Abstract
Background
Neuroendocrine differentiation (NED) is often found in colorectal cancer (CRC) and may have unique biological behavior, which has not been previously delineated. Here, we explore the relationship between CRC, NED, and clinicopathological factors. We also offer a preliminary explanation of the mechanism underlying the malignant biological behavior of NED in CRC.
Methods
Between 2013 and 2015, 394 CRC patients who underwent radical operations were selected for analysis. The relationship between NED and clinicopathological factors was analyzed. To further clarify the pivotal role of NED in CRC, we performed bioinformatic analyses and identified genes that may be involved in NED, which were obtained from in silico data from The Cancer Genome Atlas (TCGA) database. Then, we conducted functional enrichment analyses and confirmed the critical pathways for intensive study. Moreover, we detected the expression of key proteins by immunohistochemistry and analyzed the correlation of their expression with NED.
Results
The statistical analysis showed that CRC with NED was positively correlated with lymph node metastasis. Through bioinformatic analysis, we found that chromogranin A (CgA) was positively correlated with invasion and lymph node metastasis. ErbB2 and PIK3R1, which are key proteins in the PI3K-Akt signaling pathway, were closely related to NED. Furthermore, we determined that the PI3K-Akt signaling pathway likely plays a critical role in the NED of CRC.
Conclusions
CRC with NED is associated with lymph node metastasis. The PI3K-Akt signaling pathway, which is closely related to CRC, may be the mechanism promoting the malignant biological behavior of CRC with NED.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is one of the most common malignant cancers worldwide. Although its pathology is mainly adenocarcinoma, the components may differ. Adenocarcinoma with neuroendocrine differentiation (NED) is a special type of cancer. It usually contains two components: adenocarcinoma and a neuroendocrine tumor, but the main component is adenocarcinoma, with the percentage of neuroendocrine tumor cells less than 30% [1]. Neuroendocrine tumors originate from neuroendocrine cells which can secrete serotonin metabolites or polypeptide hormones. Because of these secretions, the tumor may produce changes in the biological behavior of surrounding cells [2]. Because NED is related to nerve and endocrine function, it can be diagnosed by detecting chromogranin A (CgA) and/or synaptophysin (SYP/Syn) proteins.
At present, NED has been found in many solid cancers, but research about NED in prostate cancer has been the most extensive. These studies have not only examined the poor effects of treatment and prognosis [3,4,5], but have also studied the mechanism of hormones secreted by NED cells on surrounding cells, which enhances their capacity for metastasis and invasion [6]. In vitro studies have shown that neuropeptides can stimulate androgen-independent growth [7] and increase the invasiveness of prostate cancer cells [8]. The mechanism may be related to several pathways, including PKA/CREB [9], PI3K/Akt [10], and others [11, 12]. In CRC, research has mainly focused on clinical prognosis, where conclusions have differed and are debatable [13,14,15]. Indinnimeo et al. [16] found a significant association between CgA-positivity and lymph node metastasis in human colon cancer. There are also some studies which showed that NED was associated with liver metastasis and advanced tumor events [17]. By contrast, early studies by Mori et al. [14] and Lloyd et al. [13] showed that NED did not influence patient prognosis. Therefore, the purpose of this research was to study whether NED influences the biological behavior of CRC and to conduct a preliminary exploration of the mechanisms underlying its malignant biological behavior.
In the present study, we retrospectively analyzed patients with NED. We then downloaded in silico data from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) and identified the NED (±) samples involved based on CgA and/or SYP expression. We further performed bioinformatic analysis to reveal the mechanisms that may be involved in NED. Finally, we conducted immunohistochemical staining to verify our predictions.
Methods
Study population
A total of 394 patients with primary CRC, who underwent radical operation between 2013 and 2015 at Liaoning Cancer Hospital and Institute (Shenyang, Liaoning Province, People’s Republic of China) were included in this study. All patients were at stages I, II, or III according to the Union for International Cancer Control TNM classification system. Patients who died perioperatively or with secondary malignancy and those with distant metastases such as liver or lung before the operation were excluded. Histology specimens were evaluated by two senior pathologists, and the diagnosis of CRC with or without NED was confirmed in all patients. This study was approved by the local ethics committee of Liaoning Cancer Hospital and Institute.
