishikawa cells (Fig. 4c and d). In the vitro model of a confluent monolayer of Ishikawa cells co-cultured with BeWo spheroids, HOXA10 overexpression significantly reversed the METTL3-decreased ration of BeWo spheroid attachment (Fig. 4e). Collectively, these results suggested that METTL3 impaired embryo attachment in vitro in a HOXA10-dependent manner.
HOXA10 overexpression rescues METTL3-impaired embryo attachment in vitro. a The mRNA levels of METTL3, HOXA10, ITGB3 and EMX2 in the METTL3-overexpressing Ishikawa cells with or without HOXA10 overexpression were analyzed by qRT-PCR. b The protein levels of METTL3, HOXA10, ITGB3 and EMX2 in the METTL3-overexpressing Ishikawa cells with or without HOXA10 overexpression were analyzed by Western blotting. c The levels of m6A RNA methylation in the METTL3-overexpressing Ishikawa cells with or without HOXA10 overexpression were evaluated by the m6A RNA Methylation Assay Kit. d The levels of m6A in the METTL3-overexpressing Ishikawa cells with or without HOXA10 overexpression were evaluated by dot blotting assay. e A vitro model of a confluent monolayer of Ishikawa cells co-cultured with BeWo spheroids was used to evaluate the embryo attachment. Bar 100 μm. The data are the average of three independent experiments (n=3). *P<0.05, **P<0.01, **P<0.001 versus the indicated group
Discussion
With the application of high-throughput sequencing for m6A mapping in RNA, the understanding of its internal regulatory mechanism is being revealed. At present m6A has been recognized as the most prevalent internal modification in mRNAs. The m6A modification in mRNAs could influence mRNA stability and splicing, translation efficiency, nuclear output, and selective polyadenylation. The m6A modification is maintained by three different groups of RNA binding proteins, including m6A writers, erasers and readers. Dynamic and reversible nature of m6A modification makes it play a key role in cellular communications [22,23,24,25,26,27,28,29,30,31,32]. In the current study, we found that the levels of m6A-modified RNAs and the critical methyltransferase METTL3 were significantly upregulated in the endometrial tissues of RIF. METTL3 overexpression inhibited the endometrial receptivity biomarker HOXA10 expression and impaired the embryo attachment in vitro. These results suggested that METTL3-mediated m6A modification is an important determinant of embryo implantation, and that increased METTL3 expression might contribute to the pathogenesis of RIF.
Deregulation of m6A modification has been recently implicated in endometrial diseases [36,37,38]. Jiang et al. [36] analyzed the expressions of 20 m6A regulators in 34 normal, 127 eutopic, and 46 ectopic samples of endometrium tissue from different menstrual cycle phases which were merged from public microarray datasets of endometriosis, and found that most m6A methylation regulators in endometriosis were abnormal in the eutopic vs. normal endometrium, including decreased METTL3/METTL14/RBM15/FTO and increased ALKBH5. Moreover, METTL3 expression in endometriosis was reduced in the ectopic vs. eutopic endometrium while FTO expression was elevated. Functional, co-expression, correlation analyses of proliferative phase endometrial tissues from adenomyosis vs. controls found that decreased METTL3 expression in adenomyosis led to declining total m6A levels and the downstream increased insulin-like growth factor-1 (IGF1) and D-Dopachrome Tautomerase (DDT); and it revealed that IGF1 and DDT might correlate with epithelial cell proliferation and migration, both of which are involved in the pathogenesis of adenomyosis [38]. In addition, ∼70% of endometrial tumors exhibited reductions in m6A methylation that are due to either METTL14 mutation (R298P) or decreased METTL3 expression. Reductions in m6A methylation decreased the negative AKT regulator PHLPP2 expression and increased the positive AKT regulator mTORC2 expression, which contributed to increased proliferation and tumorigenicity of endometrial cancer cells through the activation of AKT pathway [37]. In the present study, increased METTL3 expression and m6A levels were found in the endometrial tissues from RIF. Overexpression of METTL3 increased m6A methylation, and impaired embryo attachment in vitro by inhibiting the endometrial receptivity biomarker HOXA10 expression. These studies suggest that m6A methylation is involved in the pathogenesis of endometrial diseases, including endometriosis, adenomyosis, endometrial cancers and RIF, all of which shares some characteristics with each other. However, different status and pattern of m6A methylation and its regulators may be found in the endometrial tissues from different endometrial diseases, different menstrual cycle phases, and even from different sites in the same patient.
