Abstract
Background
Non-human primates (NHPs) are important models of medical research on obesity and cardiovascular diseases. As two of the most commonly used NHPs, cynomolgus macaque (CM) and African green monkey (AGM) own different capacities in lipid metabolism of which the mechanism is unknown. This study investigated the expression profiles of lipid metabolism-related microRNAs (miRNAs) in CM and AGM and their possible roles in controlling lipid metabolism-related gene expression.
Methods
By small RNA deep sequencing, the plasma miRNA expression patterns of CM and AGM were compared. The lipid metabolism-related miRNAs were validated through quantitative reverse-transcription (RT) polymerase chain reaction (PCR). Related-target genes were predicted by TargetScan and validated in Vero cells.
Results
Compared to CM, 85 miRNAs were upregulated with over 1.5-fold change in AGM of which 12 miRNAs were related to lipid metabolism. miR-122, miR-9, miR-185, miR-182 exhibited the greatest fold changes(fold changes are 51.2, 3.8, 3.7, 3.3 respectively; all P < 0.01). And 77 miRNAs were downregulated with over 1.5-fold change in AGM of which 3, miR-370, miR-26, miR-128 (fold changes are 9.3, 1.8, 1.7 respectively; all P < 0.05) were related to lipid metabolism. The lipid metabolism-related gene targets were predicted by TargetScan and confirmed in the Vero cells.
Conclusion
We report for the first time a circulating lipid metabolism-related miRNA profile for CM and AGM, which may add to knowledge of differences between these two non-human primate species and miRNAs’ roles in lipid metabolism.
Similar content being viewed by others
Background
As short (≈22-26 nt) endogeous RNA molecules that regulate gene expression at the post-transcriptional levels by binding to the 3′ untranslated regions (UTRs) of their target mRNAs [1], miRNAs have been described as vital regulators of several biological processes including developmental and metabolic functions. In many pathological conditions such as obesity, type 2 diabetes, cardiovascular diseases, hypertension and cancers, the concentration of miRNA in circulation has been found to be altered [2], suggesting regulatory role in maintaining metabolism homeostasis in humans [3,4,5].
Non-human primates (NHPs) have been vital for medical research due to their close evolutionary relationship, similar behavioral and physiological characteristics to humans. Two of the most commonly used NHPs are Cynomolgus macaque (Macaca fascicularis) and African green monkey (Chlorocebus Sabaeus), which can both be used in studies of neuroscience, infectious diseases and drug safety testing [6,7,8,9,10]. Differences also exist between these two species. For example, AGM has been known to be resistant to simian immunodeficiency virus which can be used as a model organism for HIV research [11, 12]. Also, AGM develops spontaneous hypertension with pathophysiological changes that mimic those of patients with essential hypertension [13, 14]. Research has also shown that genetic factors influence the atherogenic response of lipoproteins to dietary fat and cholesterol in nonhuman primates which reflects different capacities in lipid metabolism between AGM and CM [15].
This study aims to identify the differently expressed miRNAs and their potential targets related to lipid metabolism between AGM and CM. Understanding the roles of miRNAs in lipoprotein and lipid metabolism may eventually lead to development of new disease biomarkers or therapeutic strategies.
Methods
Animal care and housing
All protocols were in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals. Animals were housed in troop enclosures with an outdoor facility amd fed nonhuman primate chow per day and a combination of fresh bananas, apples, carrots 3 days per week. As shown in Additional file 1: Table S1, 16 animals were used of which 6 were used for miRNA profiling with the others used miRNA validation.
Plasma collection and lipid level analysis
All animals were fasted overnight and anesthetized with intramuscular injection (10 to 15 mg/kg). Blood samples (5 ml) of CM and AGM were collected from the femoral vein and drawn into EDTA-containing tubes held on ice. Part of the blood samples were separated into plasma and cellular fractions by centrifugation at 1000 g for 10 min. And the plasma lipid levels were analyzed by an automated chemistry analyzer (Olympus AU5800, Beckman Coulter).
