Abstract
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats-associated protein 9) shows the opportunity to treat a diverse array of untreated various genetic and complicated disorders. Therapeutic genome editing processes that target disease-causing genes or mutant genes have been greatly accelerated in recent years as a consequence of improvements in sequence-specific nuclease technology. However, the therapeutic promise of genome editing has yet to be explored entirely, many challenges persist that increase the risk of further mutations. Here, we highlighted the main challenges facing CRISPR/Cas9-based treatments and proposed strategies to overcome these limitations, for further enhancing this revolutionary novel therapeutics to improve long-term treatment outcome human health.
Similar content being viewed by others
Background
Cancer is one of the leading causes of disease-related death, increasing worldwide incidence [1]. At the same time, advancements have been achieved in the prevention and therapeutic approaches, resulting in longer lifetimes or even cures for certain patients with cancer. Unfortunately, chemotherapy and radiotherapy, the two gold stones in cancer treatment, are also painful for patients and cause severe side effects [2]. Therefore, developing innovative anti-cancer therapies with less side effects needs a comprehensive understanding of cancer biology. The most recent advancements in sequencing technology have made it possible to study the cancer genome more effectively and at a lower cost than ever before. The use of an integrated strategy that incorporates genomic and transcriptomic advancements can provide a comprehensive view of an individual’s genome. Additionally, this method is used to make valuable decisions relating to patient therapeutic options [3].
Different genomic engineering tools have been performed in cancer therapy such as ZFNs and TALENs by targeting DNA domain-binding proteins. Still, their efficacy was limited due to the inability to target epigenetic modification that arises in tumorigenesis [4]. Recently, a more flexible genome editing technique, CRISPRs linked with HNH domain protein Cas9, promises efficient, long-term safety cancer treatment [5]. The CRISPR/Cas9 system, unlike previous genome editing methods that used protein-DNA interactions to mediate sequence recognition, uses an RNA molecule to mediate binding. CRISPR loci, which are made up of alternating repeat-spacer units, and CRISPR-associated (Cas) proteins, are derived from a prokaryotic host defense system that protects against viral genomes and plasmids [6]. Based on the method of recognition and cleavage, CRISPR/Cas systems are divided into two classes, which are further divided into six types and various subtypes [7]. Class 1 systems cleave with protein complexes, whereas Class 2 systems only cleave with one protein, creating an opportunity for genome engineering [8]. However, certain targeting limitations apply to all Class 2 systems (types II, V, and VI). For example, a protospacer flanking sequence is recognized by Type VI systems, which use Cas13 to cleave RNA [9]. In addition, type II and Type V systems recognize the adjacent protospacer motif (PAM), a conserved 2–5 bp sequence [10]. For example, the Cas12a/Cpf1 protein uses a simple crRNA and recognizes a PAM directly before the protospacer, such as T-rich PAMs (TTTN) [11]. Conversely, type II Cas9 nuclease recognizes PAM sequences downstream of the protospacer [12]. The most well-characterized and broadly applied CRISPR system is the type II CRISPR/Cas9 system.
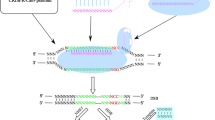
Cas9 is an RNA-guided endonuclease that recognizes and cleaves target DNAs that have template strand pairing to the guide RNA, and it requires RNA molecule known as the trans-activating crRNA (tracrRNA). TracrRNA promotes crRNA binding and processing. Moreover, a linker can join the tracrRNA and crRNA into a single molecule known as the single guide RNA used in genome editing (sgRNA) (Fig. 1).
Cas9 is an RNA-guided endonuclease that recognizes and cleaves target DNA that have template strand pairing to the guide RNA, which is composed of Crispr RNA (crRNA) and tracrRNA [13]. crRNA, which has a [17,18,19,20] nucleotide sequence that is complementary to the target DNA, and tracrRNA, which acts as a Cas nuclease binding scaffold [14].
The stages of CRISPR/Cas adaptive immunity. The three phases of the CRISPR/Cas9 system are depicted schematically. When phage DNA is injected into a bacterial cell, the Cas1–Cas2 adaptation module proteins are activated, which remove spacer-sized segments of phage DNA and channel them into the CRISPR array. The CRISPR array is transcribed, and the resulting pre-crRNA is processed at repeat sequences to form crRNAs during CRISPR RNA biogenesis. The Cas protein effectors bind individual crRNAs. Effectors programmed by suitable crRNA attach to phage DNA with sequences matching a CRISPR spacer in the cell, and the resulting R-loop complex is destroyed by Cas executor nuclease
The CRISPR/Cas9 system has been successfully applied to in vitro cancer research by inhibiting one or more oncogenic molecular pathways (Table 1). However, the in vivo use of the CRISPR/Cas9 system has faced many challenges such as the occurrence of off-targeting modifications, the possibility of causing autoimmune diseases, the identification of a proper delivery technique, and, lastly, ethical concerns. As a result, research scientists follow different procedures and investigate various bioinformatics tools to prevent, or at least reduce, these obstacles to make the CRISPR/Cas9 system more suitable for treating cancer in the human body. This review summarizes some of the main limitations of using CRISPR/Cas9 in clinical trials and some of the strategies applied in previous studies to overcome these limitations. Hopefully, this study provides a comprehensive overview of the main roadblocks to implementing this promising technique in vivo, helping future researchers focus their efforts on tackling them and making CRISPR come alive as a powerful strategy to treat cancer.
Innovative advances in CRISPR/Cas9 gene-editing technology
When Japanese scientists found several previously undiscovered tandem repeats in the E. coli genome in 1987, they didn’t report the biological relevance of those findings [15]. However, the role of these sequences remained unknown until they were termed Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) in 2002 [16]. Then, in 2005, the CRISPR loci were shown to play a significant role in adaptive immunity by three different study teams [17,18,19]. In 2007, Barrangou and his team revealed that viral gene sequences integrated by bacteria might modify the bacterium’s resistance to phages [20]. Brouns et al. in 2008 discovered that non-coding RNA produced from the CRISPR incorporating short fragments might direct the CRISPR-associated (Cas) proteins to the target-specific portion of DNA, allowing it to perform a protective function [21]. Deltcheva et al. discovered that trans-coding crRNA (tracrRNA) was related to the maturation and processing of pre-crRNA, and their research revealed new destinations for crRNA development [22]. In vivo studies in 2012 showed that mature crRNA produced two unique RNA structures when base-paired with tracrRNA, guiding CRISPR-associated protein Cas9 to create double-stranded (ds) DNA cleavage [23]. Subsequently, Cong and Mali teams made genome editing with the CRISPR/Cas9 system possible, who used two different type II Cas systems to make DNA cuts in cell cultures [24, 25]. Once the CRISPR/Cas9 technology was developed, many CRISPR/Cas9-based tools for gene editing at the DNA and RNA levels were created by 2020, with fast advancements in the technology since [26, 27] (Fig. 2).
Overview of CRISPR/Cas9-based genome editing
CRISPR is a response of the bacterial and archaea immune system to protect themselves from virus infections [28]. Approximately half of the bacteria have a CRISPR/Cas system [29, 30] a defense mechanism that allows the bacterial cell to memorize, recognize and beat recurrently infecting agents [31]. In this system, short guide CRISPR RNAs (crRNA) interfere with invading nucleic acids in a sequence-specific manner. CRISPR/Cas is composed of a genomic locus termed CRISPR that contains harsh repeating elements separated by unique sequences (spacers), which derive from Mobile genetic factors like phages, plasmids, or transposons. An AT-rich region is typically found at the beginning of Cas genes, which encode Cas proteins [32]. Nowadays, according to the structure and function of the Cas protein, the CRISPR/Cas systems can be divided into two classes (class I, class II), which are further categorized into six types (type I–VI) [33]. Class I consists of multiprotein complexes responsible for the cleavage of nucleic acid.
