Abstract
Background
This study was carried out to discover the underlying role that HOXA11 plays in lung squamous cancer (LUSC) and uncover the potential corresponding molecular mechanisms and functions of HOXA11-related genes.
Methods
Twenty-three clinical paired LUSC and non-LUSC samples were utilized to examine the level of HOXA11 using quantitative real-time polymerase chain reaction (qRT-PCR). The clinical significance of HOXA11 was systematically analyzed based on 475 LUSC and 18 non-cancerous adjacent tissues from The Cancer Genome Atlas (TCGA) database. A total of 102 LUSC tissues and 121 non-cancerous tissues were available from Oncomine to explore the expressing profiles of HOXA11 in LUSC. A meta-analysis was carried out to further assess the differential expression of HOXA11 in LUSC, including in-house qRT-PCR data, expressing data extracted from TCGA and Oncomine databases. Moreover, the enrichment analysis and potential pathway annotations of HOXA11 in LUSC were accomplished via Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). The expression of hub genes and according correlations with HOXA11 were assessed to further explore the biological role of HOXA11 in LUSC.
Results
HOXA11 expression in LUSC had a tendency to be upregulated in comparison to adjacent non-cancerous tissues by qRT-PCR. TCGA data displayed that HOXA11 was remarkably over-expressed in LUSC compared with that in non-LUSC samples, and the area under curves (AUC) was 0.955 (P < 0.001). A total of 1523 co-expressed genes were sifted for further analysis. The most significant term enriched in the KEGG pathway was focal adhesion. Among the six hub genes of HOXA11, including PARVA, ILK, COL4A1, COL4A2, ITGB1, and ITGA5, five (with the exception of COL4A1) were significantly decreased compared with the normal lung tissues. Moreover, the expression of ILK was negatively related to HOXA11 (r = − 0.141, P = 0.002).
Conclusion
High HOXA11 expression may lead to carcinogenesis and the development of LUSC. Furthermore, co-expressed genes might affect the prognosis of LUSC.
Similar content being viewed by others
Background
Non-small cell lung cancer (NSCLC) has caused the most frequently cancer-related deaths among all types of malignancy in humans worldwide, accompanied by a high incidence [1,2,3]. NSCLC is responsible for the majority of the primary lung cancer cases, including large cell carcinoma, lung adenocarcinoma (LUAD), and lung squamous cancer (LUSC). Among the three histological subtypes, LUSC is the most common type in developing countries [4,5,6,7], and patients with lung cancer are still facing a low overall 5-year survival rate [8, 9]. Molecular targeted therapy has achieved curative efficacy in clinical in LUSC. For example, EGFR targeted therapy had a modest effect in advanced LUSC patients [10]. However, the number of applicable patients is limited [11]. Thus, there is an urgent need to identify more underlying high-performance targets in LUSC.
Cumulative evidence has demonstrated that HOX genes, which belong to the large family of homeodomain genes, work to regulate growth processes, such as organogenesis and body patterning [12, 13]. Humans have HOX genes in four clusters (HOXA, HOXB, HOXC, and HOXD) [14, 15]. Cumulative studies have reported that HOX genes are expressed in healthy human lungs and play a crucial role in their development [12]. The HOXA11 cluster is located on chromosome 7p15-7p14.2, and 12 genes are involved in the cluster, including EVX1 and 11 HOX genes [16, 17]. A number of studies have been carried out to define the function of HOXA genes in malignant cancers. An increasing number of reports suggest that HOXA11 has been implicated in several malignant tumors, such as gastric cancer [18], renal cell carcinoma [19], NSCLC and lung adenocarcinoma [17, 20], and breast cancer [21]. Thus far, the expression level of HOXA11 and its potential mechanisms in LUSC have not been clarified.
In the present study, we attempted to identify the association between clinical parameters and HOXA11 expression to gain a comprehensive understanding of the role of HOXA11 in LUSCs. The mechanisms of HOXA11 co-expressed genes were mined by bioinformatics analysis.
Methods
Selection of clinical LUSC tissue samples
Clinical samples were collected from 23 LUSC patients who had been pathologically identified at the Department of Pathology, First Affiliated Hospital of the Guangxi Medical University (Nanning, Guangxi, China), from January 2012 to February 2014. The clinicopathological features of the patients are shown in Table 1. The Ethical Committee of the First Affiliated Hospital of Guangxi Medical University approved the present research. All participating clinical doctors and patients signed written informed consents.