Bioinformatic analysis of CgA and SYP expression in CRC
Data collection and analysis
We downloaded a total of 568 CRC and 44 normal tissues from TCGA (https://portal.gdc.cancer.gov). The data included RNA-seq profiles and clinicopathological information. We further compared the expression of CgA and SYP between normal and malignant samples. The overall survival and progression-free survival of patients expressing high and low levels of the two markers were also acquired by applying the Kaplan–Meier method (“limma,” “survival,” and “survminer” packages in R software).
Pathological and functional enrichment analyses of CgA and SYP
To reveal the potential mechanisms of CgA and SYP, we further calculated the expression of the two markers in different pTNM stages through R software (“ggpubr” package). Then, we divided the CRC samples into two sets based on the expression of CgA and SYP. We analyzed differentially expressed genes (DEGs) with a threshold log2 fold change (FC) > 2.0 and P < 0.01. The top 100 DEGs were also drawn as a heatmap. Moreover, we conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of DEGs using R software.
Immunohistochemistry
We selected three key proteins of the PI3K-Akt signaling pathway, including ErbB2, PIK3R1, and AKT3, to verify the pathway predictions. After dewaxing, the sections were subjected to antigen retrieval at high temperature and pressure for 3 min, washed with water, incubated with 3% hydrogen peroxide at room temperature for 10 min, washed again three times, and then placed in phosphate-buffered saline solution. We blocked the sections with 5% fetal bovine serum for 30 min, then added the first antibody for overnight incubation at 4 °C. The next day, the sections were washed three times, then incubated with a secondary antibody at room temperature for 30 min and washed three times. We then incubated the sections for 1–3 min with diaminobenzidine chromogenic solution, lightly stained the nuclei with hematoxylin, and differentiated the sections with 0.5% hydrochloric acid ethanol for 2 s. After three washes, the sections were dehydrated with graded alcohol, made transparent, and sealed with neutral gum. The antibodies used in the study were raised against CgA (dilution 1:1000, GT211407; Gene Tech, CHINA), SYP (dilution 1:1000, GT206507; Gene Tech), ErbB2 (dilution 1:1000, GT224507; Gene Tech), PIK3R1 (dilution 1:1000, ER64588; HUABIO), and AKT3 (dilution 1:1000, ER62638; HUABIO).
Based on the 2010 World Health Organization classification system, we examined two neuroendocrine biomarkers, CgA and SYP. Specimens in which CgA and/or SYP were present in 2–30% of immunoreactive cells were classified as NED [1]. All immunohistochemical sections were evaluated by two senior pathologists.
Statistics
All statistical analyses were performed using SPSS (version 22.0; IBM, USA). Pearson’s chi-square and Fisher’s exact test were used to investigate correlations between clinicopathological factors and NED, as well as the relationship between NED and predictive proteins. Correlations with P values < 0.1 were entered into the next step of multivariate analysis using logistic regression. P values < 0.05 were considered statistically significant.
Results
General information
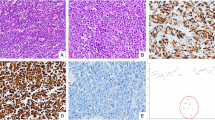
A total of 394 patients (234 males and 160 females) were included in the study. Among them, 55 (14.0%) were NED(+) patients including 33 CgA(+) (Fig. 1A) and 26 SYP(+) (Fig. 1B) patients, and four patients were positive for both. The mean age was 61.9 years (28–84 years). Of these patients, 43 had stage I, 121 had stage II, and 230 had stage III cancer, which included 222 cases of colon cancer and 172 cases of rectal cancer.
A Neuroendocrine tumor cells in CRC detected by immunostaining for CgA (magnification x 100). B Neuroendocrine tumor cells in CRC detected by immunostaining for SYP (magnification x 100). C Relative expression of CgA in CRC and corresponding non-cancerous tissues from TCGA. ***P < 0.001. D Relative expression of SYP in CRC and corresponding non-cancerous tissues from TCGA. ***P < 0.001. E Kaplan–Meier curve analysis of the correlation of CgA expression and overall survival or progression-free survival from TCGA. F Kaplan–Meier curve analysis of the correlation of SYP expression and overall survival or progression-free survival from TCGA. CgA, chromogranin A; CRC, colorectal cancer; SYP, synaptophysin; TCGA, The Cancer Genome Atlas
Clinicopathological factors and NED
Univariate analysis showed that lymph nodes retrieved (P = 0.044), depth of invasion (P = 0.074), and lymph node metastasis (P = 0.020) were closely correlated with NED. Multivariate analysis showed that only lymph node metastasis (P = 0.022, odds ratio = 2.091) was closely correlated with NED, and NED(+) patients were more prone to lymph node metastasis, while lymph nodes retrieved and depth of invasion had no significant correlation with NED (Table 1).