In contrast, in class II, only a single protein, Cas9, is used to read, identify and cleave the DNA target sequence [33]. In CRISPR technology, a single protein method is more effective than a multiprotein approach, hence the class II system is more often used, especially in research [10]. Figure 1 illustrates the details of the Type II CRISPR/Cas9 system. For instance, deactivated Cas9 can be utilized to target the epigenome by inhibiting the enzymatic activity of HNH domains without causing sequence disruption [34]. The guide RNA is composed of two core parts; the first is required to bind the RNA to the Cas protein, and the second part, called a spacer, consists of about 20 nucleotides and is responsible for identifying and binding to the targeted site [35]. Furthermore, the PAM sequence is a short DNA sequence usually between 2 and 6 nucleotides that is also required to identify the exact target site on the DNA, and it is located three base pairs from the site where the DNA will be cut, and the mutation will be introduced [10] (Fig. 1).
Anticancer application of CRISPR/Cas9 gene editing and clinical trials
Cancer initiation and spread are mediated by mutations and dysregulation of a variety of genes [36] such as oncogenes, tumor suppressor genes, and stem cell-associated genes, chemo-resistant genes and metabolic genes. Cancer treatment’s primary goal is to halt cancer cell growth and development by repairing mutations and restoring dysregulated gene expression. Since its inception, the CRISPR/Cas9 gene-editing method has been widely used in cancer research, with promising results. Georgiadis et al. recently demonstrated that fratricide-resistant T cells can be generated by removing and replacing the TCR/CD3 and CD7 with lentiviral-mediated production of CARs specific for the CD3 or CD7[37]. Table 1 lists some of the target genes, tumors, and studies that show the effectiveness of CRISPR/Cas9 in correcting these alterations. Based on promising pre-clinical results, the CRISPR/Cas9 system can be used in clinical settings to target cancer-causing genes (Fig. 3). The efficacy of CRISPR-based cancer therapeutics is now being investigated in a number of clinical trials (Table 2).
CRISPR/Cas9-mediated treatment has the potential to cure a variety of diseases. The number of diseases that CRISPR is now used to treat is rising by the day. The CRISPR/Cas9 system has been used to generate many disease-based models for many important human diseases, including viral diseases, neurological diseases, cancer, ocular disease, blood diseases, and cardiovascular diseases and disorders, as well as other complex genetic human diseases, according to data from clinical trials released recently
The programmed cell death-1 (PD-1) protein expression is being targeted by several of these clinical studies. For example, a monoclonal antibody against PD-1 called pembrolizumab exhibits anti-tumor activity in Non-Small Cell Lung Cancer (NSCLC), suppressing the immune system’s ability to produce PD-1 and PD-L1 (programmed death-ligand 1), dramatically improves patients’ survival rate [38]. Because the FDA has approved PD-1 inhibitors for cancer immunotherapy, PD-1 is an intriguing target for immunotherapy. In addition, CRISPR/Cas9 has been used in patients to begin targeting PD-1 (NCT02793856). They used CRISPR/Cas9 to suppress PD-1 expression in metastatic cells from NSCLC patients. The cells were cultured and modified before being reintroduced into the patient [39].
PD-1 knockout-engineered immune cells to treat metastatic NSCLC will be tested for safety in a dosages trial. Additional trials targeting PD-1 expression in T-cells are currently done in other types of cancer such as renal, bladder, and prostate cell malignancies [40]. Similarly, PD-1 deletion has been used in T-cells in phase II clinical trials for esophagus cancer (NCT03081715). Furthermore, the ability of CRISPR gene editing for cancer immunotherapy to persist for up to 9 months, suggests that immunogenicity is low under these settings and demonstrates the practicality of CRISPR gene editing for cancer immunotherapy [41]. Clinical experiments are also using CRISPR/Cas9 to create chimeric antigen receptor (CAR) T cells.
The first-in-human trial was conducted by scientists from the university of Pennsylvania applying CRISPR/Cas9 genome-edited NY-ESO-1 TCR cells for cancer patients [42] including advanced multiple myeloma (MM) myxoid/round cell liposarcoma (MRCL), and synovial sarcoma (NCT03399448). They showed that T cells were proven to be safe, viable, and long-lasting [42]. Furthermore, using CRISPR to eliminate endogenous TCR and PD-1 might improve tumor rejection activity [40]. Additionally, the allogeneic CAR T-cells targeted to the CD19 antigen were produced by combining the lentivirus-delivered CAR receptors and electroporation-delivered CRISPR RNA to alter the natural TCR and B2M genes. For patients with leukemia, this strategy may help avoid the host’s immune system and hence avoid graft-versus-host-disease complications. Consequently, additional CRISPR clinical trials (phase III) used CRISPR-edited CAR T-cells with dual specificity for CD19 and CD20/CD22, which can identify and destroy CD19-negative malignant cells by identification of CD20/CD22 (NCT03398967). This may have been a helpful adjunctive treatment for an extensive range of the population. In another work, Chen et al. applied CRISPR/Cas13a to disrupt human papillomavirus16/18 E6/E7 mRNAs using an emerging programmed CRISPR technology. They revealed that HPV 16/18 E6/E7 mRNA was successfully and selectively knocked down using a modified CRISPR/Cas13a system, causing growth suppression and cell death in HPV 16 and 18 positive SiHa and HeLa cell lines, but not in the HPV -negative C33A cells [43]. An additional CRISPR clinical study has been planned to test new medications and determine their effectiveness (NCT03332030). In this study, patients with Neurofibromatosis type 1 (NF1) were used to create an induced pluripotent stem cell bank (iPSC) (NF1). NF1 is a common neurocutaneous disease that frequently develops tumors of both benign and malignant types [44]. The main method that used in vivo and in vitro CRISPR/Cas9 study to treat diseases showed in Figs. 3 and 4.
To identify a particular target drug for NF1, CRISPR/Cas9 was used to create NF1 homozygous (NF1-/-) and NF1 heterozygous (NF1+/-) cell lines, as well as NF1 wild type (NF1+/+). The discovery of NF1-targeted therapies may be aided by the opposite or alleviated characteristics. Despite promising clinical trial results, more research is needed to ensure that CRISPR/Cas9 is a safe and effective method of treating human cancers [45]. On the other hand, CRISPR Cas9 indirectly can be used in cancer therapy to find out the drug-resistance mutation in a short period of time. For example, through applying CRISPR Cas9, only in 40 min can determine the FLT3-F691L with a sensitivity of 0.1% [46].
Challenges of CRISPR/Cas9
Even though the previous explanation suggests that CRISPR/Cas9 is a promising approach, this editing system still has a number of limitations and risks that make it challenging to use in clinical trials due to its recent discovery and use in humans. Immunogenicity, off-targeting, polymorphism, delivery method, and ethics are only several major concerns with the CRISPR/Cas9 system highlighted with the list of strategies that has been developed and can be used to overcome those limitations (Fig. 5).
Autoimmune response against endogenous Cas9 protein
The Cas9 protein is one of the three main components of the structure of the CRISPR system, and it has a fundamental role in binding double-stranded DNA, paired with the mRNA guide, and cutting it at a specific site, expressly 3 bases before the PAM sequence [104].This protein derives from Streptococcus pyogenes, a bacterium that is the cause of many common infections in humans. It is recognized by the body as an antigen, developing an immune response against it [105]. Similarly, the existence of a pre-existing immune response to the homologous Cas9 protein in Staphylococcus aureus has been reported [106]. Indeed, both Staphylococcus aureus and Streptococcus pyogenes, from which the main Cas9 proteins are obtained, SaCas9 and SpCas9, have infected humans for a long time [106]. Thus, the human immune system recognizes these proteins as foreign and develops an immune response against them upon injection, which leads to fast degradation of the Cas9 protein, preventing it from performing the gene-editing function [107].
