Abstract
Background
Osteoporosis, characterized by low bone mineral density (BMD) and high bone fracture risk, is prevalent in Thai menopausal women. Genetic factors are known to play a key role in BMD. Low density lipoprotein receptor-related protein 5 (LRP5), a co-receptor in the Wnt/beta-catenin pathway, is involved in many aspects of bone biology. As coding single nucleotide polymorphisms (cSNPs) of LRP5, including A1330V (rs3736228), and Asian-related Q89R (rs41494349) and N740N (rs2306862), are associated with lowered BMD, this study aimed to determine the relationship between these LRP5 polymorphisms and BMD in 277 Thai menopausal women.
Results
Only rs3736228 deviated from the Hardy–Weinberg equilibrium of allele frequency (p = 0.022). The median, range and p value for the BMD related to each SNP parameter were compared (Mann–Whitney U test). Significant differences were observed between wild-type and risk alleles for both rs3736228 (total radial, p = 0.011; and radial 33, p = 0.001) and rs2306862 (radial 33: p = 0.015) SNPs, with no significant difference for rs41494349 SNP. Linkage disequilibrium was strong for both rs3736228 and rs2306862 SNPs. Haplotype analysis identified high CC frequency in both normal and osteopenia/osteoporosis groups, with a significant odds ratio for carrying the TT haplotype; however, this was non-significant after adjusting for age. Multivariate binary logistic regression analysis performed for rs3736228 showed that individuals with a body mass index <25 kg/m2 had an increased risk of osteoporosis for each decade, but the polymorphism had no effect.
Conclusions
This study did not identify LRP5 polymorphisms as a risk factor for osteoporosis in Thai menopausal women. Further studies with larger sample sizes are needed to further clarify the role of LRP5 as a genetic determinant of osteoporosis.
Similar content being viewed by others
Background
Osteoporosis, a common metabolic bone disease, is characterized by low bone mineral density (BMD) and high risk of bone fracture [1]. During 40–50 years of age, women tend to lose bone mass significantly during the menopausal period because of greater bone resorption. In Thailand, the prevalence of osteoporosis in the lumbar spine and femoral neck of women aged 40–80 years is 19.8 % and 13.6 %, respectively. These figures are less than 5 % in women under 50 years of age, but increase more than 50 % in those older than 70 years of age [2]. Among factors which have an influence on osteoporosis, genetic factors play an essential role in bone density regulation. To date, individual genome-wide association studies (GWASs) and meta-analyses of GWASs have discovered more than 70 genes/loci to be associated with BMD phenotypes [3], particularly ZBTB40, GPR177, FGFRL1, MEF2C, ESR1, WNT16, TNFRSF11B, SOX6, LRP5, AKAP11, and FOXL1 [4, 5]. Further, candidate gene association studies have suggested additional loci, e.g., VDR [6], COL1A1 [7], TGFB1 [8] and PTH [9].
