Abstract
Background
The purpose of this study was to evaluate the effect of single-nucleotide polymorphisms (SNPs) of the GCKR and G6PC2 genes on risk for type 2 diabetes and the SNP-SNP and haplotype-based interactions between these genes.
Methods
Subjects of this nested case-control study were selected from a prospective cohort residing in the rural area of Luoyang city in China. Cases (n = 538) were individually matched with controls. Six SNPs in the GCKR and G6PC2 genes were selected and genotyped using an SNPscan™ kit. Stratified Cox proportional hazards regression models were used to generate odds ratios (ORs) and 95% confidence intervals (CI) for different genotype models for the risk of T2DM. Generalized multifactor dimensionality reduction (GMDR) was used to analyze the interactions between two genes with among six SNPs. The linkage disequilibrium (LD) analysis and the haplotype analysis were carried out by SHEsis online.
Results
We found that the C allele of rs780094 was associated with increased risk for T2DM in Han Chinese population. However, the rs492594-C allele in G6PC2 was associated with a decreased risk of T2DM. We also found a significant SNP-SNP interaction between rs2293572 and rs492594, and the CCCCGC and CGCCCA haplotypes significantly increased the risk of T2DM, however, the CCCCCA haplotype had lower susceptibility to T2DM.
Conclusion
The results suggest that the GCKR and G6PC2 genes may contribute to the risk of T2DM independently and/or in an interactive manner in the Han Chinese population.
Similar content being viewed by others
Background
Type 2 diabetes mellitus (T2DM) is a common but complex multifactorial chronic disease, accounting for > 95% of diabetes worldwide. [1] It is estimated that every year over 3.8 million people are dying of T2DM and its complications in worldwide. [2] The prevalence of T2DM presents a trend of sustainable growth over the past few decades. The International Diabetes Federation reported that the number of people with diabetes aged 20–79 years was expected to reach 642 million by 2040. [3] These observations show that T2DM is a major worldwide health problem.
With the rapidly rising prevalence of T2DM, a more systematic understanding of the natural history of the disease and its potential risk factors is urgently needed. Both genetic and environmental factors contribute to the occurrence and pathophysiology of T2DM. However, knowledge of the contribution of genetic factors to T2DM risk is still limited. The level of fasting plasma glucose (FPG) is the core factor that correlated with the risk of T2DM and various cardiovascular diseases. [4] There is strong evidence suggesting that hyperglycemia plays a key role in a concentration-dependent manner for both micro- and macro-vascular complications in diabetes. [5] Glucokinase regulatory protein (GCKR) and glucose-6-phosphatase catalytic subunit 2 (G6PC2) are both recognized glucose metabolism-related genes. Studies show that single-nucleotide polymorphisms (SNPs) of the GCKR and G6PC2 genes are associated with FPG and T2DM incidence, although the conclusions are inconsistent in different regions. [6,7,8,9] Moreover, the SNPs associated with T2DM explain only a part of the heritability. Mechanisms such as gene-gene or gene-environment interactions may account for this missing heritability. [10, 11]. There have been reports of the relationship between the GCKR and G6PC2 genes and FPG and T2DM, but the association between the interaction of these two genes and T2DM risk has not been reported.
Therefore, the aim of the present study was to detect associations between SNPs in GCKR and G6PC2 and the risk for T2DM in a rural adult Chinese population, as well as to determine if gene-gene interactions modify the risk of T2DM incidence.
Methods
Study design and population
Subjects of this nested case-control study were selected from participants of a prospective cohort residing in the rural area of Luoyang city in China. The first phase of this cohort study was conducted from July to August of 2007 and July to August of 2008 on 20,194 subjects, and follow-up examinations were conducted from July to August 2013 to July and October 2014 to identify newly developed diseases. Details of this cohort study have been published elsewhere. [12, 13] T2DM was defined as FPG ≥7.0 mmol/L or the use of insulin or oral hypoglycemic agents, and/or a self-reported history of T2DM. [14] Subjects were > 25 and < 75 years old and of Northern Chinese ancestry. We excluded participants who had body mass index (BMI) < 18.5 Kg/m2; were pregnant, handicapped, or mentally illness; and had cancer or were unable or unwilling to participate. During the follow-up, we recruited 550 subjects (196 men and 354 female) who developed T2DM. Controls were matched in a 1:1 ratio to cases by age (within 2 years), sex, and village. Because the genotype of SNPs of part of objects have not been detected, 538 incident cases and 538 controls were selected in the end.