Strategies to overcome immunogenicity
Several strategies have been proposed to overcome limits posed by immunogenicity against Cas9. Here, we are giving an overview of the main ones offered; (i) implementing the CRISPR/Cas system for gene editing early in a lifetime; (ii) targeting immune-privileged organs (Fig. 6).
Gene editing in early lifetime
Even before birth, various types of disease can be detected in children, and preventing or treating those diseases will save thousands of lives worldwide. The CRISPR/Cas system has been successfully applied in treating various types of inherited diseases in children, such as cystic fibrosis, thalassemia, and sickle cell anemia, Mucopolysaccharidosis type IVA [108,109,110,111,112,113]. Furthermore, CRISPR/Cas9 can inhibit different molecular pathways of various common types of cancer in children, such as neuroblastoma and lymphoma [114, 115]. Moreover, treating these defects by CRISPR Cas system after diagnosed can be done before the infant is immunized with anti-Cas protein.
Targeting immune-privileged organs
Another practical approach to overcoming the risk of autoimmune disease is gene editing by CRISPR Cas9 techniques in those organs recognized as immune-privileged organs. An Immune privileged organ can be defined as a site in the body where a graft tissue can be implanted without being rejected by the organism due to an immunological reaction formed against it [116]. Examples of immune privilege organs are eyes [117], brain [118], placenta, fetus [119], and testicles [120].
Many congenital eye disorders lead to blindness and other defects in the eyes, such as Leber congenital amaurosis type 10, retinal dystrophy caused by a mutation in the CEP290 [121]. Fortunately, many studies proved that the eyes are one of the immune-privileged organs that can successfully imply CRISPR Cas9 on it and edit a particular mutation there [122]. For example, Jain et al. employed CRISPR-Cas9 genome editing in human TM cells and in a POAG animal model to reduce the expression of mutant MYOC, resulting in a reduction in the stress on the ER [123].
CRISPR offers an excellent opportunity for scientists to reach high gene editing efficiency in fetuses and embryos, as the immune system has not yet reached maturity. Nevertheless, Because of the substantial danger of embryo off-targeting associated with its use in vivo, it is illegal in many countries. For example, CRISPR/Cas9’s off-targeting rate was 16% in a study aiming to target the POU5F1 gene in embryos [124]. Correspondingly, due to the cleavage of both alleles, off-target cleavage of Cas9 causes chromosomal loss and hemizygous indels [125]. These findings show that chromosomal content can be manipulated. Still, it requires other skills and strategies to reduce the high risk of off-targeting and loss of DNA fragments.
Additionally, testicles are another immuno-privileged organ that the gene editor can target to correct the mutated genes and deactivate oncogene in cancer patients [120]. These genes can be identified and reverted to their normal function through CRISPR Cas system. Sun et al. found that male fertility genes in mice can be dispensable for further fecundity by knocked out through CRISPR/Cas9 [126]. Furthermore, in mice, CRISPR/Cas9-mediated gene editing uncovered 30 testis-enriched genes not required for male fertility [127].
Likewise, brain is another immune privileged organ, and several studies were performed in vivo without immune tolerance. Normalized FMR-1 gene expression was achieved by CRISPR/Cas9-mediated deletion of the CGG repeat in hiPSCs from fragile X syndrome patients, a change that was sustained even after differentiation into neural progenitor cells (NPCs) and mature neurons; in addition, hypermethylation of the CpG sites upstream of FMR-1 was reversed [128].
Off-targeting
Another main concern about using CRISPR/Cas9 in recent years is having a high number of off-targeting [129,130,131]. When implying the CRISPR Cas9 system in a complex genomic species such as mammalians, the gRNA might bring to a wrong target due to similarities within the genome, which may lead to further mutations being introduced in undesired genomic locations [132]. In recent years, many bioinformatics tools have been developed to help predict and reduce off-target modifications. These should be further improved to enable researchers to use them effectively in the development of new therapies.
Strategies to overcome off-targeting
The main strategies that have been successfully performed in previous studies can be classified into three main groups; (I) bioinformatics tools to design more accurate gRNA and predict off-targeting; (II) use of Cas9 nickases; (III) add anti-CRISPR proteins.
Bioinformatics tools
Bioinformatics tools play a crucial role in analyzing, predicting, and determining the CRISPR Cas system. Bioinformatics tools allowed Francisco Mojica to discover that the system previously found in bacteria also existed in archaea [133]. Further, bioinformatics tools help scientists design more efficient gRNAs, detect the accurate editing site within the whole genome, and evade off-targeting percentage probability (Table 3) [134]. Studies have shown that the gRNA is responsible for most of the off-targeting [135]. For example, many studies have shown a direct correlation between gRNA length and the number of off-targeting; thus, finding the perfect size of the gRNA is essential to reduce the off-targeting probability [136]. Such as reducing the length of gRNA to less than 20 nucleotides have a significant role in lowering off-targeting by about 5000 folds in the same efficiency of the longer gRNA [34, 122]. According to another study, most of the mismatches occur within the last three nucleotides placed at the opposite side of the PAM sequence, thus removing these nucleotides and maintaining the length of gRNA about 17 nucleotides crucial role in the reduction of off-targeting [137]. On the other hand, gRNAs shorter than 15 base pairs are not safe as they would lose the specificity and could not bind the right target inside the nucleus [138].
Cas9 nickases
Another practical approach to reducing the number of off-targeting is mutating in one nuclease domain in just one strand of the DNA by CRISPR nickase, which crucial to create nick that quickly repaired in the cells nickase [155]. Cas9 nickase has a different breaking mechanism than the normal Cas9 protein; in particular, it breaks down just one strand of the DNA, and they use double adjacent gRNAs rather than sgRNAs (Fig. 7). Therefore, editing genes by using Cas9 nickase reduces further damage in the target DNA, and it has a significant role in reducing the number of off-targeting [156]. Furthermore, it was shown that paired nicking could reduce the risk of off-targeting by 50 to 1500 folds in cell lines, and in mouse zygotes, it allows the gene knockout without any effect on cleavage efficiency [155].
Nickase systems consisting of one or two nickases. H840 and D10 are two amino acids found in the Cas9 endonuclease protein that are involved in the cutting of one DNA strand by the enzyme. The RuvC domain contains the amino acid H840, while the HNH domain has the amino acid D10. The non-targeted strand is cleaved by Cas9 H840A, while the gRNA-targeting strand is cleaved by Cas9 D10A. Cas9 can only cut the strand complementary to the gRNA in a single nickase; however, a pair of sgRNA-Cas9n complexes can nick both strands at once (paired nickases). Additional concerns for gRNA design when using paired nickases include creating a 5’ overhang, the spacing between the two gRNAs, and the relative position of the two gRNA target sites
Anti-CRISPR proteins
Inactivation of Cas9 protein after targeting its site may also reduce the number of off-targeting [134]. It has been proven that the number of off-targeting is correlatively increased as long as the Cas9 protein is expressed in the human tissue culture [137]. Deactivation of Cas9 protein can be obtained through using anti-CRISPR proteins (Acr) [157]. Acr proteins are produced in both bacterial and human cells and allow to disable CRISPR function [158]. Moreover, more than 50 anti-CRISPR proteins have been discovered so far, synthesized by viruses as a defense system against prokaryotic cells [159]. The first Acr protein discovered that deactivates the CRISPR type I system in P. aeruginosa, while the other Acr proteins can act on different types of CRISPR, such as types II, III, and V [31]. Acr proteins are about 52 to 333 amino acids, meaning they are tiny molecules and diverse with no sequence overlap with other proteins [159, 160]. Also, each Acr protein has a specific and unique sequence free of conserved sequences, which increases their diversity [161]. Having a small size and a unique genomic sequence make the recognition of Acr difficult by standard homology-based methods. Therefore, these proteins can target their aimed sequences before being recognized. Furthermore, using a different mechanism is one of the successful keys used by Acr to deactivate the CRISPR/Cas9 system [162]. For example, AcrIIA4 binds to both Cas9 and sgRNA rather than binding with just one of them [161]. The efficiency of Acr depends on three main mechanisms, which are the crRNA concentration, DNA binding obstruction, and DNA cleavage inhibition [163]. When the viral genome is injected into the phage, its Acr proteins in a small concentration make the host cells immunosuppression and prepare the bacteriophage for future infections by the phase [164]. Conversely, having a high concentration of Acr proteins and vulnerable bacteriophage disables the function of the CRISPR system from the infected bacteria [164]. Moreover, Acr proteins have a stronger binding affinity with CRISPR; thus it is required a small concentration disable the function of the CRISPR system. On the other hand, anti-CRISPR-associated (Aca) proteins work oppositely to Acr proteins by preventing the transcription of anti-CRISPR proteins [163]. Therefore, the CRISPR system can be improved by using Aca proteins to suppress Acr proteins. Also, the use of Acr proteins that imply phage instead of antibiotics may overcome the issue of drug resistance[165].