The Wnt signaling pathway has an essential role in the regulation of many cellular processes, such as cell growth, differentiation, migration, polarity and apoptosis. In addition, the canonical Wnt/beta-catenin pathway is particularly involved with bone biology [10]. The transmembrane protein, low density lipoprotein receptor-related protein 5 (LRP5), acts as a co-receptor for the Wnt signaling pathway and it is widely expressed in many tissues including bone. In bone, it expresses by osteoblasts of the endoosteal and trabecular bone surface [11]. Analysis of two monogenic disorders having extreme bone phenotypes reveals that LRP5 as a modulator of bone metabolism. Osteoporosis-pseudoglioma (OPPG), an autosomal recessive disease characterized by bone thinning and blindness due to abnormal eye development has been linked to loss of function mutation in LRP5 [12]. OPPG-causing frameshift or nonsense mutations in LRP5 extracellular domain have been identified and mutant LRP5 protein can be synthesized but not secreted. Dominant negative secreted form of LRP5 can reduce bone thickness in mouse calvarial explants [12]. In contrast to OPPG, an autosomal dominant high bone mass (HBM) traits results from a gain of function mutation in LRP5. Genetic studies of two unrelated families with increased BMD (spinal Z scores ≥ 5) identify a single missense mutation of LRP5 resulting in amino acid substitution of a conserved glycine by valine at residue 171 (G171V) [13, 14]. Serum markers of affected individuals show increased bone formation but normal bone resorption [13]. Level of known target of Wnt signaling, the extracellular matrix protein fibronectin is also increased. Cell expressing LRP5V171 can abolish action of endogenous Wnt inhibitor, Dkk-1 resulting in increased LRP5 function and excessive bone formation [13]. Mice carrying both types of mutations have high and low bone mass phenotypes resembled that of human [11, 15]. Previous qualitative trait locus (QTL) work suggests that chromosome 11 q12–13 region where LRP5 located, may have important role in general BMD variance [16]. A cohort of British Caucasian adults covering broad range BMD studied by Koay et al demonstrates that LRP5 polymorphisms is a genetic determinant of normal BMD variation [17].
Several coding single nucleotide polymorphisms (cSNPs) of LRP5 have been reported. The most frequently studied and likely functional polymorphism of LRP5, A1330V (rs3736228) in exon 18, has been shown to affect BMD and increase fracture risk in various groups, including postmenopausal women [18–21]. It locates in LDL-repeat domain of LRP5 protein which may be responsible for mediating the receptor-ligand interaction [22]. HEK293T cells containing the LRP5-V1330 coexpressed with Wnt3a has reduced transcriptional activity measuring by TCF-Lef reporter assay as compared to wild type allele implying the functional significance of A1330V variant [19]. Another two LRP5 cSNPs, Q89R (rs41494349) in exon 2 and N740N (rs2306862) in exon 10 are chosen because of their association with low BMD in Asian population [23–25]. Q89R is localized on the first of four beta-propellers, while N740N is on the third one [26]. Structural analysis of mutation occurred in the first propeller domain of LRP5 suggest changing in local hydrophobic environment and subsequently possibly affecting interaction of LRP5 with other proteins [14]. Although the function of each LRP5 protein domain is still uncertain, these 3 cSNPs could have influence on LRP5 biological function and BMD. Therefore, LRP5 genetic variations may have an effect on osteoporosis within the Thai population. Thus, we studied the relationship of LRP5 polymorphisms (rs3736228, rs41494349 and rs2306862) and BMD in Thai menopausal women.
Results
Basic characteristics, including age, body mass index (BMI) and BMD of menopausal women, are shown in Table 1. Among the genotype distribution of LRP5 polymorphisms studied, those of rs41494349 and rs2306862 did not deviated significantly from Hardy–Weinberg Equilibrium (HWE), which is in contrast with rs3736228 (p = 0.022) (Table 2). Neither rs41494349 nor rs2306862 SNP was associated with risk of osteopenia/osteoporosis in terms of dominant or recessive model (Table 3). However, the genetic model of dominance for rs3736228 was assumed due to none of its homozygous (T/T) variant present.
From Table 4, it can be observed that the wild-type group of rs3736228 was older than the risk allele group (CC vs CT, 58 vs 55, respectively, p = 0.002). However, BMI was comparable between these groups. From the various sites examined, the Mann–Whitney U test revealed a statistically significant difference between total radial and radial 33 BMD for rs3736228 and rs2306862. For rs3736228, the total radial BMD of the individuals or group carrying the risk allele (CT) was significantly higher than the wild-type group (CC), p = 0.011. In addition, the radial 33 BMD was higher in the individuals or group carrying the risk allele of rs3736228 (CT, p = 0.001) and rs2306862 (CT + TT, p = 0.015). However, there were no statistically significant differences between BMD values of the wild-type (AA) and risk (AG + GG) allele carrier group of the rs41494349 SNP.