Total RNA isolation
Per the manufacturer’s instructions, we extracted total RNA with the miRNeasy FFPE Tissue Kit (QIAGEN, Shanghai, China). In addition, we detected the purity and concentration of total RNA using NanoDrop 2000 (ThermoScientific, USA).
qRT-PCR assay
The 10 μl reaction system was set up so that the prepared total RNA could be reverse transcribed using a reverse transcription kit (ABI, Life Technologies, USA) based on the manufacturer’s instruction. Fluorochrome SYBR Green I Master was used for a 20-μl real-time fluorescence PCR system. The specific primers were as follows: HOXA11 forward primer: 5′-TGGTCCCTGCTCCTCTAAC-3′, reverse primer: 5′-GGCTCAATGGCGTACTCTC-3′ [22]. GAPDH (internal control) forward primer: 5′-TGCACCACCAACTGCTTA-3′, reverse primer: 5′-GGATGCAGGGATGATGTTC-3′. The expression difference was calculated using the 2−△Cq method [23, 24].
Data mining and analyzing
All clinicopathological parameters related to LUSC and mRNA (level 3) expression in LUSC were carefully downloaded from the TCGA Data Portal website (http://cancergenome.nih.gov). Based on the HOXA11 expression in LUSC, GraphPad Prism was applied to obtain the scatter diagram. In addition, SPSS was carried out to acquire receiver operating characteristic curves (ROCs) as well as overall survival (OS) and disease-free survival (DFS) curves. Meanwhile, the available data of HOXA11 expression in LUSC was mined in Oncomine (https://www.oncomine.org). Furthermore, we collected in-house qRT-PCR and data from public databases to gain insight into the differential expression of HOXA11 in LUSC using an integrative meta-analysis. The standard mean difference (SMD) was pooled from all studies to determine the expression level of HOXA11 in LUSC.
Screening co-expressed genes of HOXA11
Co-expressed genes of HOXA11 in LUSC were collected from MEM (http://biit.cs.ut.ee/mem), cBioPortal (http://www.cbioportal.org), and GEPIA (http://gepia.cancer-pku.cn) for further evaluation. Genetic alterations of HOXA11, including amplification, deep deletion, and mRNA upregulation were additionally acquired from cBioPortal.
Enrichment analysis and pathway annotation
Gathered genes were analyzed using bioinformatics. The enrichment of functions and signaling pathways of the target genes were analyzed using The Database for Annotation, Visualization and Integrated Discovery v6.8 (DAVID), FunRich, and Cytoscape. The String database (http://www.string-db.org) was applied to construct the protein-protein interaction (PPI) network for the hub gene identification. Moreover, hub genes were selected to obtain their expression and correlation with HOXA11 in LUSC. The immunohistochemistry results of the six hub genes in LUSC were retrieved from the Human Protein Atlas (HPA) database.
Statistical analysis
Statistical analysis was conducted using SPSS 22.0. All data are shown as mean ± standard deviation (SD). An independent samples’ t test was adopted to examine the differences between cancer tissues and normal lung tissues. A one-way analysis of variance (ANOVA) was employed for analyzing differences of HOXA11 expression in various pathological gradings from in-house qRT-PCR data, as well as terms of M category, N category, race, and statuses of recurrence from the TCGA database. The relationships between the co-expressed genes and HOXA11 were assessed using the Pearson rank correlation, and the AUC was counted. To achieve an in-depth understanding of the prognostic value of HOXA11, we also used Kaplan-Meier curves to determine the survival time, including the OS and DFS. A two-sided P value < 0.05 was considered statistically significant.
Results
HOXA11 expression and clinicopathological features in LUSC using qRT-PCR
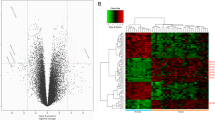
The expression and clinicopathological features of HOXA11 in LUSC are displayed in Table 1. There was no significant correlation between HOXA11 expression and all clinical parameters. However, the expression of HOXA11 was upregulated in LUSC compared to in non-cancerous tissues, and the AUC of the TNM stage was 0.831 (P = 0.008) (Fig. 1a, b).