Bioinformatic analysis of CgA and SYP expression in CRC
Firstly, we compared the expression of CgA and SYP between malignant tumors and corresponding non-cancerous tissues. The results are shown in Fig. 1C, D, indicating that CgA and SYP were upregulated in CRC (P < 0.001). Furthermore, we performed the Kaplan-Meier curve analysis to reveal the significance of expression in overall survival and progression-free survival. As shown in Fig. 1E, the expression of CgA was negatively correlated with overall survival (P = 0.003) and progression-free survival (P = 0.003); however, there was no statistically significant difference in SYP expression (Fig. 1F).
Pathological analyses of CgA and SYP in CRC
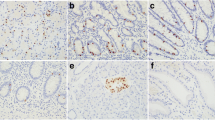
To further clarify the pivotal role of NED in CRC, we further analyzed the relationship between the expression of the two genes and the pTNM stage. As shown in Fig. 2A, B, the expression of CgA was positively correlated with the depth of invasion (P = 0.01) and lymph node metastasis (P = 0.03). The expression of SYP was positively correlated with pT stage (Fig. 2D, P = 0.0039), but was not different between pN0 and pN1+N2 stages (Fig. 2E, P = 0.13). We further found that CgA and SYP had no correlation with metastasis (Fig. 2C, P = 0.7 and Fig. 2F, P = 0.87).
The correlation between CgA and SYP expression and pTNM stage of CRC. A The analyses of CgA expression in pT1+T2 stage and pT3+T4 stage. B The analyses of CgA expression in pN0 stage and pN1+N2 stage. C The analyses of CgA expression in pM0 stage and pM1 stage. D The analyses of SYP expression in pT1+T2 stage and pT3+T4 stage. E The analyses of SYP expression in pN0 stage and pN1+N2 stage. F The analyses of SYP expression in pM0 stage and pM1 stage. CgA, chromogranin A; CRC, colorectal cancer; SYP, synaptophysin
KEGG pathway analyses of CgA and SYP
Since CgA and SYP were closely related to clinicopathological features, this prompted us to further explore their molecular mechanisms in CRC. The two groups of DEGs were obtained based on the upregulation and downregulation of CgA and SYP, and the top 20 DEGs were mapped as heatmaps in Fig. 3A, B. Then, we performed KEGG pathway analyses of CgA and SYP. The results are shown in Fig. 4A, B and indicated that multiple cancer-related pathways were enriched. Interestingly, the “PI3K-Akt signaling pathway” and “neuroactive ligand-receptor interaction” co-occurred in both results and had the most genes involved. Because previous studies have confirmed that the PI3K-Akt signaling pathway-regulated NED in CRC, we tried to determine which genes played a role in these pathways. We performed intersection analysis on the DEGs of CgA and SYP, compared to the genes enriched in the PI3K-Akt signaling pathway. Finally, we identified three genes (PIK3R1, ErbB2, and AKT3) which intersected. The Venn diagram is shown in Fig. 5.