RNA isolation and small RNA deep sequencing
Total plasma RNA was harvested from the rest blood samples with the TRI Reagent BD (Sigma Aldrich) and the RNeasy mini kit (Qiagen) according to the manufacturers’ instructions. In detail, 250 μL EDTA-containing plasma was transferred to an Eppendorf tube and mixed thoroughly with TRI reagent, incubated for 5 min at room temperature, and subsequently mixed with 140 μL chloroform. The aqueous phase containing the RNA was carefully removed, and RNA was precipitated by addition of 100% ethanol. The mixture was applied to an RNeasy Mini spin column and washed several times, and RNA was eluted by addition of 25 μL RNase-free water. RNA was stored at − 80 °C until further processing. Total RNA (4 to 8 μg) was size fractionated and the small RNAs (< 300 nucleotides) were isolated and 3′ extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent-dye staining. Hybridization was performed overnight on a microfluidic chip provided by Shanghai KangChen biotechnology company. The hybridization buffer was 6 × SSPE (0.9 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25% formamide at 34 °C. After RNA hybridization, tag-conjugating Cy3 and Cy5 dyes were circulated through the microfludic chip for dye staining. Scanning was performed with the Axon GenePix 4000B microarray scanner. GenePix pro version 6.0 was used to read image raw intensity. The intensity of the the green signal was calculated after background subtraction. The median normalization method was used to acquire normalized data. The threshold value for significance used to define upregulation or downregulation of miRNAs was a fold change > 1.5, with a value of P < 0.05 calculated by the t test.
Inhibition of endogenous miRNAs
Locked nucleic acid (LNA)-modified anti-miRs (Exiqon) were used for the inhibition of endogenous miRNAs in African green monkey-derived Vero cells [16]. Vero cells were maintained in DMEM supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, nonessential amino acids, and 10% fetal bovine serum (FBS) (Invitrogen). Cells were subcultured at 80% confluency for propagation. Vero cells were subcultured onto 96 well plates and transfected at > 90% confluency with LNA-modified anti-miRs which were transfected at a final concentration of 10 nM by Lipofectamine 3000 (Invitrogen).
Stem-loop miRNA quantitative RT-PCR
First, each mature miRNA was extended and reverse transcribed by a sequence specific stem-loop primer using MMLV reverse transcriptase (Takara). Then the reverse transcribed miRNA was quantified by a fluorescently labeled hybridization probe using the strand replacement reaction. and quantitative PCR was performed by an ABI PRISM7500 system (Applied Biosystems). The PCR conditions were as follows: 95 °C 3 min, 40 cycles of 95 °C1 2 s, 62 °C4 0 s. miRNAs and related target genes implicated in the regulation of lipid metabolism were obtained through the literature [2]. The RT primers and the primer sets specific for each miRNA amplification designed according the mature miRNA sequence obtained from the miRbase database and shown in Additional file 2: Table S2. Selected miRNAs were further quantified with TaqMan quantitative RT-PCR. mRNA expression levels of the lipid metabolism-related genes predicted to be targeted by miRNAs inhibited by anti-miR transfection were measured by qPCR using SYBR green chemistry [17]. Primer sequences for the target genes are listed in Additional file 3: Table S3.
Western blot analysis
Anti-miR-treated vero cells were lysed in protein sample buffer containing 50 mM Tris-HCl, 150 mM NaCl. 5 mM EDTA, 0.2 mM sodium orthovanadate, 1% Triton X-100, 1% sodium deoxycholate, and 1% sodium dodecyl sulfate; the buffer was also supplemented with aprotinin (2 μg/ml), pepstatin A (0.7 μg/ml), leupeptin (0.5 μg/ml), and PMSF (1 mM). For each sample, 30 μg total protein was electrophoresed through a 10% sodium dodecyl sulfate-polyacrylamide gel and transfered onto a nitrocellulose membrane.The membrane was blocked by pre-incubation with 5% skim milk, incubated with the specific rabbit anti-CPT1A (ThermoFisher) and mouse anti-GAPDH (ThermoFisher) overnight at 4 °C, which were detected with horseradish peroxidase-conjugated secondary antibody (Pierce) and visualized with a chemiluminescent substrate (Pierce).