Screening before the treatment
Pre-existing mutations in genes like TP53 and KRAS may raise the risk of additional mutations during CRISPR Cas cancer therapy [166]. And the two primary ways for dealing with this problem are screening before using the CRISPR Cas system and monitoring the patient after injection.
Polymorphism in cancer
Unlike other genetic diseases such as Duchenne Muscle, Dystrophy, and cystic fibrosis, cancer relies on several mutations [167,168,169,170]. Moreover, dysregulation of the multiple genes leads to cancer most of the time. For example, mutations happen in approximately 190 codons in the human TP53 gene, and around 25% of the mutations occur in eight codons [171]. Hence, editing a single mutated nucleotide is not enough in most cases that are widely performed in gene therapy [170]. Correcting mutated nucleotide by knocking-in is much more challenging in CRISPR Cas9 since it is more precise than knocking out, which creates alterations, as knocking in, all of the cancer-causing genes takes longer and needs multi-guide RNA [172]. However, by CRISPR Cas9, knocking in is potentially helpful in many ways, such as studying particular gene variation to find out the gene regulation [172].
Correcting or editing the mutated nucleotides of tumor suppressor genes is one of the approaches that should be thought about to obtain the desired result in cancer therapy by knock-in in the mutated gene (Fig. 8). CRISPR/Cas9 technology targeted these tumor-suppressor genes to inhibit or reduce tumorigenesis by restoring the activities of tumor-suppressor genes [34]. However, as in cancer, there is plenty of mutations in tumor-suppressor genes, it requires a higher number of gRNA, and there is a higher risk of off-targeting. On the other hand, the CRISPR Cas system can disrupt the nucleotides located in the active site of the protein to suppress the activity of oncogenes, such as KRAS in pancreatic cancer and ATM in neuroendocrine cancer by deleting their inactivation sequences (Table 1) [34, 173]. On the other side, in TNBC cells, the deactivation of CXCR7 and the co-knockout of CXCR4 and CXCR7 have been shown to inhibit the expression of oncogenes and may have a potential target in TNBC treatment [52]. For the Cas system to be effective in knocking out oncogenes, the proper gRNA must be designed to target the binding site of oncogenes and prevent protein-protein interaction, which is an essential step in the molecular pathway of cancer progression [52].
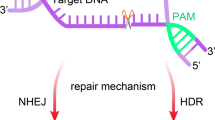
Mechanisms of DNA repair outcomes of genome editing. Typically, DNA double-strand (ds) breaks caused by CRISPR/Cas9 are repaired via either homology-directed repair (HDR) or non-homologous end joining (NHEJ), depending on the circumstances. Exogenous ‘repair templates’ can be introduced into the genome by HDR, whereas NHEJ creates random insertions and deletions (indels) that can disrupt coding areas or catalyse genome rearrangements. The preference for HDR or NHEJ after DNA damage can be increased by small compounds that interfere with each system and so bias the cell toward one or the other after DNA damage
Strategies
We described two primary solutions for managing polymorphism issues: CHyMErA and bioinformatics techniques to investigate the protein interaction site and forecast the results.
Performing CHyMErA
To edit many targets in a single mammalian cell, CRISPR may be utilized with various kinds and procedures, such as the CHyMErA (Cas hybrid for multiplexed editing and screening applications) method. CHyMErA depends upon two Cas proteins, Cas9 and Cas12a nucleases, rather than just the standard CRISPR/Cas9 gene editing (Fig. 9) [174]. Exons may be deleted using CHyMErA, which is helpful for the high deletion of gene sequences. As a result, employing CHyMErA to target multiple sites is one of the novel approaches to overcoming cancer polymorphism [175].
DNA editing platform CHyMErA is a combinatorial system. Cell lines harbouring nuclear SpCas9 and LbCas12a, as well as a hgRNA expression cassette, provide the basis of the CHyMErA system. Cas12a gRNAs are fused with Cas9 and expressed under a single U6 promoter in hgRNAs. This process is completed by Cas12a, which identifies the direct repeat sequence and cuts upstream of it to release functional Cas9 and Cas12 gRNAs that can be loaded onto their respective nucleases for directed combinatorial genome editing
Detecting protein interaction site
Bioinformatics tools can have a crucial role in predicting and obtaining the desired results in knocking out. For example, different databases can be used for finding out the interaction site of the proteins, such as InterPred [176]. By using this platform, amino acids located in the active site of the proteins can be detected, and then gRNA is designed based on it. Moreover, different databases can be used for predicting the result of CRISPR knockout (Table 3).
In conclusion, knock-in and Knock-out for oncogenes and tumor suppressor genes are critical in gene editing using CRISPR Cas9. However, knocking in to edit a particular nucleotide should be performed more precisely. Indeed, having more than one mutation in cancer cells required performing other techniques such as ChyMErA, which can target multi targets by binding two cutting DNA enzymes Cas9 and Cas12a. On the other hand, creating one mutation in the protein active site of the oncogenes is enough to suppress its role, so performing Knocking out in cancer therapy to suppress oncogene is much more practical.
The delivery challenges
Choosing a proper safe, and precise delivery technique to carry the CRISPR system into the tumor site, especially in vivo, and targeting the right sequence inside the nucleus is another challenge that should be considered. CRISPR/Cas9 technologies are delivered through different approaches, such as viral, physical, and extracellular vesicle-base system delivery techniques [177]. Additionally, each method is used for a specific purpose and has its limitations. Thus, the main challenges while choosing the right vector are packaging, delivery, and targeting the right site [178, 179]. For example, viral vectors are used widely in both in vivo and in vitro, but it has many limitations, such as immune response and insertional limitation [177]. For instance, after implying viral vector in vivo, it exposes continuously for a long time and increases the risk of mutations and off-targeting [180].
Viral delivery vectors
Adeno-associated viruses (AAV) such as adenovirus and lentivirus have been utilized successfully in other research in vivo [181] and they do not cause any other diseases in humans, only a very few immune responses [178, 182]. On the other hand, the main disadvantage of AAV has a tiny packaging size, so more than one AAV is required to carry all the CRISPR systems such as gRNA and Cas protein [182]. Additionally, the maximum size that a single AAV vector can deliver is about 4.7 kbp, while the genomic size of SpCas9 alone is around 4.3 kbp [178]. Thus, more than one victor is necessary to hold all the systems.