Data on demographic and anthropometric characteristics were collected by a standard interviewer-administered questionnaire. Anthropometric data included body weight, body height, BMI, WC, waist-height ratio and blood pressure. Blood pressure were measured by using an electronic sphygmomanometer.
Biochemical measurements
All blood samples were collected from subjects in the morning after an overnight fast for measuring FPG, total cholesterol (TC), triglycerides (TG), and high-density lipoprotein cholesterol (HDL-C), as well as genotyping. FPG was measured using an oxidase enzymatic method, whereas TC, TG, and HDL-C were measured using an automatic biochemical analyzer (Hitachi, Tokyo, Japan). The concentration of low-density lipoprotein cholesterol (LDL-C) was calculated using the Friedewald formula. [15]
Selection and genotyping of SNPs
The tag SNPs rs780094, rs2293572, rs12603206 and rs492594, rs16856187, rs13387347 for GCKR and G6PC2 were selected for the Chinese population from the International HapMap project by use of the exact criteria of a minor allele frequency (MAF) > 0.01 and r2 ≥ 0.8. Genomic DNA was extracted from peripheral blood by use of a blood genome DNA purification kit (Yaneng BIO, Shenzhen, China). Genotype polymorphisms were identified by SNPscan™ kit (Genesky Biotechologies Inc., Shanghai, China). This kit was developed using patented SNP genotyping technology by Genesky Biotech Co., Ltd., involving the technology of double ligation and multiplex fluorescence polymerase chain reaction. The minimum call rate was 97.8%. To verify reproducibility, genotyping was reanalyzed based on 50 duplicates randomly selected from 1076 specimens and the concordance rate was more than 99%. All subjects underwent genotyping at baseline.
Statistical analysis
Baseline data are summarized as the median with interquartile range for quantitative variables for data with non-normal distribution and number (percentage) for categorical variables. The Mann-Whitney Wilcoxon test was used to assess the significance of differences in quantitative variables and the chi-square test for categorical variables. The Hardy-Weinberg Equilibrium (HWE) was calculated by Pearson’s chi-square statistic test. Stratified Cox proportional hazards regression models were used to generate odds ratios (ORs) and 95% confidence intervals (CI) of different genotype models for the risk of T2DM adjusting for the baseline data including BMI, BMI-change, smoking and drinking status, and family history of diabetes. The interactions between the two genes for six SNPs were analyzed by statistical analysis and were performed using generalized multifactor dimensionality reduction (GMDR). SNP-SNP interaction analysis used unconditional logistic regression. Data were analyzed using SPSS v21.0 for Windows (SPSS Inc., Chicago, IL). The linkage disequilibrium (LD) analysis and the haplotype analysis were carried out by SHEsis online (http://analysis.bio-x.cn/myAnalysis.php). [16] Two-sided p < 0.05 was considered statistically significant.
Results
Characteristics of study participants
Baseline characteristics of participants by cases and controls are shown in Table 1. There was no significant difference between the two groups in smoking, drinking, physical activity and the level of LDL-c. However, compared with controls, the values of BMI, waist-height ratio, FPG, family history of diabetes and the level of TC, TG, were significant higher and the level of HDL-c were lower among cases at the beginning of the study.
All SNPs for GCKR and G6PC2 were in Hardy-Weinberg equilibrium (P > 0.05). Genotypic and allelic distributions of SNPs for GCKR and G6PC2 at baseline are given in Additional file 1: Table S1. There was no difference in the distribution of SNPs of them (P > 0.05).