Verification of HOXA11 expression in TCGA and Oncomine
As shown in Table 2, a cohort of 475 LUSC and 18 non-cancerous adjacent lung tissues was obtained from TCGA database. The results demonstrated that the expression of HOXA11 was noted higher in LUSC (5.531 ± 2.054) than in adjacent lung tissues (1.209 ± 0.813) (P < 0.001) (Fig. 2a). The AUC of the HOXA11 expression of LUSC for diagnosis was 0.955 (P < 0.001) (Fig. 2b). The expression of HOXA11 did not significantly differ in OS (P = 0.795) and DFS (P = 0.864) (Fig. 2c, d). As shown in Table 3, in the five studies of HOXA11 expression available at Oncomine, Hou et al. [25] and Garber et al. [26] found that HOXA11 expression in LUSC was significantly over-expressed (1.118 ± 0.211 vs 0.972 ± 0.135, P = 0.002; 0.752 ± 0.312 vs 0.451 ± 0.146, P = 0.006). The ROC curve analysis showed that the AUCs were 0.717 and 0.781 respectively, which were both significant (P < 0.05) (Fig. 3a, b, f, and g). Wachi et al. [27] and Bhattacharjee [28] illustrated that there were no noticeable differences between the HOXA11 expression in LUSC and adjacent normal tissues, but the expressions of HOXA11 was also upregulated (0.538 ± 0.044 vs 0.507 ± 0.045, P = 0.314; 4.141 ± 3.527 vs 2.796 ± 3.070, P = 0.228) (Fig. 3c, d). Meanwhile, the AUCs of Wachi’s and Bhattacharjee’s were estimated. There was no significant value of the AUC in the study of Wachi (AUC = 0.720, P = 0.251) (Fig. 3h). As for the study of Bhattacharjee, the AUC was 0.753 (P = 0.009) (Fig. 3i). In contrast, Talbot et al. [29] showed that HOXA11 was significantly downregulated (1.336 ± 0.161 vs 1.503 ± 0.236, P = 0.003), with an AUC of 0.744 (Fig. 3e, j). Additionally, as shown in Fig. 4a, scatterplots of pooled data demonstrated a remarkably higher level of HOXA11 in LUSC (4.700 ± 2.596 vs. 1.159 ± 1.217, P < 0.001). The AUC of pooled data was estimated to be 0.873 with P < 0.001 (Fig. 4b). The forest plots also revealed an increased HOXA11 in LUSC compared to non-cancerous lung tissues (SMD = 0.820, 95% CI = 0.594–1.046, P < 0.001) (Fig. 4c).
Data analysis of five studies extracted from Oncomine. a HOXA11 expressed higher in LUSC in Hou’s study (P = 0.002). b HOXA11 was higher in LUSC tissues in Garber’s study (P = 0.007). c HOXA11 expressed insignificant in Wachi’s study (P = 0.318). d HOXA11 showed no significant results in Bhattacharjee’s study (P = 0.224). e HOXA11 decreased in LUSC in Talbot’s study (P = 0.003). f The AUC of HOXA11 for diagnosing LUSC in Hou’s study was 0.717 (P = 0.001). g The AUC of HOXA11 for diagnosing LUSC in Garber’s study was 0.781 (P = 0.047). h The AUC of HOXA11 for diagnosing LUSC in Wachi’s study was 0.720 (P = 0.251). i The AUC of HOXA11 for diagnosing LUSC in Bhattacharjee’s study was 0.753 (P = 0.009). j The AUC of HOXA11 for diagnosing LUSC in Talbot’s study was 0.744 (P = 0.001)
Achievement of co-expressed genes
As shown in Venn, 1340 genes were obtained from MEM, 7 from cBioPortal, and 200 co-expression genes from GEPIA, respectively (Fig. 5a). Three genes, HOXA10, HOXA13, and HOXC10, were intersected in various platforms. Alterations of HOXA11 were indicated via cBioPortal, which showed that HOXA11 expression was upregulated in 27 LUSC patients, deleted in 2 LUSC patients, and amplified in 8 LUSC patients (Fig. 5b).