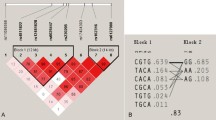
Because a greater radial 33 BMD was found in both rs3736228 and rs2306862 SNP risk allele carrier groups compared with their respective wild-type groups, it was of interest to determine whether a genotypic additive effect was present. Linkage disequilibrium (LD) analysis was conducted and a high level of LD was observed (D’/r2 = 0.9294/0.7549). Haplotype analysis was performed to observe the haplotypic effect of these two genotypes on causing osteopenia/osteoporosis using radial 33 BMD T-score cut-off. In both the normal and osteopenia/osteoporosis groups, CC haplotype was found at high frequency. The odds ratio of carrying the TT haplotype was found to be 0.48 (95 % CI = 0.26–0.88, p = 0.018) in comparison to the more common CC haplotype (Table 5). However, after adjusting for age, the TT haplotype was not found to be statistically significant: OR = 0.62 (95 % CI = 0.32–1.20, p = 0.160).
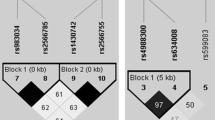
Although we did not detect an association between osteopenia/osteoporosis and haplotypic effect of the two SNPs, rs3736228 and rs2306862, they belonged to a big LD block comprising other variants in HapMap genotype data of Han Chinese in Beijing, China (CHB) and Japanese in Tokyo, Japan (JPT) population (Fig. 1). Haploview LD map of LRP5 showed almost similar LD structure in both populations (Additional file 1 Figure S1). Both variants were in complete LD in CHB (block 6; D’/r 2 = 1/0.913) and JPT despite located in non-adjacent blocks (block 4 and 6; D’/r 2 = 1/0.941).
Linkage disequilibrium plot between LRP5 SNPs showing region the rs3736228 and rs2306862 located using HapMap populations determined by Haploview 4.2 program. Each black triangle depicts haplotype blocks. LD is reported as D’. Bright red represents D’ = 1 and LOD ≥ 2, blue represents D’ = 1 and LOD < 2, pink represents D’ < 1, and LOD ≥ 2, and white represents D’ < 1 and LOD < 2. The r 2 values are shown in blocks. The rs3736228 and rs2306862 are indicated by boxes. CHB, Han Chinese in Beijing, China; JPT, Japanese in Tokyo, Japan
Univariate analysis was performed to determine if any factors contributed to osteoporosis, represented by total radial BMD T-score cut-off [27]. The factors tested were the rs3736228 genotype and those affecting osteoporosis risk, age and BMI. The combined effect of these factors was analyzed using binary logistic regression analysis (Table 6). For individuals with a BMI of less than 25 kg/m2, for every decade older, there was an increased risk of osteoporosis. The rs3736228 polymorphism had no effect.
Discussion
LRP5, a co-receptor for the Wnt signaling pathway, is an important regulator for bone homeostasis [28]. Polymorphisms of LRP5 have been demonstrated to have an influence on BMD and to be associated with an increased risk of osteoporosis. We investigated the effect of genetic variation of LRP5 on BMD in Thai menopausal women.
Departure from HWE was detected in the rs3736228 genotype distribution. The most common cause of deviation from HWE is genotyping error [29]. However, for SNP it tends to be due to a deficit of heterozygotes, making allele dropout the most frequent cause of genotyping error [30]. On the contrary, with respect to our results however, it was likely due to a lack of homozygotes and the number of heterozygotes being higher than expected. This is in agreement with the Lewis phenotyping study showing that the cause of departure from HWE was a deficit of homozygotes [31].