Association of genetic variants of GCKR and G6PC2 with the risk for T2DM
Table 2 reports the association between six SNPs and the risk for T2DM, along with their adjusted ORs. As subjects with rs2293572-GG genotype was uncommon, we combined the GC and GG genotypes in the analysis. Stratified Cox proportional hazards regression analysis under three different genetic models found that the C allele of rs780094 was significantly associated with an increased risk of T2DM after adjustment for BMI, BMI-change, smoking, drinking, and family history of diabetes (adjusted OR = 1.779, 95% CI 1.028–3.078, p = 0.040). In addition, the multivariable-adjusted ORs for each T-allele of rs780094 was 1.334 (95% CI 1.014–1.755). For SNPs of G6PC2, the GC genotype and dominant model (CC + GC vs. GG) of rs492594 were significantly associated with a decreased risk for T2DM (adjusted OR = 0.567, 95% CI = 0.362–0.888, p = 0.013; 0.601, 95% CI = 0.394–0.916, p = 0.018).
However, the other SNPs (GCKR rs1260326, rs2293572 and G6PC2 rs13387347, rs16856187) were not found to be associated with T2DM (p > 0.05) in this population.
Haplotype analyses
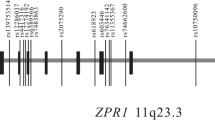
The SHEsis online program was used to analyze the degree of linkage disequilibrium of the six SNPs and haplotypes in this study. The results showed that there was linkage equilibrium between the GCKR and G6PC2 gene, as shown in Fig. 1. For SNPs of the GCKR gene, 3 haplotypes each with a frequency greater than 3% were detected due to linkage disequilibrium, and 3 haplotypes for 3 SNPs of the G6PC2 gene were also detected. Haplotype analysis showed that there were no significant differences in frequency distribution between the 2 groups for both the GCKR and G6PC2 genes (P > 0.05) as shown in Additional file 1: Table S2.
SNP-SNP and haplotype-based interaction
The generalized multifactor dimensionality reduction (GMDR) v0.9 was applied in this research to detect the interaction of the 6 selected SNPs in GCKR and G6PC2. Table 3 provided the best model testing by GMDR. In all of the models the combination of GCKR rs2293572 and G6PC2 rs492594 formed the best model with a statistically significant p value of 0.011, a maximum testing balanced accuracy of 55.0% and the biggest CV consistency (10/10) after adjusting the covariables including age, sex, smoking, drinking, BMI, BMI-change and family history of diabetes.
To obtain the ORs and 95% CIs for the interaction effect of rs2293572 and rs492594 on T2DM risk under different genetic models, we conducted SNP-SNP interaction by using unconditional logistic regression adjusted variables including covariables (age, sex, BMI, BMI-change, smoking, drinking, family history of diabetes), each with two SNPs and their interaction term. As shown in Table 4, the interaction was significant under additive-additive conditions (OR = 1.695, 95% CI 1.125–2.552; p = 0.012). The parameter of the partial interaction term could not be estimated in the logistic regression model because the recessive model of rs2293572 was not established. Moreover, no statistically significant interaction was found for other SNP-SNP pairs (data no shown).
In order to further explore the effect of gene-gene interaction, we constructed haplotypes analysis use SHEsis online. The haplotypes of these 2 genes from left to right were the alleles of rs780094, rs2293572, rs1260326, rs492594, rs16856187 and rs13387347. Ten haplotypes, each with a frequency greater than 3% were detected. Haplotype comparison analysis indicated that CCCCGC and CGCCCA haplotypes significantly increased the risk of T2DM (OR = 1.366, 95% CI 1.034–1.806, p = 0.028 and 1.817, 95% CI 1.261–2.618, p = 0.001, respectively). However, the CCCCCA haplotype correlates with lower susceptibility to T2DM (p = 0.000) (Additional file 1: Table S3 and Fig. 2).
Discussion
The human GCKR and G6PC2 genes are both located on chromosome 2. Glucokinase (GCK) plays a central role in the sensing of glucose in pancreatic beta-cells and parenchymal cells of liver. [17] GCKR can competitively inhibit GCK, which plays a major role in the regulation of insulin secretion and glycogen metabolism and is considered as a potential susceptibility gene for T2DM. [18] G6PC2 is another important glucose related metabolism gene, principally expressed in the beta cells of pancreatic islets. [19, 20] GCKR and G6PC2 encode different enzymes that may jointly regulate glucose homeostasis, effectively establishing the glucose set point. In the current study, we selected the loci of rs780094, rs2293572, rs1260326, rs492594, rs16856187 and rs13387347 from the GCKR and G6PC2 genes and evaluated the association between these loci and T2DM as well as the interaction between the GCKR and G6PC2 genes, which have never been reported either for the Chinese or the global population.