Bioinformatics analysis
All analysis was based on the number of 1523 co-expression genes. The Gene Ontology enrichment analysis comprised three categories: a biological process (BP), a molecular function (MF), and a cellular component (CC). The most valuable 10 pathways of each category are presented in Fig. 6a, c, and 6e, including different kinds of functional relationship graphs (Fig. 6b, d, and f). For the KEGG pathways, the 10 most significant pathways are shown in Table 4. The PPI network is displayed (Fig. 7); three pairs of hub genes with the highest combined scores (PARVA, ILK, COL4A1, COL4A2, ITGB1, and ITGA5) were collected from the PPI network (Table 5).
Top 10 significant pathways and GO enrichment analysis. a Graph of the 10 most significant pathways of BP category. b Enrichment analysis of BP, each node means one different function and more significant ones are filled in with a deeper color. c Top 10 significant terms in the CC category. d Enrichment analysis of the CC category; each node means one different function, and more significant ones are filled in with a deeper color. e Ten most valuable annotations of the MF category. f Enrichment analysis of the CC category; each node means one different function, and more significant ones are filled in with a deeper color
Expression and correlation of hub genes with HOXA11
Compared with non-cancerous lung tissues, the hub genes ILK, PARVA, COL4A2, ITGB1, and ITGA5 were significantly downregulated in LUSC (Table 6, Fig. 8a–f). Moreover, correlations between hub genes and HOXA11 were analyzed, and the gene ILK was negatively correlated with HOXA11 in LUSC (r = − 0.141, P = 0.002) (Fig. 8g–l). The HPA database indicated that there were lower levels of the six hub genes in NSCLC tissues: ILK (Antibody CAB004041), PARVA (Antibody HPA005964), COL4A1 (Antibody CAB001695), ITGB1 (Antibody CAB003434), ITGA5 (Antibody CAB009008), and COL4A2 (Antibody CAB010751) (Fig. 9a–f).
Hub genes’ expression in LUSC and correlations with HOXA11. a ILK was lower in LUSC tissues than in non-cancerous tissues (P < 0.001). b The gene PARVA was significantly overexpressed in normal tissues (P < 0.001). c The levels of COL4A1 in different tissues showed no significance (P = 0.061). d The hub gene ITGB1 revealed higher levels in normal tissues (P < 0.001). e ITGA5 was significantly decreased in LUSC tissues (P < 0.001). f COL4A2 upregulated in non-LUSC tissues (P < 0.001). g ILK and HOXA11 showed a negative correlation (r = − 0.141, P = 0.002). h Correlations between PARVA and HOXA11 showed no significance (P = 0.645). i Correlations between COL4A1 and HOXA11 showed no significance (P = 0.337). j Correlations between ITGB1 and HOXA11 showed no significance (P = 0.936). k Correlations between ITGA5 and HOXA11 showed no significance (P = 0.501). l Correlations between COL4A2 and HOXA11 showed no significance (P = 0.248)
The protein level of the selected hub genes in LUSC tissues from The Human Protein Atlas. a ILK (Antibody CAB004041) expression in LUSC tissues. b PARVA (Antibody HPA005964) expression in LUSC tissues. c COL4A1 (Antibody CAB001695) expression in LUSC tissues. d ITGB1 (Antibody CAB003434) expression in LUSC tissues. e ITGA5 (Antibody CAB009008) expression in LUSC tissues. f COL4A2 (Antibody CAB010751) expression in LUSC tissues
Discussion
HOX genes may play a central role in regulating gene expression, differentiation, and receptor signaling. Members of HOX family can also encode DNA-binding transcription factors [30, 31]. A growing body of studies observed that HOXA11 expression was downregulated in different tumor types. In glioblastoma, the decreased level of HOXA11 was confirmed as a significant marker of poorer prognosis. [32]. Moreover, Bai et al. suggested that the down-expressed HOXA11 gene may play an essential role in carcinogenesis by promoting gastric cancer development, which may be helpful to forecast the malignant behaviors of gastric cancer [33]. HOXA11 has been notably over-expressed in epithelial ovarian cancers [34]. Data from the current study, TCGA, Hou’s study [25], along with Garber’s