Heatmap of DEGs between upregulation and downregulation of CgA or SYP. A Heatmap of the top 25 DEGs between upregulation and downregulation of CgA (log2 FC > 2, P < 0.01). B Heatmap of the top 25 DEGs between upregulation and downregulation of SYP (log2 FC > 2, P < 0.01). The left vertical axis shows clusters of DEGs and right vertical axis represents gene names. Red represents upregulated genes and blue represents downregulated genes. CgA, chromogranin A; DEGs, differentially expressed genes; FC, fold change; SYP, synaptophysin
KEGG pathway enrichment network diagram for DEGs. A KEGG pathways enrichment analysis of CgA. B KEGG pathways enrichment analysis of SYP. “Count” represents the number of genes. CgA, chromogranin A; DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes; SYP, synaptophysin
Immunohistochemical staining for verification
We randomly selected 33 NED(+) patients and 34 NED(−) patients for immunohistochemistry. We selected three key proteins of the PI3K-Akt signaling pathway, including ErbB2, PIK3R1, and AKT3, for further immunohistochemical verification. After statistical analysis of these three proteins, we found that ErbB2 was positive in 14 of 33 NED(+) patients and positive in six of 34 NED(−) patients (Fig. 6A). We also found that PIK3R1 was positive in 30 of 33 NED(+)patients and positive in 19 of 34 NED(−) patients (Fig. 6B). The statistical analysis showed that ErbB2 and PIK3R1 were closely correlated with NED (Table 2).
Discussion
Adenocarcinoma with NED has been studied in CRC in many previous studies, but findings have been controversial. Some studies have shown no significant correlation between NED and prognosis [13, 18, 19], while other studies have drawn the opposite conclusion, suggesting that NED was significantly associated with the prognosis [20,21,22]. Furthermore, some other studies have shown that patients with NED have a higher incidence of lymph node metastasis [16] and liver metastasis [17]. In our previous study, we found that patients with NED were associated with lymph node metastasis in poorly differentiated CRC [23]. In the present study, we extended our analysis to include various differentiated adenocarcinomas and have reached the same result. We firmly believe that NED is closely related to lymph node metastasis.
What is the mechanism underlying the relationship between NED and lymph node metastasis? It is already known that NED cells secrete hormone substances in autocrine or paracrine loops and a previous study has shown that biogenic amines and polypeptide hormones play an important role in the growth regulation of normal and neoplastic intestinal epithelia [24]. Therefore, we hypothesized that NED cells may secret certain neurohormonal substances which stimulate growth and the metastatic capacity of CRC cells. In order to preliminarily confirm this hypothesis, we predicted the proteins interacting with NED using biological function analysis and verified their presence in CRC tissues by immunohistochemistry. We found ErbB2 and PIK3R1, which are key proteins in the PI3K-Akt signaling pathway, were closely correlated with NED. In previous studies, the PI3K-Akt signaling pathway was found to be closely related to the proliferation and invasion of CRC [25,26,27,28]. Therefore, we speculated that NED cells may enhance the invasion of CRC by activating the PI3K-Akt signaling pathway, so as to promote malignant biological behavior (Fig. 7). However, confirming this hypothesis will require further studies on cell function and mechanisms.
Considering the different diagnostic criteria of NED used in previous studies, the results of these studies may not be reliable [29,30,31,32]. To ensure the accuracy of our results, we re-stained all cases and use the latest recognized method for the diagnosis of NED [33, 34]. Nevertheless, there still are some limitations in our study. First, because of the nature of retrospective studies, selective bias may inevitably exist. Second, our examination of the mechanisms involved may be insufficient, as we have only conducted a preliminary exploration at the protein level, according to the results of the bioinformatic analysis. Therefore, further studies will be needed to confirm these results.
Conclusions
In CRC, patients with NED were positively correlated with lymph node metastasis. The PI3K-Akt signaling pathway, which is closely related to the proliferation and invasion of CRC, may be the mechanism promoting the malignant biological behavior of lymph node metastasis in CRC with NED.
Availability of data and materials
Not applicable.
Abbreviations
- NED:
-
Neuroendocrine differentiation
- CRC:
-
Colorectal cancer
- TCGA:
-
The Cancer Genome Atlas
- CgA:
-
Chromogranin A
- SYP/Syn:
-
Synaptophysin
- DEGs:
-
Differentially expressed genes
- FC:
-
Fold change
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Kleist B, Poetsch M. Neuroendocrine differentiation: the mysterious fellow of colorectal cancer. World J Gastroenterol. 2015;21:11740–7.
Li Z, Chen CJ, Wang JK, Hsia E, Li W, Squires J, et al. Neuroendocrine differentiation of prostate cancer. Asian J Androl. 2013;15:328–32.
Hu CD, Choo R, Huang J. Neuroendocrine differentiation in prostate cancer: a mechanism of radioresistance and treatment failure. Front Oncol. 2015;5:90.