Statistical analysis
Quantile normalized values for miRNA in each sample were plotted as fold change in comparison with the mean expression of each miRNA within the control group. Significance was assessed by the t test. Statistical analyses were performed using GraphPad Prism version 5.01.
Results
Expression profiles of plasma miRNAs in CM and AGM
As shown in Additional file 1: Table S1, 16 animals were used of which 6 were used for miRNA profiling with the others used miRNA validation. There were no significant differences for weights and plasma lipid levels between the CM and AGM group. We then profiled the plasma miRNA expression in 3 CM and 3 AGM. As shown in Fig. 1, 162 miRNAs exhibited different expression with over 1.5-fold change between CM and AGM. GO analysis for biological process (BP) demonstrated that more genes reside in the categories of metabolic processes (Fig. 2). Among the 85 upregulated miRNAs in AGM, 12 were related to lipid metabolism, while 3 out of 77 downregulated miRNAs were related to lipid metabolism. miR-122 and miR-370 own the greatest upregulated and donwregulated fold changes (P < 0.05), respectively. miRNAs with over 2-fold change related to lipid metabolism are shown in Table 1.
Validation of miRNA expression in CM and AGM
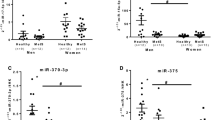
To validate the profiling data, miRNAs in Table 1 were further analyzed via quantitative RT-PCR between plasma samples from 5 CM and 5 AGM. Results showed a 48.0-fold increase for hsa-miR-122-5p (P < 0.01), a 3.0~4.0-fold increase for hsa-miR-9-5p, hsa-miR-185-5p, hsa-miR-182-5p (all P < 0.05), a 2.0~3.0-fold increase for hsa-miR-181-5p, hsa-miR-144-3p, hsa-miR-10b-5p, hsa-miR-148a-3p, hsa-miR-130b-5p (all P < 0.05), and an ≈9.5 decrease for hsa-miR-370-3p (P < 0.01) (Fig. 3).
Independent validation of differential expression of miRNA in PBMCs of CM and AGM. Quantitative reverse-transcription polymerase chain reaction for miRNAs in an independent validation set of 5 CMs and 5 AGMs. The relative expression of each miRNA in AGM compared with CM was normalized to U6 expression. The P values were calculated by 2-sided Student t test. * stands for P < 0.05 while ** stands for P < 0.01
Effects of selected miRNAs on the lipid metabolism-related target genes
The potential lipid metabolism-related genes targeted by the above miRNAs were selected and analyzed (Table 2). The potential miRNA-target pairs were examined by inhibiting these endogenous miRNAs in Vero cells individually using LNA-modified anti-miRs. Anti-miR treatment may cause the increase on the abundance of mRNA encoded by lipid metabolism-related genes if the predicted target mRNA is indeed suppressed by the endogenous miRNA via mRNA degradation. As shown in Fig. 4, the validated target of hsa-miR-122-5p, hsa-miR-185-5p, hsa-miR-181a-5p, hsa-miR-10b-5p, hsa-miR-370-3p was AGPAT1, LDLR, IDH1, ABCG1, CPT1A, respectively. ABCA1 was the target of both hsa-miR-144-3p and hsa-miR-130b-5p. The validated targets of hsa-miR-148a-3p included SIK1 and CPT1A. The ABCA1 target mRNA was downregulated after anti-miR treatment, suggesting unconventional effects of hsa-miR-10b-5p. Also, we tested whether the effects of anti-miR treatment on the target genes were similar both at mRNA level and at the protein level. As shown in Fig. 4, anti-miR-370 treatment caused upregulation of CPT1A at the protein level. Further, we tested the effects of every anti-miR treatment on unpredicted targets. As shown in Fig. 4 and Fig. 5, anti-miR-370 treatment caused upregulation of ACAT1 while anti-miR-181a treatment caused upregulation of CPT1A, indicating the indirect targets of miRNA may exist.