study [26], all illustrated that the corresponding mRNA expression of HOXA11 is significantly over-represented in LUSC compared with adjacent tissues. The enhanced expression of HOXA11 might be a diagnostic target to use for distinguishing LUSC from healthy controls based on the ROC (AUC = 0.955, P < 0.001) from TCGA. Unfortunately, the expression detected by qRT-PCR revealed no significance. However, HOXA11 tends to be upregulated in LUSC compared to normal lung tissues. Interestingly, Talbot reported that the levels of HOXA11 were reduced in patients with LUSC. In light of these previous studies, HOXA11 expression seems to be higher in LUSC than in healthy controls. More LUSC specimens need to be collected to study the expression levels of HOXA11 in LUSC in the future. Among the co-expressed genes of HOXA11, a total of three genes, HOXA10, HOXA13, and HOXC10, were finally intersected, which all belong to the HOX gene family. HOXA10 is involved in gene expression, regulation, morphogenesis, and differentiation in ovarian carcinoma. HOXA10 and HOXA11 might be associated with primary tumors and specific histological subtypes [34]. HOXA13 regulates gene expression and differentiation. HOXA10, HOXA11, and HOXA13 might be useful targets to further mine the molecular pathogenesis of HOXA11 in early-stage lung adenocarcinoma [35]. Moreover, HOXC10 may act as the accelerator for original activation. In previous studies, HOXC10 has been reported to be associated with the increased invasion of malignancies [36, 37]. These three co-expressed genes and HOXA11 might play several pivotal roles in LUSC, such as the subtype differentiation of lung cancer, the regulation of LUSC progression, and the development of efficient therapeutic strategies.
The pathway of focal adhesion has been extensively reported to be the most significant pathway in various diseases and signaling pathways as well. It regulates diverse cellular functions that were once activated by focal adhesion kinase (FAK), including adhesion, proliferation, migration, and survival [38,39,40]. A pattern of enhanced expression of FAK in lung carcinomas has been reported, which is related to nodal involvement and the deterioration of advanced disease stages [41, 42]. Therefore, FAK protein expression may help in predicting the aggressive behavior of LUSC. Meanwhile, FAK might be pursued as a promising therapeutic target for LUSC.
Six hub genes were analyzed, PARVA, ILK, COL4A1, COL4A2, ITGB1, and TIGA5. Except COL4A1 gene, the rest of the hub genes were all significantly decreased in LUSC compared to adjacent healthy controls. COL4A1 and COL4A2 both belong to the type IV collagen gene family. Both can act as inhibitors of angiogenesis and tumor growth. In LUSC, they can control vascular invasion and inhibit the size of tumors. There is more reason to believe that the upregulated HOXA11 gene plays a carcinogenic role in LUSC. HOXA11 might depress COL4A1 and COL4A2 expression levels in LUSC. ITGB1 has been recognized in the processing of metastatic diffusion of tumor cells, and ITGA5 may promote tumor invasion. In lung cancer, higher expression of ITGA5 may be correlated with a shorter survival time. Meanwhile, previous studies have indicated that modulating the ILK signaling pathway by PARVA made it more vulnerable to metastasis in lung adenocarcinoma [43]. These hub genes might also similarly interact with each other via various signaling pathways in LUSC. Thus, we speculate that LUSC patients’ prognosis might be better and their survival time might be longer than for patients with other subtypes of lung cancer. The mechanisms and functions between hub genes and HOXA11 in LUSC remain elusive and need to be validated in the future.
Conclusion
The findings of the present study indicate that HOXA11 high expression might lead to the occurrence and development of LUSC. Meanwhile, co-expressed HOXA11 genes may influence the prognosis of LUSC, and the gene ILK may have the complete reverse functions in LUSC compared with HOXA11. Hub genes need to be further analyzed to ensure their mechanisms and functions in LUSC.