The results show that BMI, waist-height ratio and the levels of FPG, TC, TG and HDL-C of the T2DM group were significantly higher than those of non-T2DM at baseline group. These findings are similar to a study on Southern Han Chinese ancestry [21] In addition, the present study found that the T2DM risk was significantly higher in carriers with the C allele of rs780094 in GCKR. The GC genotype and dominant model of rs492594 in G6PC2 were significantly associated with a decreased risk of T2DM. The conclusions were in line with a large scale meta-analyses by Wang et al. which indicated that GCKR rs780094 variants contributed to high cross-ethnicity risk for development of T2DM, with OR values (95% CI) of 1.08 (1.05–1.12) [6]. On the other hand, a study of Southern Han Chinese demonstrated that rs780094 was significantly associated with T2DM. [21] Hence, GCKR is thought to increase the risk of T2DM by regulating glucose levels. G6PC2 rs16856187 was also found to be associated with T2DM in a Southern Han Chinese population [22], and rs560887 was associated with T2DM in Caucasians [23]. Nevertheless, our study did not detect a significant difference in these genotype and allele frequencies between T2DM and control groups. Besides, we also found that the rs492594 of G6PC2 had a protective role in the pathogenesis of T2DM, nevertheless, this effect was inverted when investigating the interaction between SNP-SNP, which was not detected in other studies. These contradictory results may be affected by sample size and different types of research design. The matched nested case-control study can combine the advantages of prospective and case-control designs, effectively avoiding the potential reverse causality and confounders that are more likely to occur in cross-sectional studies, which can increase the credibility of the results.
This study is the first to identify a significant interactive effect between the rs2293572 polymorphism in GCKR and the rs492594 in the G6PC2 gene on T2DM risk. It is interesting to note that although the genetic variants in the rs2293572 of GCKR gene did not have noticeable effects on T2DM, their interplay with genetic variants in another gene were found to have a greater effect. A carrier of the rs2293572 G allele in GCKR and the rs492594 C allele in G6PC2 on average has a 1.65-fold higher risk of T2DM compared with a noncarrier under the additive model. By taking into account the epistatic interactions between potential risk loci, genetic variants, which might otherwise have remained undetected, were identified successfully here.
The role of genes in the pathogenesis of T2DM is complex. Genetic susceptibility is always inherited in the form of haplotypes, and there is more powerful statistical efficacy to identify a complex association between a pair of SNPs and a trait using haplotypes than with a univariate analysis or the interaction term. [24,25,26] In order to further probe the interaction effect, we conducted an analysis of haplotypes of the GCKR and G6PC2 genes. Our results suggest that the interaction of the haplotypes CGC (GCKR gene) and GCC and CAT (G6PC2 gene) increased the susceptibility to T2DM, while the presence of CCC (GCKR gene) and CAT (G6PC2 gene) lowered the risk of T2DM. However, the mechanism of the interaction between GCKR and G6PC2 genes remains unclear. There is still a need for further study at the molecular level on whether there is an association between gene regulation and expression.
The strengths of our study include its prospective study design with comprehensive evaluation of known and underlying confounding factors, blood samples, and GCKR and G6PC2 genotypes in a well-characterized population. However, several limitations for this study should be considered. First, just 6 SNPs within the GCKR and G6PC2 gene were chosen, and more SNPs should be included in the further studies. Second, gene-environment interaction should be investigated in future studies. Third, the results obtained in the current study should be checked in future studies with a larger sample size and in different nationalities.