Effect of anti-miR transfections in vero cells on the abundance of mRNAs encoded by lipid metabolism-related genes. Vero cells were transfected with the indicated LNA-modified anti-miRs or scambled anti-miRs (10 nmol/L) for 48 h. mRNA abundance for predicted targets genes was measured by real-time reverse transcription polymerase chain reaction (PCR) and expressed as % change relative to cells treated with scrambled anti-miR on the same quantitative PCR plate.n = 3–6.*P < 0.05 vs scrambled anti-miR
Discussion
As regulators of several physiological processes including developmental and metabolic functions, miRNAs play central role in regulating lipoprotein metabolism and cholesterol homeostasis [18,19,20,21]. Many pathological conditions such as familial hypercholesterolemia, cardiovascular diseases, obesity, type 2 diabetes occur when the circulation miRNA concentration alters. miRNAs are stable in plasma and may be suitable as biomarkers in cancers and other diseases [22, 23].
NHPs shares similar characteristics in many aspects to humans when they are used in models of neuroscience, infectious diseases and drug safety testing. And differences also exist between these NHP species like CM and AGM. AGM are resistant to simian immunodeficiency virus and can be used as a model spontaneous hypertension. Besides, genetic differences between CM and AGM influence the diet responsiveness [15]. Although draft genome sequences and transcriptome reconstruction and annotation for CM and AGM have been published [24,25,26], comparison of miRNA profiling data between these two species lacks.
In this study, we compared the circulating miRNA profile between CM and AGM. Several miRNAs related to lipid metabolism exhibited expression with different levels. miR-122 was significantly upregulated with the greatest fold change in AGM. As the most highly expressed miRNA in the liver, miR-122 has been found to regulate liver lipid metabolism [27,28,29]. Knockdown of miR-122 in mice decreased expression of genes important for cholesterol biosynthesis, whereas adenoviral overexpression of miR-122 increased cholesterol biosynthesis [30]. miR-122 was found to be downregulated in patients with nonalcoholic fatty liver disease (NAFLD) and knockdown of miR-122 in HepG2 cells recapitulated the lipogenic gene expression profile observes in patients with NAFLD [31]. Thus, we conclude that upregulation of miR-122 renders AGM more capable, compared to CM, in regulating lipoprotein metabolism and cholesterol homeostasis. The validated target of miR-122 in vero cells was AGPAT1 which may cause effect on TG biosynthesis and lipid storage. miR-370 has been shown be significantly downregulated in AGM and target CPT1A which facilitates the transfer of long-chain fatty acids across the mitochondrial membrane for β-oxidation. Previous research also demonstrate that miR-370 functions in cholesterol biosynthesis by increasing the level of miR-122 [32]. Whether miR-370 exerts an effect on upregulation of miR-122 in AGM still needs investigation.
Other upregulated miRNAs in AGM include miR-185, miR-181a, miR-10b, miR-144, miR-130b,miR-148a with the validated targets of LDLR, IDH1, ABCG1, ABCA1, SIK1 and CPT1A which function in LDL clearance, regulation of lipid biosynthesis and β-oxidation, HDL biogenesis and cholesterol efflux.
Conclusion
The present study demonstrates that AGM possess a significantly different miRNA expression profile from CM. The differently expressed miRNAs and their potential targets related to lipid metabolism were investigated, which provide clues that may eventually lead to development of new disease biomarkers or therapeutic strategies.
Abbreviations
- ABCA1:
-
ATP-binding cassette subfamily A member 1
- ABCG1:
-
ATP-binding cassette subfamily G member 1
- ACAT1:
-
acetyl-CoA acetyltransferase 1
- AGPAT1:
-
1-acylglycerol-3-phosphate O-acyltransferase 1
- CPT1A:
-
carnitine O-palmitoyltransferase 1
- FASN:
-
fatty acid synthase
- FBXW7:
-
F-box and WD-40 domain protein 7
- IDH1:
-
isocitrate dehydrogenase 1
- LDLR:
-
low-density lipoprotein receptor
- SIK1:
-
salt-inducible kinase 1
References
Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123(6):1133–46. https://doi.org/10.1016/j.cell.2005.11.023 PMID:16337999.