Non-viral delivery vectors
Besides that, non-viral delivery vectors are another approach, such as lipid nanoparticles and inorganic nanoparticles [183]. In addition, a non-viral delivery approach, like nanoparticle-based delivery, allow for more frequent administration of gene therapy with lower risk of immunogenicity, less exposure to nuclease and more accurate targeting [183, 184]. Furthermore, non-viral vectors have a greater capacity than viral vectors without integrating in the carried genome [185]. On the other hand, extracellular vesicle-based systems have been utilized in both in vivo and in vitro systems successfully and, compared to the other system, are safer and cheaper [186]. Another challenge is the delivery and efficiency percentage of the CRISPR system into the targeted size, especially in cancer therapy with total editing efficiency [178]. And according to Yin et al. (2014), the total delivery efficiency is 1 out of 250 in targeting liver cells using hydrodynamic injection [187] (Table 4).
Strategies
One of the effective ways to overcome the packaging challenge is splitting the Cas9 protein into two AAV (AAV-split-Cas9) vectors instead of one [188]. As previously explained, large size vectors increase the risk of off-targeting and mutation [189]. As a result, employing a smaller Cas9 protein and splicing it into two AAV vectors is critical for reducing off-targeting and increasing delivery efficiency [190]. Another option that can be used to reduce the risk of off-targeting associated with delivery techniques is the use of ribonucleoprotein (RNP) complexes, such as recombinant CRISPR-Cpf1 Ribonucleoprotein (CRISPR-Cpf1-RNP) suppressed off-target activity in mouse cells [191]. Furthermore, according to their results, Mout and his colleagues applied Cas9-RNP methods, which efficacy around 95% in cultured cells [192]. This approach also degrades after 24 to 48 h of injection [182] (Table 4). Thus, the risk of further mutations and off-targeting that occur due to the continuous expression of viral vectors is reduced significantly [177] .
Ethical issues and CRISPR/Cas9 technology
Human genetic alterations have long been a source of ethical debate; CRISPR/Cas9-mediated genome editing has provided a new perspective. Considering the unpredictability and broad-reaching effects of this technology’s appealing applications, a thorough examination of its ethical and societal implications is required. The conceptions of various members of society, such as the public and religious academics, are fundamental.
Current ethical standpoint
The application scope of CRISPR/ Cas9 is expanding at an incredible rate. Switching genes on or off to investigate how they work or causing mutations in cells to learn why and how they become malignant, are some of the opportunities it has opened up in molecular biology research. Gene editing can be used to create resistant crops and stronger police dogs, for example [210, 211]. Another highly contentious concept would edit the human genome permanently to eliminate disease-causing mutations or even improve or introduce desired features in offspring by inserting helpful genes and this is debatable [212]. Non-reproductive cell genome modifications are not heritable, whereas germ cell modifications can be passed down to the next generation. As a result, the attractive uses of this approach raise ethical, moral, and safety concerns [213]. Human germline modification using CRISPR/Cas9-based gene editing has raised concerns about threats to human safety and dignity, as well as the potential for genocide. There was an effort to halt human genome research until a national or global agreement on society’s acceptance of this new technology was reached [214].
Morality concepts
Morality concepts, particularly in biomedicine, are based on empirical research and entail evaluating potential risk-benefit ratios, to maximize the latter while decreasing the former. It is vital to assess the spectrum of conceivable outcomes, the likelihood of each occurring, and the various arguments for the outcomes of any one while making moral decisions. There are at least three major causes for ethical concerns concerning CRISPR genome engineering technology. Concerns have been raised about the power and technical limitations of CRISPR technology in the first concept. These drawbacks include a lack of on-target editing efficiency [215], incomplete editing (mosaicism) [216, 217], and inaccurate on-target or off-target editing [218, 219]. CRISPR experiments with animals and human cell lines have revealed these limitations. Technology, on the other hand, is evolving at a tremendous speed. The second concern is for the transformed species’ long-term survival: if they will be influenced indefinitely and whether the edited genes will be passed down through generations, perhaps influencing them in unanticipated ways.
Making precise predictions regarding the future of a modified creature and estimating potential hazards and advantages may be difficult, if not impossible, given the aforementioned technical constraints and the intricacies of biological systems. As a result, the uncertainty created by these circumstances makes precise risk/benefit assessments difficult, making moral decision-making more difficult [220]. Finally, even if the genome is altered as planned and the necessary functional output is achieved on time, the complicated link between genetic information and biological phenotypes is not fully understood, according to the skeptical viewpoint. As a result, depending on the circumstances, the biological impact of altering a gene in germline and/or somatic cells may be unknown. The intricate regulatory actions of many genes govern many biological features [221]. As a result, “designing” a biological phenotype at the organismal level is difficult, if not impossible.
Strategies
On the Brightside, it has the potential to make a significant difference in terms of health and wellbeing if used properly [222]. There are several reasons why this technology can be used correctly, although patient safety is one of the most important. One of the most compelling arguments in favor of allowing the use of this technology is the need to protect patients [212]. When germline editing research is applied in a clinical setting to avoid the inheritance of a specific genetic condition, it may alleviate the sorrow and anxiety that parents encounter in the life of the possibility that their child may be born with that genetic disease [214, 223]. Recently, Bioengineer Feng Zhang of MIT and Harvard has modified the Cas9 enzyme to limit mutations outside its target region [224]. Furthermore, the error rate of CRISPR/Cas9 might be further decreased to a safe range if further modifications are introduced [225]. Considering this, CRISPR/Cas9 mediated genome editing safety concerns might be overcome to some extent. Overall, CRISPR/Cas9 technology’s risk profile varies depending on the design. After overcoming some ethical and safety problems, some are approved or predicted to be used soon. On the Brightside, it may significantly improve health and quality of life, but still, it relies on how this technique is used.
Conclusion and future perspectives
The therapeutic genome editing field has made tremendous progress in recent years, progressing from essential investigation to preclinical development and into human trials, in particular for ex vivo HSC and T cell editing and also for in vivo liver genome editing, as a result of the relatively efficient delivery methodologies developed for these systems. However, several considerable challenges need to be addressed before the biomedical promise of genome editing can be fully realized. First, delivery has always been one of the most, if not the most, formidable problems in the gene therapy field. Importantly, HDR and even knockout efficiencies are currently low in many tissues, so higher delivery efficiency is needed to compensate. There should be an international law to ensure that gene editing does not harm humanity, and experiments should be restricted in a health care system.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Abbreviations
- CRISPR/Cas9:
-
clustered regularly interspaced short palindromic repeats-associated protein9
- ZFNs:
-
Zinc-finger nucleases
- TALEN:
-
Transcription activator-like effector nucleases
- TTTN:
-
T-rich PAMs
- tracrRNA:
-
trans-activating crRNA
- crRNA:
-
CRISPR RNAs
- PD-1:
-
programmed cell death-1
- PD-L1:
-
programmed death-ligand 1
- MRCL:
-
myxoid/round cell liposarcoma
- POAG:
-
Primary open angle glaucoma
- hiPSCs:
-
Human induced pluripotent stem cells
- NPCs:
-
neural progenitor cells
- gRNA:
-
Guide RNA
- Acr:
-
anti-CRISPR proteins
- CXCR:
-
Chemokine receptors
- CHyMErA:
-
Cas hybrid for multiplexed editing and screening applications
- AAV:
-
Adeno-associated viruses
- RNP:
-
ribonucleoprotein
- CRISPR-Cpf1-RNP:
-
CRISPR-Cpf1 Ribonucleoprotein
- HDR:
-
homology-directed repair
- NHEJ:
-
non-homologous end joining
- HSCs:
-
hematopoietic stem cells
References
Lortet-Tieulent J, Georges D, Bray F, Vaccarella S. Profiling global cancer incidence and mortality by socioeconomic development. Int J Cancer. 2020;147(11):3029–36.