Conclusions
In conclusion, the results suggest that the C allele of rs780094 in GCKR was associated with an increased risk for T2DM. However, the rs492594-C allele in G6PC2 was associated with a decreased risk of T2DM in this Han Chinese population. We also found a significant SNP-SNP interaction between rs2293572 and rs492594, and that participants with the CCCCGC and CGCCCA haplotypes had a significantly increased the risk of T2DM, while the CCCCCA haplotype conferred lower susceptibility to T2DM. The associations of gene-gene with incident of T2DM might infer potential mechanisms underlying the pathogenesis for T2DM.
Abbreviations
- BMI:
-
Body mass index
- CI:
-
confidence intervals
- FPG:
-
Fasting plasma glucose
- G6PC2:
-
Glucose-6-phosphatase catalytic subunit 2
- GCKR:
-
Glucokinase regulatory protein
- GMDR:
-
Generalized multifactor dimensionality reduction
- HDL-C:
-
High-density lipoprotein cholesterol
- HWE:
-
Hardy-Weinberg Equilibrium
- LD:
-
Linkage disequilibrium
- LDL-C:
-
Low-density lipoprotein cholesterol
- OR:
-
Odds ratio
- SNPs:
-
Single-nucleotide polymorphisms
- T2DM:
-
Type 2 diabetes mellitus
- TC:
-
Total cholesterol
- TG:
-
Triglycerides
References
Nolan CJ, Damm P, Prentki M. Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet. 2011;378:169–81.
Yoon U, Kwok LL, Magkidis A. Efficacy of lifestyle interventions in reducing diabetes incidence in patients with impaired glucose tolerance: a systematic review of randomized controlled trials. Metabolism. 2013;62:303–14.
Ogurtsova K, da Rocha Fernandes JD, Huang Y, Linnenkamp U, Guariguata L, Cho NH, Cavan D, Shaw JE, Makaroff LE. IDF diabetes atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract. 2017;128:40–50.
Tirosh A, Shai I, Tekes-Manova D, Israeli E, Pereg D, Shochat T, Kochba I, Rudich A. 3-normal fasting plasma glucose levels and type 2 diabetes in young men. N Engl J Med. 2005;353:1454–62.
Iqbal N, Rubenstein AH. Does lowering of blood glucose improve cardiovascular morbidity and mortality? Clin J Am Soc Nephrol. 2008;3:163–7.
Wang H, Liu L, Zhao J, Cui G, Chen C, Ding H, Wang DW. Large scale meta-analyses of fasting plasma glucose raising variants in GCK, GCKR, MTNR1B and G6PC2 and their impacts on type 2 diabetes mellitus risk. PLoS One. 2013;8:e67665.
Li H, Xu R, Peng X, Wang Y, Wang T. Association of glucokinase regulatory protein polymorphism with type 2 diabetes and fasting plasma glucose: a meta-analysis. Mol Biol Rep. 2013;40:3935–42.
Xu M, Lv X, Xie L, Huang X, Huang Y, Chen Y, Peng K, Wang P, Wang W, Qi L, et al: Discrete associations of the GCKR variant with metabolic risk in a Chinese population: longitudinal change analysis. Diabetologia 2016, 59:307–315.
Mohas M, Kisfali P, Jaromi L, Maasz A, Feher E, Csongei V, Polgar N, Safrany E, Cseh J, Sumegi K, et al. GCKR gene functional variants in type 2 diabetes and metabolic syndrome: do the rare variants associate with increased carotid intima-media thickness? Cardiovasc Diabetol. 2010;9:79.
Langenberg C, Sharp SJ, Franks PW, Scott RA, Deloukas P, Forouhi NG, Froguel P, Groop LC, Hansen T, Palla L, et al. Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med. 2014;11:e1001647.
Qi Q, Wu Y, Li H, Loos RJ, Hu FB, Sun L, Lu L, Pan A, Liu C, Wu H, et al: Association of GCKR rs780094, alone or in combination with GCK rs1799884, with type 2 diabetes and related traits in a Han Chinese population. Diabetologia 2009, 52:834–843.
Zhang M, Zhao Y, Sun H, Luo X, Wang C, Li L, Zhang L, Wang B, Ren Y, Zhou J, et al. Effect of dynamic change in body mass index on the risk of hypertension: results from the rural Chinese cohort study. Int J Cardiol. 2017;238:117–22.