Krauss DJ, Hayek S, Amin M, Ye H, Kestin LL, Zadora S, et al. Prognostic significance of neuroendocrine differentiation in patients with Gleason score 8-10 prostate cancer treated with primary radiotherapy. Int J Radiat Oncol Biol Phys. 2011;81:e119–25.
Krauss DJ, Amin M, Stone B, Ye H, Hayek S, Cotant M, et al. Chromogranin A staining as a prognostic variable in newly diagnosed Gleason score 7-10 prostate cancer treated with definitive radiotherapy. Prostate. 2014;74:520–7.
Uchida K, Masumori N, Takahashi A, Itoh N, Kato K, Matusik RJ, et al. Murine androgen-independent neuroendocrine carcinoma promotes metastasis of human prostate cancer cell line LNCaP. Prostate. 2006;66:536–45.
Jongsma J, Oomen MH, Noordzij MA, Romijn JC, van Der Kwast TH, Schröder FH, et al. Androgen-independent growth is induced by neuropeptides in human prostate cancer cell lines. Prostate. 2000;42:34–44.
Hoosein NM, Logothetis CJ, Chung LW. Differential effects of peptide hormones bombesin, vasoactive intestinal polypeptide and somatostatin analog RC-160 on the invasive capacity of human prostatic carcinoma cells. J Urol. 1993;149:1209–13.
Deeble PD, Cox ME, Frierson HF Jr, Sikes RA, Palmer JB, Davidson RJ, et al. Androgen-independent growth and tumorigenesis of prostate cancer cells are enhanced by the presence of PKA-differentiated neuroendocrine cells. Cancer Res. 2007;67:3663–72.
Martín-Orozco RM, Almaraz-Pro C, Rodríguez-Ubreva FJ, Cortés MA, Ropero S, Colomer R, et al. EGF prevents the neuroendocrine differentiation of LNCaP cells induced by serum deprivation: the modulator role of PI3K/Akt. Neoplasia. 2007;9:614–24.
Unno K, Chalmers ZR, Pamarthy S, Vatapalli R, Rodriguez Y, Lysy B, et al. Activated ALK cooperates with N-Myc via Wnt/β-catenin signaling to induce neuroendocrine prostate cancer. Cancer Res. 2021;81:2157–70.
Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29:536–47.
Lloyd RV, Schroeder G, Bauman MD, Krook JE, Jin L, Goldberg RM, et al. Prevalence and prognostic significance of neuroendocrine differentiation in colorectal carcinomas. Endocr Pathol. 1998;9:35–42.
Mori M, Mimori K, Kamakura T, Adachi Y, Ikeda Y, Sugimachi K. Chromogranin positive cells in colorectal carcinoma and transitional mucosa. J Clin Pathol. 1995;48:754–8.
Bernick PE, Klimstra DS, Shia J, Minsky B, Saltz L, Shi W, et al. Neuroendocrine carcinomas of the colon and rectum. Dis Colon Rectum. 2004;47:163–9.
Indinnimeo M, Cicchini C, Memeo L, Stazi A, Provenza C, Ricci F, et al. Correlation between chromogranin-A expression and pathological variables in human colon carcinoma. Anticancer Res. 2002;22:395–8.
Shinji S, Naito Z, Ishiwata T, Tanaka N, Furukawa K, Suzuki H, et al. Neuroendocrine cell differentiation of poorly differentiated colorectal adenocarcinoma correlates with liver metastasis. Int J Oncol. 2006;29:357–64.
Secco GB, Campora E, Fardelli R, Lapertosa G, De Lucchi F, Gianquinto D, et al. Chromogranin-A expression in neoplastic neuroendocrine cells and prognosis in colorectal cancer. Tumori. 1996;82:390–3.
Cho YB, Yang SS, Lee WY, Song SY, Kim SH, Shin HJ, et al. The clinical significance of neuroendocrine differentiation in T3-T4 node-negative colorectal cancer. Int J Surg Pathol. 2010;18:201–6.
Atasoy P, Ensari A, Demirci S, Kurşun N. Neuroendocrine differentiation in colorectal carcinomas: assessing its prognostic significance. Tumori. 2003;89:49–53.