In this study, we found that heterozygous risk allele carriers, the T-allele of rs3736228 (CT), had a significantly higher total radial BMD than those carrying homozygous C-allele carriers (CC), p = 0.011 (N = 210 vs 67, respectively). In addition, the risk allele carriers of rs3736228 and rs2306862 had a significantly higher radial 33 BMD than those with the wild-type allele (p = 0.001 (N = 210 vs 67) and p = 0.015 (N = 193 vs 68), respectively). However, the wild-type allele carrier group of rs3736228 was older than the risk allele group. Aging is an important confounding factor in osteoporosis, and an increase of age makes people more prone to developing osteoporosis, coinciding with a lower BMD. Therefore, it is likely that aging contributes to the lower radial BMD found in the wild-type group of rs3736228, as observed from logistic regression analysis. In postmenopausal women, age is reported to be the second most important predictor of BMD variation [32].
These findings are contradictory to previous studies in LRP5 SNPs, whereby subjects with a risk allele have a lower BMD. The rs3736228 is associated with low BMD in various ethnic groups [18–21]. Furthermore, the association of rs2306862 and rs41494349 with BMD has been reported predominantly within the Asian population [23–25]. However, we failed to demonstrate the association of rs41494349 with BMD in our study.
The strong LD found between rs3736228 and rs2306862 is in agreement with a study by Mizuguchi et al [23]. In this study, the researchers identified an LD block between intron 7 and exon 18 of the LRP5 gene in the Japanese women. This included rs2306862 on exon 10 and rs3736228 on exon 18. Haplotype blocks distribution generated by Haploview gave similar LD patterns between these two SNPs in CHB and JPT populations of HapMap. Although the TT haplotype showed a protective effect against osteopenia/osteoporosis, it disappeared after adjusting for age. This is likely because in the rs3736228 SNP, the wild-type allele carrier group was older than the risk allele group.
Our rs3736228 heterozygous risk allele carriers had a higher total radial BMD compared with the wild-type, which is in contrast to the majority of previous studies of LRP5 polymorphisms, in which the rs3736228 risk allele carrier is associated with a reduced BMD [18–21]. However, our finding is consistent with two previous studies of rs3736228 in men. Kiel et al. showed that men with a reduced physical activity level carrying the risk allele of rs3736228 have a higher spine BMD, whereas Kruk et al found that men with no reported physical activity level have a high BMD at all hip sites [33, 34].
Both the Kiel and Kruk research groups suggest involvement of physical activity and mechanical loading in modulating the effect of LRP5 on bone remodeling balance in response to physical exercise or on BMD determination. The mechanism by which the different alleles of rs3736228 polymorphism interact through physical activity to modify bone or BMD is unknown. However, LRP5 and Wnt/beta-catenin signaling have been identified as essential components of mechanically induced signal transduction [35, 36]. Loading increases Wnt signaling pathway expression and Wnt/beta-catenin target genes; however, there is no response to mechanical loading in mice with deletion of Lrp5 [37, 38].
Estrogen receptor alpha (ER-alpha) is involved in the bone adaptation process to mechanical loading [39]. Moreover, the LRP5 and ER-alpha pathway are reported to interact during mechanical loading [40]. One study investigating gene-exercise interactions on BMD, showed an association between physical activity and ER-alpha polymorphism on BMD modulation at the loaded bone sites [41]. Furthermore, an interleukin-6-association study showed that its genetic variant influences cortical bone resorption during exercise [42]. Accordingly, it is possible that LRP5 could have an effect on BMD in response to physical activity. Although no study has directly assessed the role of LRP5 polymorphism, it remains to be determined if this polymorphism has an interaction with mechanical loading via physical activity or exercise, finally resulting in BMD change.
Our study has several limitations. First, because the DNA used for genotyping was derived from stored specimens, the sample size was fixed. Second, the available BMD data was much smaller in comparison with the genotyping data. Third, age is an important confounding factor for osteoporosis, which significantly different between the wild-type and risk allele carrier groups of rs3736228. Therefore, we cannot avoid or exclude the confounding factor effect of age. Fourth, our data deviated from HWE because of the higher incidence of heterozygotes found than expected. Taken together, this leads to difficulties in statistical analysis; giving rise to false positives or negatives and even non-significant results. Lastly, other potential confounders, such as years since menopause, physical activity, smoking, alcohol consumption and others, were not assessed.