Embryo implantation is a subtle and complicated process that requires accurate communication between high-quality embryos and receptive endometrium under the action of maternal hormones and their downstream molecules [1,2,3]. During ∼6 days after ovulation, ovarian estrogen and progestin cooperatively induces the morphological and physiological changes of epithelial cells in the endometrium and secretion of various cytokines. These transformations cause the uterus to be receptive to blastocyst implantation [4, 5]. The transcription factor homologue HOXA10 has emerged as an important and well-characterized biomarker of endometrial receptivity. The expression of HOXA10 in the uterus depends on the stage of menstrual cycle, which is significantly increased in the mid-secretory phase, corresponding to the implantation time and the increase of progesterone level [8, 9]. Both estrogen and progestin independently and synergically elevated the expression of HOXA10 in endometrium [8]. In turn, HOXA10 regulates endometrial acceptance and decidualization activation or compression by downstream markers specific to the window of implantation [10,11,12, 18,19,20,21]. Abnormal expression of HOXA10 and its downstream target genes leading to decreased endometrial receptivity are closely related to female infertility in the patients with gynecological diseases, such as endometriosis [11, 20, 21], adenomyosis [39, 40], and hydrosalpinx [41]. The importance of maternal HOXA10 expression in embryo implantation has been demonstrated by a targeted disruption of the Hoxa10 gene in mice. Female mice with deletion of Hoxa10 gene were infertile due to endometrial receptivity defects [42]. Small ubiquitin like-modifier 1 (SUMO1) inhibited HOXA10 protein stability and transcriptional activity via sumoylation at the evolutionarily conserved lysine 164 residue in the endometrium of women with RIF, which impairs endometrial receptivity and embryo implantation [19]. In our study, we found that wild-type METTL3, but not mutated METTL3 (D395A and W398A), decreased the expression of HOXA10 due to increases in the m6A methylation of HOXA10 mRNA in Ishikawa cells. Enforced expression of HOXA10 in Ishikawa cells effectively rescued the impairment of METTL3 on the embryo attachment in vitro. Jiang et al. [19] found that increased SUMO1-modified HOXA10 expression without changes of HOXA10 expression was detected in the mid-secretory endometrium of women with RIF; however, increased m6A content in total RNA with decreased HOXA10 expression was found in our study. The difference may be due to individual difference and limitation of sample size.
Conclusions
In conclusion, increased m6A content in total RNA with high METTL3 expression was found in the mid-secretory endometrium of women with RIF compared with that of the control fertile women. METTL3 catalyzed the m6A methylation of HOXA10 mRNA and repressed the expression of HOXA10 leading to the impairment on embryo attachment in vitro. However, global RNA m6A methylation in the endometrium from women with RIF during the window of implantation is not restricted to METTL3/HOXA10, and further studies are required to investigate the function of m6A methylation in endometrial receptivity and embryo implantation.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Fazleabas AT, Kim JJ, Srinivasan S, Donnelly KM, Brudney A, Jaffe RC. Implantation in the baboon: endometrial responses. Semin Reprod Endocrinol. 1999;17:257–65.
Nimbkar-Joshi S, Rosario G, Katkam RR, Manjramkar DD, Metkari SM, Puri CP, et al. Embryo-induced alterations in the molecular phenotype of primate endometrium. J Reprod Immunol. 2009;83:65–71.
Simón C, Martín JC, Pellicer A. Paracrine regulators of implantation. Baillieres Best Pract Res Clin Obstet Gynaecol. 2000;14:815–26.
Bergh PA, Navot D. The impact of embryonic development and endometrial maturity on the timing of implantation. Fertil Steril. 1992;58:537–42.
Strowitzki T, Germeyer A, Popovici R, von Wolff M. The human endometrium as a fertility-determining factor. Hum Reprod Update. 2006;12:617–30.
Moustafa S, Young SL. Diagnostic and therapeutic options in recurrent implantation failure. F1000Res. 2020;9:F1000 Faculty Rev-208.
Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, et al. Molecular cues to implantation. Endocr Rev. 2004;25:341–73.
Taylor HS, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J Clin Invest. 1998;101:1379–84.
Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14:1328–31.
Daftary GS, Troy PJ, Bagot CN, Young SL, Taylor HS. Direct regulation of beta3-integrin subunit gene expression by HOXA10 in endometrial cells. Mol Endocrinol. 2002;16:571–9.
Kim JJ, Taylor HS, Lu Z, Ladhani O, Hastings JM, Jackson KS, et al. Altered expression of HOXA10 in endometriosis: potential role in decidualization. Mol Hum Reprod. 2007;13:323–32.
Troy PJ, Daftary GS, Bagot CN, Taylor HS. Transcriptional repression of peri-implantation EMX2 expression in mammalian reproduction by HOXA10. Mol Cell Biol. 2003;23:1–13.
Germeyer A, Savaris RF, Jauckus J, Lessey B. Endometrial beta3 integrin profile reflects endometrial receptivity defects in women with unexplained recurrent pregnancy loss. Reprod Biol Endocrinol. 2014;12:53.
Liu N, Zhou C, Chen Y, Zhao J. The involvement of osteopontin and β3 integrin in implantation and endometrial receptivity in an early mouse pregnancy model. Eur J Obstet Gynecol Reprod Biol. 2013;170(1):171–6.
Marron K, Harrity C, Dunne H, Shkrobot L, Kennedy J. Cytometric assessment of uterine receptivity via epithelial β3 integrin expression. Reprod Biomed Online. 2019;39(2):294–303.