Abbreviations
- ANOVA:
-
Analysis of variance
- BP:
-
Biological process
- CC:
-
Cellular component
- COL4A1:
-
Collagen type IV alpha 1 chain
- COL4A2:
-
Collagen type IV alpha 2 chain
- DFS:
-
Disease-free survival
- FAK:
-
Focal adhesion kinase
- GO:
-
Gene Oncology
- HOXA11:
-
Homeobox A11
- ILK:
-
Integrin-linked kinase
- ITGA5:
-
Integrin subunit alpha 5
- ITGB1:
-
Integrin subunit beta 1
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cancer
- MF:
-
Molecular function
- NSCLC:
-
Non-small cell lung cancer
- OS:
-
Overall survival
- PARVA:
-
Parvin alpha
- PPI:
-
Protein-protein interaction
- ROC curve:
-
Receiver operating characteristic curve
- SD:
-
Standard deviation
- TCGA:
-
The Cancer Genome Atlas
References
Xiong W, Zhao Y, Xu M, Guo J, Pudasaini B, Wu X, Liu J. The relationship between tumor markers and pulmonary embolism in lung cancer. Oncotarget. 2017;8:41412–21.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
Sztankay M, Giesinger JM, Zabernigg A, Krempler E, Pall G, Hilbe W, et al. Clinical decision-making and health-related quality of life during first-line and maintenance therapy in patients with advanced non-small cell lung cancer (NSCLC): findings from a real-world setting. BMC Cancer. 2017;17:565.
Han Z, Wang T, Han S, Chen Y, Chen T, Jia Q, Li B, Li B, Wang J, Chen G, et al. Low-expression of TMEM100 is associated with poor prognosis in non-small-cell lung cancer. Am J Transl Res. 2017;9:2567–78.
Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84:13–22.
Zhang M, Gao C, Yang Y, Li G, Dong J, Ai Y, Ma Q, Li W. MiR-424 promotes non-small cell lung cancer progression and metastasis through regulating the tumor suppressor gene TNFAIP1. Cell Physiol Biochem. 2017;42:211–21.
Coudray C, Fouret G, Lambert K, Ferreri C, Rieusset J, Blachnio-Zabielska A, et al. A mitochondrial-targeted ubiquinone modulates muscle lipid profile and improves mitochondrial respiration in obesogenic diet-fed rats. Br J Nutr. 2016;115:1155–66.
Wang S, Pan H, Liu D, Mao N, Zuo C, Li L, Xie T, Huang D, Huang Y, Pan Q, et al. Excision repair cross complementation group 1 is a chemotherapy-tolerating gene in cisplatin-based treatment for non-small cell lung cancer. Int J Oncol. 2015;46:809–17.
Zhang M, Li G, Wang Y, Wang Y, Zhao S, Haihong P, Zhao H, Wang Y. PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis. Sci Rep. 2017;7:10255.
Memon AA, Zhang H, Gu Y, Luo Q, Shi J, Deng Z, Ma J, Ma W. EGFR with TKI-sensitive mutations in exon 19 is highly expressed and frequently detected in Chinese patients with lung squamous carcinoma. Onco Targets Ther. 2017;10:4607–13.
Weeden CE, Solomon B, Asselin-Labat ML. FGFR1 inhibition in lung squamous cell carcinoma: questions and controversies. Cell Death Discov. 2015;1:15049.
Golpon HA, Geraci MW, Moore MD, Miller HL, Miller GJ, Tuder RM, Voelkel NF. HOX genes in human lung: altered expression in primary pulmonary hypertension and emphysema. Am J Pathol. 2001;158:955–66.
Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42:421–9.
Sucic M, Boban D, Markovic-Glamocak M, Bogdanic V, Nemet D, Labar B, et al. Relation between urinary cytology abnormalities and cyclosporine a therapy in bone marrow transplant recipients. Ren Fail. 1998;20:613–20.
Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–92.
Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–32.
Hwang JA, Lee BB, Kim Y, Park SE, Heo K, Hong SH, et al. HOXA11 hypermethylation is associated with progression of non-small cell lung cancer. Oncotarget. 2013;4:2317–25.
Cui Y, Gao D, Linghu E, Zhan Q, Chen R, Brock MV, et al. Epigenetic changes and functional study of HOXA11 in human gastric cancer. Epigenomics. 2015;7:201–13.
Wang L, Cui Y, Sheng J, Yang Y, Kuang G, Fan Y, Jin J, Zhang Q. Epigenetic inactivation of HOXA11, a novel functional tumor suppressor for renal cell carcinoma, is associated with RCC TNM classification. Oncotarget. 2017;8:21861–70.
Li Q, Chen C, Ren X, Sun W. DNA methylation profiling identifies the HOXA11 gene as an early diagnostic and prognostic molecular marker in human lung adenocarcinoma. Oncotarget. 2017;8:33100–9.