Guo J, Zheng P, Wang R, Tan X, Yang W, Xiao S, et al. Prognostic relevance of neuroendocrine differentiation in colorectal cancer: a population-based, propensity score matching study. Int J Colorectal Dis. 2020;35:2185–95.
Herold Z, Dank M, Herold M, Nagy P, Rosta K, Somogyi A. Histopathological chromogranin A-positivity is associated with right-sided colorectal cancers and worse prognosis. Cancers (Basel). 2020;13:67.
Chen Y, Liu F, Meng Q, Ma S. Is neuroendocrine differentiation a prognostic factor in poorly differentiated colorectal cancer. World J Surg Oncol. 2017;15:71.
Johnson LR. Regulation of gastrointestinal mucosal growth. Physiol Rev. 1988;68:456–502.
Wan ML, Wang Y, Zeng Z, Deng B, Zhu BS, Cao T, et al. Colorectal cancer (CRC) as a multifactorial disease and its causal correlations with multiple signaling pathways. Biosci Rep. 2020;40:BSR20200265.
Li N, Bu X, Wu P, Wu P, Huang P. The "HER2-PI3K/Akt-FASN Axis" regulated malignant phenotype of colorectal cancer cells. Lipids. 2012;47:403–11.
Narayanankutty A. PI3K/Akt/ mTOR pathway as a therapeutic target for colorectal cancer: a review of preclinical and clinical evidence. Curr Drug Targets. 2019;20:1217–26.
Papadatos-Pastos D, Rabbie R, Ross P, Sarker D. The role of the PI3K pathway in colorectal cancer. Crit Rev Oncol Hematol. 2015;94:18–30.
Syversen U, Halvorsen T, Mårvik R, Waldum HL. Neuroendocrine differentiation in colorectal carcinomas. Eur J Gastroenterol Hepatol. 1995;7:667–74.
Pifano M, Garona J, Capobianco CS, Gonzalez N, Alonso DF, Ripoll GV. Peptide agonists of vasopressin V2 receptor reduce expression of neuroendocrine markers and tumor growth in human lung and prostate tumor cells. Front Oncol. 2017;7:11.
Jansson D, Gould VE, Gooch GT, Rittenhouse HG, Shin SS, Manderino GL, et al. Immunohistochemical analysis of colon carcinomas applying exocrine and neuroendocrine markers. APMIS. 1988;96:1129–39.
Kobayashi H, Koizumi N, Takagi T, Fukumoto K. The case of a long-surviving patient with colorectal neuroendocrine carcinoma with invasion of the jejunum and lymph node metastases after operation. Gan To Kagaku Ryoho. 2015;42:2218–20.
Klöppel G, Couvelard A, Perren A, Komminoth P, McNicol AM, Nilsson O, et al. ENETS Consensus Guidelines for the Standards of Care in Neuroendocrine Tumors: towards a standardized approach to the diagnosis of gastroenteropancreatic neuroendocrine tumors and their prognostic stratification. Neuroendocrinology. 2009;90:162–6.
Klimstra DS, Modlin IR, Adsay NV, Chetty R, Deshpande V, Gönen M, et al. Pathology reporting of neuroendocrine tumors: application of the Delphic consensus process to the development of a minimum pathology data set. Am J Surg Pathol. 2010;34:300–13.
Acknowledgements
We would like to thank Dr. Yongxi Song at First Hospital of China Medical University for critical revisions.
Funding
This work was supported by Shenyang Young and Middle-aged Scientific and Technological Innovation Talents Support Project (RC190487), Health Development Promotion Project (BJHA-CRP-019), and Cultivation Fund of National Natural Science Foundation of Liaoning Cancer Hospital (2021-ZLLH-12).
Author information
Authors and Affiliations
Contributions
Yue Chen and Yu Liang collected documents and wrote the paper. Deyu Sun managed the design and modified the paper. Lianqun Cao and Xinxin Dong did the experiment and assisted in writing discussion section. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the local ethics committee of Liaoning Cancer Hospital and Institute.
Consent for publication
All presentations had consent for publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, Y., Liang, Y., Cao, L. et al. Neuroendocrine differentiation: a risk fellow in colorectal cancer. World J Surg Onc 21, 89 (2023). https://doi.org/10.1186/s12957-023-02952-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-023-02952-8