Conclusions
From our findings and limitations, it seems that LRP5 polymorphisms are not a risk factor for osteoporosis in Thai menopausal women. However, this research is not conclusive and further studies with a larger sample size, and that address all the aforementioned limitations, will help clarify the role of LRP5 as a genetic determinant of osteoporosis.
Methods
Study population
The original research project was the collaboration between the Department of Obstetrics and Gynecology, Faculty of Medicine, Ramathibodi hospital and the Department of Tropical Nutrition and Food Science, Faculty of Tropical Medicine with EC approval from Ramathibodi hospital since 2009. Data and blood collection was performed at the Ramathibodi hospital and the experiments were performed at the Faculty of Tropical Medicine. In the present study, we used stored specimen left from that original project with permission and EC approval.
In this study, subjects included 277 menopausal Thai women who attended the menopause post-operation follow-up clinic at the Department of Obstetrics and Gynecology, Ramathibodi Hospital, Bangkok, Thailand. Inclusion criteria used were: (1) women with 45 years of age or older; (2) not having serious disease history; (3) not receiving vitamin/mineral for bone supplement; and (4) willing to participate in the study. All subjects were in good health and gave informed consent to participate in the study. Exclusion criteria included any known diseases that could affect bone metabolism such as hyperthyroidism, hyperparathyroidism, rheumatoid arthritis, rickets and osteomalacia, hypogonadism, type I diabetes mellitus, and Cushing’s syndrome. Physical examinations were conducted by the same physician. The study protocol was approved by the Ethics committees of the Faculty of Tropical Medicine and Faculty of Medicine (Ramathibodi Hospital), Mahidol University, Bangkok, Thailand.
BMD measurement
Bone mineral density (BMD, g/cm2) was assessed at the lumbar spine (L2–L4), femoral neck, total radius, radial 33, and total hip using dual-energy X-ray absorptiometry (DEXA) (Lumar Prodigy, Lunar, USA) by a single, experienced technician.
DNA extraction and genotyping
Genomic DNA was extracted from EDTA-treated peripheral blood samples using the Flexi Gene DNA kit (Qiagen, Hilden, Germany). Genomic DNA was stored at −70 °C for further analysis. Genotyping of LRP5 SNP variation was conducted using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method.
The forward and reverse primers for A1330V (rs3736228) SNP were designed as 5′-GACTGTCAGGACCGCTCACACG-3′ and 5′-AAGGTTTTCAGAGCCCCTAC-3′, respectively. The PCR product was cut by DraIII (New England Biolabs, Beverly, CA) and separated by 6 % polyacrylamide gel electrophoresis. DNA from a subject homozygous for the C allele appeared as a band of 143 bp in length relative to the 100 bp size marker. The T allele constructs cut site for DraIII, and DNA from a subject homozygous for the T allele, appeared as bands of 119 and 24 bp, respectively [25].
The forward and reverse primers for Q89R (rs41494349) SNP were designed as 5′-CTCTGGGCATAGTGCTCCATC-3′ and 5′-CCGGAGATGACCACGTTCTG-3′, respectively. The PCR product was cut by AvaI (New England Biolabs, Beverly, CA) and separated by 6 % polyacrylamide gel electrophoresis. DNA from a subject homozygous for the A allele appeared as a band of 308 bp in length relative to the 100 bp size marker. The G allele constructs cut site for AvaI, and DNA from a subject homozygous for the G allele, appeared as bands of 257 and 51 bp, respectively.