Xia B, Shan M, Wang J, Zhong Z, Geng J, He X, et al. Homeobox A11 hypermethylation indicates unfavorable prognosis in breast cancer. Oncotarget. 2017;8:9794–805.
Martino V, Bianchera A, Reia L, Bussolati O, Fazzina R, Marino F, et al. Down-regulation of HOXA4, HOXA7, HOXA10, HOXA11 and MEIS1 during monocyte-macrophage differentiation in THP-1 cells. Mol Med Rep. 2009;2:241–4.
Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19–24.
Chen G, Kronenberger P, Umelo IA, Teugels E, De Greve J. Quantification of epidermal growth factor receptor T790M mutant transcripts in lung cancer cells by real-time reverse transcriptase-quantitative polymerase chain reaction. Anal Biochem. 2010;398:266–8.
Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312.
Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–9.
Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–8.
Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790-13795.
Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, P O-charoenrat, et al: Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071.
Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–71.
Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl). 2014;92:811–23.
Se YB, Kim SH, Kim JY, Kim JE, Dho YS, Kim JW, et al. Underexpression of HOXA11 is associated with treatment resistance and poor prognosis in glioblastoma. Cancer Res Treat. 2017;49:387–98.
Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D, et al. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014;14:79.
Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11:531–7.
Kang JU. Characterization of amplification patterns and target genes on the short arm of chromosome 7 in early-stage lung adenocarcinoma. Mol Med Rep. 2013;8:1373–8.
Xiong W, Zhou Q, Liu G, Liu XS, Li XY. Homeodomain-containing gene 10 inhibits cell apoptosis and promotes cell invasion and migration in osteosarcoma cell lines. Tumour Biol. 2017;39:1010428317697566.
Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67:10163–72.
Woo JK, Jung HJ, Park JY, Kang JH, Lee BI, Shin D, et al. Daurinol blocks breast and lung cancer metastasis and development by inhibition of focal adhesion kinase (FAK). Oncotarget. 2017;8:57058–71.
Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68.
Golubovskaya VM. Targeting FAK in human cancer: from finding to first clinical trials. Front Biosci (Landmark Ed). 2014;19:687–706.
Ji HF, Pang D, Fu SB, Jin Y, Yao L, Qi JP, Bai J. Overexpression of focal adhesion kinase correlates with increased lymph node metastasis and poor prognosis in non-small-cell lung cancer. J Cancer Res Clin Onco. 2013;139:429–35.
Carelli S, Zadra G, Vaira V, Falleni M, Bottiglieri L, Nosotti M, et al. Up-regulation of focal adhesion kinase in non-small cell lung cancer. Lung Cancer. 2006;53:263–71.
Huang AH, Pan SH, Chang WH, Hong QS, Chen JJ, Yu SL. PARVA promotes metastasis by modulating ILK signaling pathway in lung adenocarcinoma. PLoS One. 2015;10:e0118530.
Acknowledgements
The study was supported by the fund of Guangxi Medical University Students Innovative Project (WLXSZX17035) and the Innovation Project of Graduate Education (YCSW2018103).
Funding
The study was supported by the fund of Guangxi Medical University Students Innovative Project (WLXSZX17035). The fund was used for the clinical sample collection, in-house qRT-PCR experiments, and the language editing of the manuscript. The fund of Innovation Project of Graduate Education (YCSW2018103) was used for the language editing and the publication fee.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Author information
Authors and Affiliations
Contributions
GQZ and XYG collected data from public datasets and analyzed the data. YQ performed the statistical analysis. TQG and YZ carried out the clinical samples gathering and in-house qRT-PCR experiments. GC participated in the design of the study and language modification. RZ and TTZ drafted the manuscript and analyzed the GO and KEGG terms. WJM and ZBF conceived of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University. Ethics, consent, and permissions: Written informed consents were signed by all the patients involved to ensure their approval of the data used in this research.
Consent for publication
All the patients signed the agreement consents for publishing their individual clinical data.
Competing interests
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, R., Zhang, Tt., Zhai, Gq. et al. Evaluation of the HOXA11 level in patients with lung squamous cancer and insights into potential molecular pathways via bioinformatics analysis. World J Surg Onc 16, 109 (2018). https://doi.org/10.1186/s12957-018-1375-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-018-1375-9