Daftary GS, Taylor HS. EMX2 gene expression in the female reproductive tract and aberrant expression in the endometrium of patients with endometriosis. J Clin Endocrinol Metab. 2004;89(5):2390–6.
Zhu Y, Luo M, Huang H, Du X, Chen D, Xing Q, et al. HOXA10, EMX2 and TENM1 expression in the mid-secretory endometrium of infertile women with a Müllerian duct anomaly. Reprod Biomed Online. 2016;32(4):388–93.
Bagot CN, Troy PJ, Taylor HS. Alteration of maternal Hoxa10 expression by in vivo gene transfection affects implantation. Gene Ther. 2000;7:1378–84.
Jiang R, Ding L, Zhou J, Huang C, Zhang Q, Jiang Y, et al. Enhanced HOXA10 sumoylation inhibits embryo implantation in women with recurrent implantation failure. Cell Death Discov. 2017;3:17057.
Yan Q, Huang C, Jiang Y, Shan H, Jiang R, Wang J, et al. Calpain7 impairs embryo implantation by downregulating β3-integrin expression via degradation of HOXA10. Cell Death Dis. 2018;9:291.
Zhu LH, Sun LH, Hu YL, Jiang Y, Liu HY, Shen XY, et al. PCAF impairs endometrial receptivity and embryo implantation by down-regulating β3-integrin expression via HOXA10 acetylation. J Clin Endocrinol Metab. 2013;98:4417–28.
Uddin MB, Wang Z, Yang C. The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol Cancer. 2021;20:61.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, , et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816-3831.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–56.
Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–8.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335–45.
Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol. 2019;317:C762–75.
Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat Commun. 2019;10:1898.
Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y, et al. RNA m6A Methyltransferase METTL3 Promotes The Growth Of Prostate Cancer By Regulating Hedgehog Pathway. Onco Targets Ther. 2019;12:9143–52.
Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Xue P, Fan W, Diao Z, Li Y, Kong C, Dai X, et al. Up-regulation of PTEN via LPS/AP-1/NF-κB pathway inhibits trophoblast invasion contributing to preeclampsia. Mol Immunol. 2020;118:182–90.
Yao Y, Yang Y, Guo W, Xu L, You M, Zhang YC, et al. METTL3-dependent m6A modification programs T follicular helper cell differentiation. Nat Commun. 2021;12:1333.
Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M, et al. Exploring diagnostic m6A regulators in endometriosis. Aging (Albany NY). 2020;12:25916–38.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Zhai J, Li S, Sen S, Opoku-Anane J, Du Y, Chen ZJ, et al. m6A RNA Methylation Regulators Contribute to Eutopic Endometrium and Myometrium Dysfunction in Adenomyosis. Front Genet. 2020;11:716.
Fischer CP, Kayisili U, Taylor HS. HOXA10 expression is decreased in endometrium of women with adenomyosis. Fertil Steril. 2011;95:1133–6.
Wang J, Huang C, Jiang R, Du Y, Zhou J, Jiang Y, et al. Decreased Endometrial IL-10 Impairs Endometrial Receptivity by Downregulating HOXA10 Expression in Women with Adenomyosis. Biomed Res Int. 2018;2018:2549789.
Kong C, Sun L, Zhang M, Ding L, Zhang Q, Cheng X, et al. miR-133b Reverses the Hydrosalpinx-induced Impairment of Embryo Attachment Through Down-regulation of SGK1. J Clin Endocrinol Metab. 2016;101:1478–89.
Satokata I, Benson G, Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374:460–3.
Acknowledgements
We would like to thank all the patients for their participation in this study.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20201158), Qingmiao Talents Training Program of Changzhou Health Commission (CZQM2020095 and CZQM2020103), Great Science and Technology Project of Changzhou Municipal Health Commission (ZD201921 and ZD202016).
Author information
Authors and Affiliations
Contributions
PX, Wenbo Zhou, WF, LX, BY and LC conceived and designed this study. PX, JJ, CK, Wei Zhou, JZ, XH, HY and QH recruited the patients and collected endometrial samples. PX, Wenbo Zhou, WF, CK, BZ performed the experiments. PX, Wenbo Zhou, WF, JJ, CK, BZ analyzed the data. LX, BY and LC supervised the research. PX, Wenbo Zhou and WF wrote the original draft. LX, BY and LC edited the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All the endometrial samples were collected with written informed consent of the patients who were recruited from in vitro fertilization unit of Reproductive Medicine Center of the Affiliated Changzhou Maternity and Child Health Care Hospital of Nanjing Medical University, and approval was obtained from the Scientific Research Ethics Committee of Changzhou Maternity and Child Health Care Hospital for this study (2020103).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xue, P., Zhou, W., Fan, W. et al. Increased METTL3-mediated m6A methylation inhibits embryo implantation by repressing HOXA10 expression in recurrent implantation failure. Reprod Biol Endocrinol 19, 187 (2021). https://doi.org/10.1186/s12958-021-00872-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12958-021-00872-4