The forward and reverse primers for N740N (rs2306862) SNP were designed as 5′-CTACTGGGCCGACACTGGGATTAA-3′ and 5′-ACAGCTCTAATCACCGAGGG-3′, respectively. The PCR product was cut by AseI (New England Biolabs, Beverly, CA) and separated by 6 % polyacrylamide gel electrophoresis. DNA from a subject homozygous for the C allele appeared as a band of 237 bp in length relative to the 100 bp size marker. The T allele constructs cut site for AseI, and DNA from a subject homozygous for the T allele appeared as bands of 216 and 21 bp, respectively [25].
Results have been either randomly duplicated checked and performed direct sequencing. Resolution of 24 bp (A1330V), 51 bp (Q89R) and 21 bp (N740N) cleaved products cannot be resolved in 6 % PAGE and are invisible on the gel. Therefore, only DNA bands with larger size for each SNP can be seen. Gel depicted 3 SNPs genotyping is shown below in Fig. 2.
Statistical analysis
The SPSS v.15.0 program for Windows (SPSS, Chicago, IL) was used to analyze the median, range and p value (Mann–Whitney U test) between groups. Departure from Hardy–Weinberg Equilibrium (HWE) of allele frequency for each SNP was tested by the Chi-square test. Linkage disequilibrium (LD) and haplotype analysis were performed using the SNPStat web tool program [43]. LD plot of CHB and JPT populations genotype data from HapMap phase 3 (June 15, 2016) was generated using Haploview 4.2 program [44].
Univariate analysis was performed to determine any factor that had an effect on osteoporosis, represented by total radial BMD T-score cut-off for osteoporosis. Any variable with p ≤ 0.1 in univariate analysis was further analyzed in stepwise multivariate logistic regression analysis using a backward Wald method for determining independent associated factors for osteoporosis. Backward Wald method is a stepwise entry method used to construct the logistic regression model by removing explanatory variables from the full model that including all the specified explanatory variables. All tests of significance were 2-sided, and a p-value <0.05 was considered statistically significant.
References
Kanis JA, Burlet N, Cooper C, Delmas PD, Reginster JY, Borgstrom F, et al. European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int. 2008;19:399–428.
Limpaphayom KK, Taechakraichana N, Jaisamrarn U, Bunyavejchevin S, Chaikittisilpa S, Poshyachinda M, et al. Prevalence of osteopenia and osteoporosis in Thai women. Menopause. 2001;8:65–9.
Liu YJ, Zhang L, Papasian CJ, Deng HW. Genome-wide Association Studies for Osteoporosis: A 2013 Update. J Bone Metab. 2014;21:99–116.
Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23:1923–33.
Niu T, Liu N, Zhao M, Xie G, Zhang L, Li J, et al. Identification of a novel FGFRL1 MicroRNA target site polymorphism for bone mineral density in meta-analyses of genome-wide association studies. Hum Mol Genet. 2015;24:4710–27.
Morrison NA, Qi JC, Tokita A, Kelly PJ, Crofts L, Nguyen TV, et al. Prediction of bone density from vitamin D receptor alleles. Nature. 1994;367:284–7.
Grant SF, Reid DM, Blake G, Herd R, Fogelman I, Ralston SH. Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I alpha 1 gene. Nat Genet. 1996;14:203–5.
Yamada Y, Miyauchi A, Goto J, Takagi Y, Okuizumi H, Kanematsu M, et al. Association of a polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to osteoporosis in postmenopausal Japanese women. J Bone Miner Res. 1998;13:1569–76.
Hosoi T, Miyao M, Inoue S, Hoshino S, Shiraki M, Orimo H, et al. Association study of parathyroid hormone gene polymorphism and bone mineral density in Japanese postmenopausal women. Calcif Tissue Int. 1999;64:205–8.
Krishnan V, Bryant HU, MacDougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–9.
Kato M, Patel MS, Levasseur R, Lobov I, Chang BHJ, Glass 2nd DA, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistant embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;2:303–14.
Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–23.
Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–21.
Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–9.
Babij P, Zhao W, Small C, Kharode Y, Yaworsky PJ, Bouxsein ML, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18:960–74.