Arunachalam SS, Shetty AP, Panniyadi N, Meena C, Kumari J, Rani B, et al. Study on knowledge of chemotherapy’s adverse effects and their self-care ability to manage - The cancer survivors impact. Clin Epidemiol Glob Health. 2021;11:100765.
Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66(1):75–88.
Sachdeva M, Sachdeva N, Pal M, Gupta N, Khan IA, Majumdar M, et al. CRISPR/Cas9: molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther. 2015;22(11):509–17.
Deng H-X, Zhai H, Shi Y, Liu G, Lowry J, Liu B, et al. Efficacy and long-term safety of CRISPR/Cas9 genome editing in the SOD1-linked mouse models of ALS. Commun Biol. 2021;4(1):396.
Li Y, Peng N. Endogenous CRISPR-cas system-based genome editing and antimicrobials: review and prospects. Front Microbiol. 2019;10:2471.
Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans R Soc Lond B Biol Sci. 2019;374(1772):20180087.
Mir A, Edraki A, Lee J, Sontheimer EJ. Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol. 2018;13(2):357–65.
Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR-Cas systems and their applications. Int J Mol Sci. 2020;21(3).
Gleditzsch D, Pausch P, Müller-Esparza H, Özcan A, Guo X, Bange G, et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 2019;16(4):504–17.
Yang Z, Edwards H, Xu P. CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab Eng Commun. 2020;10:e00112.
Leenay RT, Beisel CL. Deciphering, communicating, and engineering the CRISPR PAM. J Mol Biol. 2017;429(2):177–91.
Allen D, Rosenberg M, Hendel A. Using synthetically engineered guide RNAs to enhance CRISPR genome editing systems in mammalian cells. Front Genome Edit. 2021;2:35.
Wang S-R, Wu L-Y, Huang H-Y, Xiong W, Liu J, Wei L, et al. Conditional control of RNA-guided nucleic acid cleavage and gene editing. Nat Commun. 2020;11(1):91.
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–33.
Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–75.
Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–82.
Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151(Pt 3):653–63.
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(Pt 8):2551–61.
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–12.
Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–4.
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–7.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6.
Zhang H, Qin C, An C, Zheng X, Wen S, Chen W, et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer. 2021;20(1):126.
Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol. 2019;55:106–19.
Hryhorowicz M, Lipiński D, Zeyland J, Słomski R. CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch Immunol Ther Exp. 2017;65(3):233–40.
Ishino Y, Krupovic M, Forterre P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J Bacteriol. 2018;200(7):e00580-17.
Chevallereau A, Meaden S, van Houte S, Westra ER, Rollie C. The effect of bacterial mutation rate on the evolution of CRISPR-Cas adaptive immunity. Philosophical Transactions of the Royal Society B: Biological Sciences. 2019;374(1772):20180094.
Pinilla-Redondo R, Shehreen S, Marino ND, Fagerlund RD, Brown CM, Sørensen SJ, et al. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat Commun. 2020;11(1):5652.
Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1(6):e60.
Liu Z, Dong H, Cui Y, Cong L, Zhang D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb Cell Fact. 2020;19(1):1–14.
Hazafa A, Mumtaz M, Farooq MF, Bilal S, Chaudhry SN, Firdous M, et al. CRISPR/Cas9: A powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 2020;263:118525.
Peters JM, Koo BM, Patino R, Heussler GE, Hearne CC, Qu J, et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nature microbiology. 2019;4(2):244–50.
Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–41.
Georgiadis C, Rasaiyaah J, Gkazi SA, Preece R, Etuk A, Christi A, et al. Base-edited CAR T cells for combinational therapy against T cell malignancies. Leukemia. 2021.
Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–33.
Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539(7630):479.
Tian X, Gu T, Patel S, Bode AM, Lee M-H, Dong Z. CRISPR/Cas9 – an evolving biological tool kit for cancer biology and oncology. NPJ Precision Oncol. 2019;3(1):8.
Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, Tian L. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481).
Xu J, Gohil M, Stadtmauer E, Fraietta J, Gonzalez VE, Salas J, et al. Characterization of autologous T cells engineered to express NY-ESO-1 TCR with multiplexed CRISPR/Cas9 editing (NYCE T Cells). Cytotherapy. 2020;22:35-S6.
Chen Y, Jiang H, Wang T, He D, Tian R, Cui Z, et al. In vitro and in vivo growth inhibition of human cervical cancer cells via human papillomavirus E6/E7 mRNAs’ cleavage by CRISPR/Cas13a system. Antiviral Res. 2020;178:104794.
Chaka A, Ennaceur F, Tormen MA, Korbi I, Noomen F, Zouari K. Neurofibromatosis type I and stromal tumor with a multiple digestive localization. Case Rep Surg. 2021;2021:2868966.
Mo J, Anastasaki C, Chen Z, Shipman T, Papke J, Yin K, et al. Humanized neurofibroma model from induced pluripotent stem cells delineates tumor pathogenesis and developmental origins. J Clin Investig. 2021;131(1):e139807.
Liu Y, Chen Y, Huang S, Ma X, Huang X, Wang X, et al. Rapid and sensitive diagnosis of drug-resistant FLT3-F691L mutation by CRISPR detection. Front Mol Biosci. 2021;8(1032).
Dekkers JF, Whittle JR, Vaillant F, Chen H-R, Dawson C, Liu K, et al. Modeling breast cancer using CRISPR-Cas9–mediated engineering of human breast organoids. J Natl Cancer Inst. 2020;112(5):540–4.
Hannafon BN, Cai A, Calloway CL, Xu Y-F, Zhang R, Fung K-M, et al. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: evidence from a CRISPR/Cas9 deletion study. BMC Cancer. 2019;19(1):642.
Annunziato S, Lutz C, Henneman L, Bhin J, Wong K, Siteur B, et al. In situ CRISPR-Cas9 base editing for the development of genetically engineered mouse models of breast cancer. EMBO J. 2020;39(5):e102169.
Korkmaz G, Manber Z, Lopes R, Prekovic S, Schuurman K, Kim Y, et al. A CRISPR-Cas9 screen identifies essential CTCF anchor sites for estrogen receptor-driven breast cancer cell proliferation. Nucleic Acids Res. 2019;47(18):9557–72.
Chen A, Wen S, Liu F, Zhang Z, Liu M, Wu Y, et al. CRISPR/Cas9 screening identifies a kinetochore-microtubule dependent mechanism for Aurora-A inhibitor resistance in breast cancer. Cancer Commun (Lond). 2021;41(2):121–39.
Yang M, Zeng C, Li P, Qian L, Ding B, Huang L, et al. Impact of CXCR4 and CXCR7 knockout by CRISPR/Cas9 on the function of triple-negative breast cancer cells. Onco Targets Ther. 2019;12:3849–58.
Mintz RL, Lao YH, Chi CW, He S, Li M, Quek CH, et al. CRISPR/Cas9-mediated mutagenesis to validate the synergy between PARP1 inhibition and chemotherapy in BRCA1-mutated breast cancer cells. Bioeng Transl Med. 2020;5(1):e10152.
Zhao R, Kaakati R, Liu X, Xu L, Lee AK, Bachelder R, et al. CRISPR/Cas9-mediated BRCA1 knockdown adipose stem cells promote breast cancer progression. Plast Reconstr Surg. 2019;143(3):747–56.
Mendes de Almeida R, Bandarra S, Clara Ribeiro A, Mascarenhas P, Bekman E, Barahona I. Inactivation of APOBEC3G gene in breast cancer cells using the CRISPR/Cas9 system. Ann Med. 2019;51(sup1):40-.
Zhao X, Li J, Liu Z, Powers S. Combinatorial CRISPR/Cas9 screening reveals epistatic networks of interacting tumor suppressor genes and therapeutic targets in human breast cancer. Cancer Res. 2021.