Desgagné V, Bouchard L, Guérin R. microRNAs in lipoprotein and lipid metabolism: from biological function to clinical application. Clin Chem Lab Med. 2017;55(5):667–86. https://doi.org/10.1515/cclm-2016-0575 PMID:27987357.
Fernández-Hernando C, Ramírez CM, Goedeke L, Suárez Y. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol. 2013;33(2):178–85. https://doi.org/10.1161/ATVBAHA.112.300144 PMID: 23325474.
Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22(2):86–92. https://doi.org/10.1097/MOL.0b013e3283428d9d PMID: 21178770.
Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21(12):699–706. https://doi.org/10.1016/j.tem.2010.08.008 PMID: 20880716.
Carlsson HE, Schapiro SJ, Farah I, Hau J. Use of primates in research: a global overview. Am J Primatol. 2004;63(4):225–37. https://doi.org/10.1002/ajp.20054 PMID: 15300710.
Osada N, Hirata M, Tanuma R, Suzuki Y, Sugano S, Terao K, Kusuda J, Kameoka Y, Hashimoto K, Takahashi I. Collection of Macaca fascicularis cDNAs derived from bone marrow, kidney, liver, pancreas, spleen, and thymus. BMC Res Notes. 2009;2:199. https://doi.org/10.1186/1756-0500-2-199 PMID:19785770.
Ebeling M, Kung E, See A, Broger C, Steiner G, Berrera M, Heckel T, Inigue L, Alber T, Schmucki R, Biller H, Singer T, Certa U. Genome-based analysis of the nonhuman primate Macaca fascicularis as a model for drug safety assessment. Genome Res. 2011;21(10):1746–56. https://doi.org/10.1101/gr.123117.111 PMID:21862625.
Jones SM, Feldmann H, Stroher U, Geisbert JB, Fernando L, Grolla A, Klenk HD, Sullivan NJ, Volchkov VE, Fritz EA, Daddari KM, Hensley LE, Jahrling PB, Geisbert EW. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med. 2005;11(7):786–90. https://doi.org/10.1038/nm1258 PMID:15937495.
Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163(6):2347–70. https://doi.org/10.1016/S0002-9440(10)63591-2 PMID: 14633608.
Fukasawa M, Miuta T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333(6172):457–61. https://doi.org/10.1038/333457a0 PMID: 3374586.
Apetrei C, Robertson DL, Marx PA. The history of SIVS and AIDS:epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front Biosci. 2004;9:225–54 PMID: 14766362.
Renna NF, de Las Heras N, Miatello RM. Pathophysiology of vascular remodeling in hypertension. Int J Hypertens. 2013;2013:808353. https://doi.org/10.1155/2013/808353 PMID: 23970958.
Rhoads MK, Goleva SB, Beierwaltes WH, Osborn JL. Renal vascular and glomerular pathologies associated spontaneous hypertension in the nonhuman primate Chlorocebus aethiops sabaeus. Am J Physiol Regul Integr Comp Physiol. 2017;313(3):R211–8. https://doi.org/10.1152/ajpregu.00026.2017 PMID: 28659284.
Rudel LL. Genetic factors influence the atherogenic response of lipoproteins to dietary fat and cholesterol in nonhuman primates. J Am Coll Nutr. 1997;16(4):306–12 PMID: 9263179.
Mladinov D, Liu Y, Mattson DL, Liang M. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics:miR-192 targets Na+/K+-ATPase β1. Nucleic Acids Res. 2013;41:1273–83. https://doi.org/10.1093/nar/gks1228 PMID: 23221637.
Kriegel AJ, Fang Y, Liu Y, Tian Z, MIadinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta1: a novel role of miR-382. Nucleic Acids Res. 2010;38(22):8338–47. https://doi.org/10.1093/nar/gkq718 PMID: 20716515.