Koller DL, Rodriguez LA, Christian JC, Slemenda CW, Econs MJ, Hui SL, et al. Linkage of a QTL contributing to normal variation in bone mineral density to chromosome 11q12-13. J Bone Miner Res. 1998;13:1903–8.
Koay MA, Woon PY, Zhang Y, Miles LJ, Duncan EL, Ralston SH, et al. Influence of LRP5 polymorphisms on normal variation in BMD. J Bone Miner Res. 2004;19:1619–27.
Ezura Y, Nakajima T, Urano T, Sudo Y, Kajita M, Yoshida H, et al. Association of a single-nucleotide variation (A1330V) in the low-density lipoprotein receptor-related protein 5 gene (LRP5) with bone mineral density in adult Japanese women. Bone. 2007;40:997–1005.
Urano T, Shiraki M, Usui T, Sasaki N, Ouchi Y, Inoue S. A1330V variant of the low-density lipoprotein receptor-related protein 5 (LRP5) gene decreases Wnt signaling and affects the total body bone mineral density in Japanese women. Endocr J. 2009;56:625–31.
van Meurs JJ, Trikalinos TA, Ralston SH, Balcells S, Brandi ML, Brixen K, et al. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299:1277–90.
Markatseli AE, Hatzi E, Bouba I, Georgiou I, Challa A, Tigas S, et al. Association of the A1330V and V667M polymorphisms of LRP5 with bone mineral density in Greek peri-and postmenopausal women. Maturitas. 2011;70:188–93.
Hey PJ, Twells RC, Phillips MS, Nakagawa Y, Brown SD, Kawaguchi Y, et al. Cloning of a novel member of the low-density lipoprotein receptor family. Gene. 1998;216:103–11.
Mizuguchi T, Furuta I, Watanabe Y, Tsukamoto K, Tomita H, Tsujihata M, et al. LRP5, low-density-lipoprotein-receptor-related protein 5, is a determinant for bone mineral density. J Hum Genet. 2004;49:80–6.
Koh JM, Jung MH, Hong JS, Park HJ, Chang JS, Shin HD, et al. Association between bone mineral density and LDL receptor-related protein 5 gene polymorphisms in young Korean men. J Korean Med Sci. 2004;19:407–12.
Zhang ZL, Qin YJ, He JW, Huang QR, Li M, Hu YQ, et al. Association of polymorphisms in low-density lipoprotein receptor-related protein 5 gene with bone mineral density in postmenopausal Chinese women. Acta Pharmacol Sin. 2005;26:1111–6.
Balemans W, Van Hul W. The genetics of low-density lipoprotein receptor-related protein 5 in bone: a story of extremes. Endocrinology. 2007;148:2622–9.
WHO. Prevention and management of osteoporosis. WHO Tech Rep Ser. 2003;921:1–164.
Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19:179–92.
Hosking L, Lumsden S, Lewis K, Yeo A, McCarthy L, Bansal A, et al. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur J Hum Genet. 2004;12:395–9.
Sen S, Burmeister M. Hardy-Weinberg analysis of a large set of published association studies reveals genotyping error and a deficit of heterozygotes across multiple loci. Hum Genomics. 2008;3:36–52.
Hong YJ, Hwang SM, Kim TS, Song EY, Park KU, Song J, et al. Significance of Lewis Phenotyping Using Saliva and Gastric Tissue: Comparison with the Lewis Phenotype Inferred from Lewis and Secretor Genotypes. BioMed Res Int. 2014;2014:573652.
Lau HH, Ng MY, Ho AY, Luk KD, Kung AW. Genetic and environmental determinants of bone mineral density in Chinese women. Bone. 2005;36:700–9.
Kiel DP, Ferrari SL, Cupples LA, Karasik D, Manen D, Imamovic A, et al. Genetic variation at the low-density lipoprotein receptor-related protein 5 (LRP5) locus modulates Wnt signaling and the relationship of physical activity with bone mineral density in men. Bone. 2007;40:587–96.