Al-Mulhim F, Alqosaibi AI, Al-Muhnna A, Farid K, Abdel-Ghany S, Rizk H, et al. CRISPR/Cas9-mediated activation of CDH1 suppresses metastasis of breast cancer in rats. Electron J Biotechnol. 2021;53:54–60.
Behbahani RG, Danyaei A, Teimoori A, Neisi N, Tahmasbi MJ. Breast cancer radioresistance may be overcome by osteopontin gene knocking out with CRISPR/Cas9 technique. Cancer/Radiothérapie. 2021;25(3):222–8.
Wu X, Zhao Y, Xie X, Xuei X, Liu Y, Cheng L, et al. Abstract 549: Genome-wide CRISPR-Cas9 screen and RNAseq analysis identify new candidate synthetic lethality partners to PARP inhibitor in triple-negative breast cancer. Can Res. 2020;80(16 Supplement):549.
Yang L, Yu SJ, Shao ZM. Abstract P2-01-16: CRISPR-Cas9 screen identifies TMEM106A as a suppressor of breast cancer metastasis. Can Res. 2019;79:P2-01.
Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, Tian L, Gonzalez VE, Xu J, Jung IY, Melenhorst JJ, Plesa G, Shea J, Matlawski T, Cervini A, Gaymon AL, Desjardins S, Lamontagne A, Salas-Mckee J, Fesnak A, Siegel DL, Levine BL, Jadlowsky JK, Young RM, Chew A, Hwang WT, Hexner EO, Carreno BM, Nobles CL, Bushman FD, Parker KR, Qi Y, Satpathy AT, Chang HY, Zhao Y, Lacey SF, June CH. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481):eaba7365. https://doi.org/10.1126/science.aba7365. Epub 2020 Feb 6.
Cheng C, Pei X, Li S-W, Yang J, Li C, Tang J, et al. CRISPR/Cas9 library screening uncovered methylated PKP2 as a critical driver of lung cancer radioresistance by stabilizing β-catenin. Oncogene. 2021;40(16):2842–57.
Hartmann O, Reissland M, Maier CR, Fischer T, Prieto-Garcia C, Baluapuri A, et al. Implementation of CRISPR/Cas9 genome editing to generate murine lung cancer models that depict the mutational landscape of human disease. Front Cell Dev Biol. 2021;9(201).
Yu C, Luo D, Yu J, Zhang M, Zheng X, Xu G, Wang J, Wang H, Xu Y, Jiang K, Xu J, Ma X, Jing J, Shi H. Genome-wide CRISPR-cas9 knockout screening identifies GRB7 as a driver for MEK inhibitor resistance in KRAS mutant colon cancer. Oncogene. 2022;41(2):191–203. https://doi.org/10.1038/s41388-021-02077-w. Epub 2021 Oct 30.
Gunes S, Soykan MN, Sariboyaci AE, Uysal O, Sevimli TS. Enhancement of Apo2L/TRAIL signaling pathway receptors by the activation of Klotho gene with CRISPR/Cas9 in Caco-2 colon cancer cells. Med Oncol. 2021;38(12):146.
Biagioni A, Chillà A, Del Rosso M, Fibbi G, Scavone F, Andreucci E, et al. CRISPR/Cas9 uPAR Gene Knockout Results in Tumor Growth Inhibition, EGFR Downregulation and Induction of Stemness Markers in Melanoma and Colon Carcinoma Cell Lines. Front Oncol. 2021;11:1705.
Rushworth LK, Harle V, Repiscak P, Clark W, Shaw R, Hall H, Bushell M, Leung HY, Patel R. In vivo CRISPR/Cas9 knockout screen: TCEAL1 silencing enhances docetaxel efficacy in prostate cancer. Life Sci Alliance. 2020;3(12):e202000770. https://doi.org/10.26508/lsa.202000770.
Jiang FN, Liang YX, Wei W, Zou CY, Chen GX, Wan YP, et al. Functional classification of prostate cancer–associated miRNAs through CRISPR/Cas9–mediated gene knockout. Mol Med Rep. 2020;22(5):3777–84.
Palit SAL, van Dorp J, Vis D, Lieftink C, Linder S, Beijersbergen R, et al. A kinome-centered CRISPR-Cas9 screen identifies activated BRAF to modulate enzalutamide resistance with potential therapeutic implications in BRAF-mutated prostate cancer. Sci Rep. 2021;11(1):13683.
Batır MB, Şahin E, Çam FS. Evaluation of the CRISPR/Cas9 directed mutant TP53 gene repairing effect in human prostate cancer cell line PC-3. Mol Biol Rep. 2019;46(6):6471–84.
Chen J, Huang Y, Tang Z, Li M, Ling X, Liao J, et al. Genome-Scale CRISPR-Cas9 transcriptional activation screening in metformin resistance related gene of prostate cancer. Front Cell Dev Biol. 2021;8(1726).
Zhu W, Liu C, Lu T, Zhang Y, Zhang S, Chen Q, et al. Knockout of EGFL6 by CRISPR/Cas9 mediated inhibition of tumor angiogenesis in ovarian cancer. Front Oncol. 2020;10:1451.
Ouyang Q, Liu Y, Tan J, Li J, Yang D, Zeng F, et al. Loss of ZNF587B and SULF1 contributed to cisplatin resistance in ovarian cancer cell lines based on Genome-scale CRISPR/Cas9 screening. Am J Cancer Res. 2019;9(5):988–98.
Ling K, Jiang L, Liang S, Kwong J, Yang L, Li Y, et al. Correction to: Nanog interaction with the androgen receptor signaling axis induce ovarian cancer stem cell regulation: studies based on the CRISPR/Cas9 system. J Ovarian Res. 2019;12(1):11.
Xu M, Huang S, Chen J, Xu W, Xiang R, Piao Y, et al. Cytotoxic lymphocytes-related gene ITK from a systematic CRISPR screen could predict prognosis of ovarian cancer patients with distant metastasis. J Transl Med. 2021;19(1):447.
Wen D, Huang R, Xie J, Wen H, Lin S. [Construction of ACT-1 human undifferentiated thyroid cancer cell line with knockout of axis inhibition protein 1 (AXIN1) gene using CRISPR/Cas9]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020;36(5):419–24.
Bao MH-R, Yang C, Tse AP-W, Wei L, Lee D, Zhang MS, et al. Genome-wide CRISPR-Cas9 knockout library screening identified PTPMT1 in cardiolipin synthesis is crucial to survival in hypoxia in liver cancer. Cell Rep. 2021;34(4):108676.
An SB, Yang K, Kim CW, Choi SH, Kim E, Kim SD, et al. Longitudinal Imaging of Liver Cancer Using MicroCT and Nanoparticle Contrast Agents in CRISPR/Cas9-Induced Liver Cancer Mouse Model. Technol Cancer Res Treat. 2021;20:15330338211016466.
Hu W, Guo G, Chi Y, Li F. Construction of Traf3 knockout liver cancer cell line using CRISPR/Cas9 system. J Cell Biochem. 2019;120(9):14908–15.
Elkhadragy L, Regan MR, W MT, Goli KD, Patel S, Garcia K, et al. Generation of genetically tailored porcine liver cancer cells by CRISPR/Cas9 editing. Biotechniques. 2021;70(1):37–48.
Brusson M, Miccio A. Genome editing approaches to β-hemoglobinopathies. Prog Mol Biol Transl Sci. 2021;182:153–83.
Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N Engl J Med. 2021;384(3):252–60.
Modarai SR, Kanda S, Bloh K, Opdenaker LM, Kmiec EB. Precise and error-prone CRISPR-directed gene editing activity in human CD34+ cells varies widely among patient samples. Gene Ther. 2021;28(1-2):105–13.