Rayner KJ, Moore KJ. MicroRNA control of high-density lipoprotein metabolism and function. Circ Res. 2014;114(1):183–92. https://doi.org/10.1161/CIRCRESAHA.114.300645 PMID: 24385511.
Rotllan N, Price N, Patl P, Goedeke L, Fernandez-Hernando C. microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis. 2016;246:352–60. https://doi.org/10.1016/j.atherosclerosis.2016.01.025 PMID: 26828754.
Goedeke L, Wagschal A, Fernández-Hernando C, Naar AM. miRNA regulation of LDL-cholesterol metabolism. Biochim Biophys Acta 2016;1861(12PtB)::2047:–2052. https://doi.org/10.1016/j.bbalip.2016.03.007 PMID: 26968099.
DiMarco DM, Fernandez ML. The regulation of reverse cholesterol transport and cellular cholesterol homeostasis by microRNAs. Biology (Basel). 2015;4(3):494–511. https://doi.org/10.3390/biology4030494 PMID:26226008.
Chin LJ, Slach FJ. A truth serum for cancer:microRNAs have major potential as cancer biomarkers. Cell Res. 2008;18(10):983–4. https://doi.org/10.1038/cr.2008.290 PMID:18833286.
Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet. 2010;3(5):484–8. https://doi.org/10.1161/CIRCGENETICS.110.958363 PMID:20959591.
Chlorocebus aethiops Sabeus (vervet) Sequence Assembly Release. [https://www.ncbi.nlm.nih.gov/assembly/GCF_000409795.2]. Accessed 25 Mar 2014
Macaca fascicularis (cynomolgus macaque) Sequence Assembly Release. [http://www.ncbi.nlm.nih.gov/assembly/GCF_000364345.1]. Accessed 12 June 2013
Lee A, Khiabanian H, Kugelman J, Elliott O, Nagle E, Yu GY, Warren T, Palacio G, Rabadan R. Transcriptome reconstruction and annotation of cynomolgus and African green monkey. BMC Genomics. 2014;15:846. https://doi.org/10.1186/1471-2164-15-846 PMID:25277458.
Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37(1):70–7. https://doi.org/10.1093/nar/gkn904 PMID:19015151.
Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7):896–9. https://doi.org/10.1038/nature06783 PMID:18368051.
Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monica BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. https://doi.org/10.1016/j.cmet.2006.01.005 PMID:16459310.
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschi T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. https://doi.org/10.1038/nature04303.
Cheuong O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanval AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48(6):1810–20. https://doi.org/10.1002/hep.22569 PMID:19030170.
Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression the expression of microRNA-122 and Cpt1ɑ and affects lipid metabolism. J Lipid Res. 2010;51(6):1513–23. https://doi.org/10.1194/jlr.M004812 PMID:20124555.
Acknowledgments
We are grateful to Professor Jun-feng Li (Institute of Microbiology and Epidemiology, Beijing, China) for providing the Vero cell line.
Funding
This study was funded by the National Projects of Infectious Disease under Grant No.2017ZX10304402003.
Availability of data and materials
Please contact author for data requests.
Author information
Authors and Affiliations
Contributions
XZ conceived and designed the experiments. ZY and JW performed experiments. HY helped with the experiments. XZ and YF analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Sex, age, body weight, and plasma lipid information about CMs and AGMs used. 1~3 and 4~6 are CMs and AGMs used for miRNA profiling, respectively. 7~11 and 12~16 are CMs and AGMs used for miRNA validation, respectively. (DOC 44 kb)
Additional file 2:
Table S2. Sequences of the primers used in the SYBR-green-based quantitative RT-PCR validation. (DOC 48 kb)
Additional file 3:
Table S3. Primer sequences for miRNA-targeted genes. (DOC 40 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhou, Xj., Wang, J., Ye, Hh. et al. Signature MicroRNA expression profile is associated with lipid metabolism in African green monkey. Lipids Health Dis 18, 55 (2019). https://doi.org/10.1186/s12944-019-0999-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-019-0999-2