Kruk M, Ralston SH, Albagha OM. LRP5 Polymorphisms and response to risedronate treatment in osteoporotic men. Calcif Tissue Int. 2009;84:171–9.
Burgers TA, Williams BO. Regulation of Wnt/β-catenin signaling within and from osteocytes. Bone. 2013;54:244–9.
Kang KS, Robling AG. New insights into Wnt–Lrp5/6–β-catenin signaling in mechanotransduction. Front Endocrinol. 2014;5:1–5.
Robinson JA, Chatterjee-Kishore M, Yaworsky PJ, Cullen DM, Zhao W, Li C, Kharode Y, et al. Wnt/beta-catenin signaling is a normal physiological response to mechanical loading in bone. J Biol Chem. 2006;281:31720–8.
Sawakami K, Robling AG, Ai M, Pitner ND, Liu D, Warden SJ, et al. The Wnt co-receptor LRP5 is essential for skeletal mechanotransduction but not for the anabolic bone response to parathyroid hormone treatment. J Biol Chem. 2006;281:23698–711.
Lee K, Jessop H, Suswillo R, Zaman G, Lanyon L. Endocrinology: bone adaptation requires oestrogen receptor-alpha. Nature. 2003;424:389.
Kouzmenko AP, Takeyama K, Ito S, Furutani T, Sawatsubashi S, Maki A, et al. Wnt/beta-catenin and estrogen signaling converge in vivo. J Biol Chem. 2004;279:40255–8.
Suuriniemi M, Mahonen A, Kovanen V, Alen M, Lyytikainen A, Wang Q, et al. Association between exercise and pubertal BMD is modulated by estrogen receptor alpha genotype. J Bone Miner Res. 2004;19:1758–65.
Dhamrait SS, James L, Brull DJ, Myerson S, Hawe E, Pennell DJ, et al. Cortical bone resorption during exercise is interleukin-6 genotype-dependent. Eur J Appl Physiol. 2003;89:21–5.
Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5.
Acknowledgements
We did not receive funding for this study. However, we acknowledge the author RT for his financial support of this study. We thank the Department of Tropical Nutrition and Food Science and the Faculty of Tropical Medicine, Mahidol University, for material support. We would like to thank Dr. Wanida Chuenta for help in haplotype analysis and critical reading of the manuscript. We would also like to thank Dr. Sivaporn Wannaiampikul for her critical reading of the manuscript.
Funding
This study has no funding.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Authors’ contributions
AK conceived the study, performed the statistical analysis and wrote the manuscript. HH carried out the experiments. BP assisted with the manuscript draft and statistical analysis. SP & RT provided samples for genotyping. RT participated in the study design and coordination. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study protocol was approved by the Ethics committees of the Faculty of Tropical Medicine (MUTM 2012-039-01) and Faculty of Medicine (Ramathibodi Hospital, MURA 2008/996), Mahidol University, Bangkok, Thailand. Written inform consent were obtained from all subjects.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Haplotype blocks distribution in the LRP5 gene of CHB and JPT populations of HapMap generated by Haploview 4.2 program. Each black triangle depicts haplotype blocks. LD is reported as D′. Bright red represents D' = 1 and LOD ≥ 2, blue represents D' = 1 and LOD < 2, pink represents D' < 1, and LOD ≥ 2, and white represents D' < 1 and LOD < 2. The r2 values are shown in blocks. CHB, Han Chinese in Beijing, China; JPT, Japanese in Tokyo, Japan.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kitjaroentham, A., Hananantachai, H., Phonrat, B. et al. Low density lipoprotein receptor-related protein 5 gene polymorphisms and osteoporosis in Thai menopausal women. J Negat Results BioMed 15, 16 (2016). https://doi.org/10.1186/s12952-016-0059-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12952-016-0059-7