Zhang L, Wang B, Wang C, Li L, Ren Y, Zhang H, Yang X, Zhao Y, Han C, Zhou J, et al. High pulse pressure is related to risk of type 2 diabetes mellitus in Chinese middle-aged females. Int J Cardiol. 2016;220:467–71.
American Diabetes Association. Standards of medical care in diabetes. Diabetes Care. 2005;28(suppl 1):S4–S36. https://doi.org/10.2337/diacare.28.suppl_1.S4.
Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502.
Pei X, Liu L, Cai J, Wei W, Shen Y, Wang Y, Chen Y, Sun P, Imam MU, Ping Z, Fu X. Haplotype-based interaction of the PPARGC1A and UCP1 genes is associated with impaired fasting glucose or type 2 diabetes mellitus. Medicine (Baltimore). 2017;96:e6941.
Grewal AS, Sekhon BS, Lather V. Recent updates on glucokinase activators for the treatment of type 2 diabetes mellitus. Mini Rev Med Chem. 2014;14:585–602.
Brown KS, Kalinowski SS, Megill JR, Durham SK, Mookhtiar KA. Glucokinase regulatory protein may interact with glucokinase in the hepatocyte nucleus. Diabetes. 1997;46:179–86.
Wang Y, Martin CC, Oeser JK, Sarkar S, McGuinness OP, Hutton JC, O'Brien RM. Deletion of the gene encoding the islet-specific glucose-6-phosphatase catalytic subunit-related protein autoantigen results in a mild metabolic phenotype. Diabetologia. 2007;50:774–8.
Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, Dauriz M, Hivert MF, Raghavan S, Lipovich L, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6:5897.
Ling Y, Li X, Gu Q, Chen H, Lu D, Gao X. Associations of common polymorphisms in GCKR with type 2 diabetes and related traits in a Han Chinese population: a case-control study. BMC Med Genet. 2011;12:66.
Hu C, Zhang R, Wang C, Ma X, Wang C, Fang Q, Bao Y, Xiang K, Jia W. A genetic variant of G6PC2 is associated with type 2 diabetes and fasting plasma glucose level in the Chinese population. Diabetologia. 2009;52:451–6.
Shi Y, Li Y, Wang J, Wang C, Fan J, Zhao J, Yin L, Liu X, Zhang D, Li L. Meta-analyses of the association of G6PC2 allele variants with elevated fasting glucose and type 2 diabetes. PLoS One. 2017;12:e0181232.
Ken-Dror G, Humphries SE, Drenos F. The use of haplotypes in the identification of interaction between SNPs. Hum Hered. 2013;75:44–51.
Romao I, Roth J. Genetic and environmental interactions in obesity and type 2 diabetes. J Am Diet Assoc. 2008;108:S24–8.
Al-Daghri NM, Pontremoli C, Cagliani R, Forni D, Alokail MS, Al-Attas OS, Sabico S, Riva S, Clerici M, Sironi M. Susceptibility to type 2 diabetes may be modulated by haplotypes in G6PC2, a target of positive selection. BMC Evol Biol. 2017;17:43.
Acknowledgements
We thank all of the participants for their contribution to this study.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 8150120621).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
LL and WZ participated in the study design, analyzed data, and drafted the manuscript. WZ, YL, LZ, YS, DZ, XL, CW, and ZM extracted data and performed statistical analyses. WZ wrote the manuscript. YS and CW polished the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed consent was obtained from all individual participants included in the study, and the study was approved by the Ethics Committees of Zhengzhou University.
Consent for publication
All the authors provided written informed consent for the publication of the results of this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1 Genotypic and allelic distributions of single nucleotide polymorphisms (SNPs) of GCKR and G6PC2. Table S2 Associations between the GCKR and G6PC2 gene haplotypes and T2DM. Table S3 Distribution of haplotypes of the G6PC2 and GCKR genes in case and control groups. (DOCX 28 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhou, W., Li, Y., Zhang, L. et al. Gene-gene interactions lead to higher risk for development of type 2 diabetes in a Chinese Han population: a prospective nested case-control study. Lipids Health Dis 17, 179 (2018). https://doi.org/10.1186/s12944-018-0813-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-018-0813-6