Jing Z, Zhang N, Ding L, Wang X, Hua Y, Jiang M, et al. Safety and activity of programmed cell death-1 gene knockout engineered t cells in patients with previously treated advanced esophageal squamous cell carcinoma: An open-label, single-arm phase I study. J Clin Oncol. 2018;36(15_suppl):3054.
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–43.
Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348(6230):62–8.
Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–14.
Lu Y, Xue J, Deng T, Zhou X, Yu K, Huang M, et al. A phase I trial of PD-1 deficient engineered T cells with CRISPR/Cas9 in patients with advanced non-small cell lung cancer. J Clin Oncol. 2018;36(15_suppl):3050-.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28.
Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(2):123–35.
Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26(5):732–40.
Yi L, Li J. CRISPR-Cas9 therapeutics in cancer: promising strategies and present challenges. Biochim Biophys Acta. 2016;1866(2):197–207.
Kim SY, Park C, Kim HJ, Park J, Hwang J, Kim JI, et al. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology. 2015;148(1):137–47.e9.
Quan L, Chen X, Liu A, Zhang Y, Guo X, Yan S, et al. PD-1 Blockade Can Restore Functions of T-Cells in Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma In Vitro. PLoS One. 2015;10(9):e0136476.
Louis CU, Straathof K, Bollard CM, Ennamuri S, Gerken C, Lopez TT, et al. Adoptive transfer of EBV-specific T cells results in sustained clinical responses in patients with locoregional nasopharyngeal carcinoma. J Immunother. 2010;33(9):983–90.
Lloyd A, Vickery ON, Laugel B. Beyond the antigen receptor: editing the genome of T-cells for cancer adoptive cellular therapies. Front Immunol. 2013;4:221.
Su S, Hu B, Shao J, Shen B, Du J, Du Y, et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep. 2016;6:20070.
Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–63.
Zhang X, Zhang D, Thompson JA, Chen SC, Huang Z, Jennings L, et al. Gene correction of the CLN3 c.175G>A variant in patient-derived induced pluripotent stem cells prevents pathological changes in retinal organoids. Mol Genet Genomic Med. 2021;9(3):e1601.
Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, et al. CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N Engl J Med. 2019;381(13):1240–7.
Bruno B, Wäsch R, Engelhardt M, Gay F, Giaccone L, D’Agostino M, et al. European Myeloma Network perspective on CAR T-Cell therapies for multiple myeloma. Haematologica. 2021;106(8):2054–65.
Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N Engl J Med. 2021;385(6):493–502.
Stephenson AA, Flanigan KM. Gene editing and modulation for Duchenne muscular dystrophy. Prog Mol Biol Transl Sci. 2021;182:225–55.
Makarova KS, Koonin EV. Annotation and classification of CRISPR-Cas systems. Methods Mol Biol. 2015;1311:47–75.
Colque-Navarro P, Jacobsson G, Andersson R, Flock J-I, Möllby R. Levels of antibody against 11 Staphylococcus aureus antigens in a healthy population. Clin Vaccine Immunol. 2010;17(7):1117–23.
Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25(2):249–54.
Crudele JM, Chamberlain JS. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat Commun. 2018;9(1):3497.
Kotagama OW, Jayasinghe CD, Abeysinghe T. Era of Genomic Medicine: A Narrative Review on CRISPR Technology as a Potential Therapeutic Tool for Human Diseases. Biomed Res Int. 2019;2019:1369682-.
Geurts MH, de Poel E, Amatngalim GD, Oka R, Meijers FM, Kruisselbrink E, et al CRISPR-Based Adenine Editors. Correct Nonsense Mutations in a Cystic Fibrosis Organoid Biobank. Cell Stem Cell. 2020;26(4):503-10.e7.
Pavani G, Fabiano A, Laurent M, Amor F, Cantelli E, Chalumeau A, et al. Correction of β-thalassemia by CRISPR/Cas9 editing of the α-globin locus in human hematopoietic stem cells. Blood advances. 2021;5(5):1137–53.
Demirci S, Leonard A, Haro-Mora JJ, Uchida N, Tisdale JF. CRISPR/Cas9 for Sickle Cell Disease: Applications, Future Possibilities, and Challenges. Adv Exp Med Biol. 2019;1144:37-52.
Park SH, Bao G. CRISPR/Cas9 gene editing for curing sickle cell disease. Transfusion Apheresis Sci. 2021;60(1):103060.
Bertolin J, Sánchez V, Ribera A, Jaén ML, Garcia M, Pujol A, et al. Treatment of skeletal and non-skeletal alterations of Mucopolysaccharidosis type IVA by AAV-mediated gene therapy. Nat Commun. 2021;12(1):5343.
Rysenkova KD, Semina EV, Karagyaur MN, Shmakova AA, Dyikanov DT, Vasiluev PA, et al. CRISPR/Cas9 nickase mediated targeting of urokinase receptor gene inhibits neuroblastoma cell proliferation. Oncotarget. 2018;9(50):29414–30.
Valletta S, Dolatshad H, Bartenstein M, Yip BH, Bello E, Gordon S, et al. ASXL1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts. Oncotarget. 2015;6(42):44061–71.
Benhar I, London A, Schwartz M. The privileged immunity of immune privileged organs: the case of the eye. Front Immunol. 2012;3:296-.
Hong S, Van Kaer L. Immune privilege: keeping an eye on natural killer T cells. J Exp Med. 1999;190(9):1197–200.
Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix CC. CNS immune privilege: hiding in plain sight. Immunol Rev. 2006;213:48–65.
Kanellopoulos-Langevin C, Caucheteux SM, Verbeke P, Ojcius DM. Tolerance of the fetus by the maternal immune system: role of inflammatory mediators at the feto-maternal interface. Reprod Biol Endocrinol. 2003;1:121-.
Zhao S, Zhu W, Xue S, Han D. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol. 2014;11(5):428–37.
Leroy BP, Birch DG, Duncan JL, Lam BL, Koenekoop RK, Porto FBO, et al. LEBER CONGENITAL AMAUROSIS DUE TO CEP290 MUTATIONS-SEVERE VISION IMPAIRMENT WITH A HIGH UNMET MEDICAL NEED: A Review. Retina. 2021;41(5):898–907.
Chung SH, Sin T-N, Ngo T, Yiu G. CRISPR Technology for Ocular Angiogenesis. Frontiers in Genome Editing. 2020;2:29.
Jain A, Zode G, Kasetti RB, Ran FA, Yan W, Sharma TP, et al. CRISPR-Cas9-based treatment of myocilin-associated glaucoma. Proc Natl Acad Sci U S A. 2017;114(42):11199–204.
Alanis-Lobato G, Zohren J, McCarthy A, Fogarty NME, Kubikova N, Hardman E, et al. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc Natl Acad Sci U S A. 2021;118:22.
Zuccaro MV, Xu J, Mitchell C, Marin D, Zimmerman R, Rana B, et al. Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell. 2020;183(6):1650–64.e15.
Sun J, Lu Y, Nozawa K, Xu Z, Morohoshi A, Castaneda JM, et al. CRISPR/Cas9-based genome editing in mice uncovers 13 testis- or epididymis-enriched genes individually dispensable for male reproduction†. Biol Reprod. 2020;103(2):183–94.
Lu Y, Oura S, Matsumura T, Oji A, Sakurai N, Fujihara Y, et al. CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice†. Biol Reprod. 2019;101(2):501–11.
Park CY, Halevy T, Lee DR, Sung JJ, Lee JS, Yanuka O, et al. Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep. 2015;13(2):234–41.
Lessard S, Francioli L, Alfoldi J, Tardif J-C, Ellinor PT, MacArthur DG, et al. Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci. Proceedings of the National Academy of Sciences. 2017;114(52):E11257-E66.
Li D, Zhou H, Zeng X. Battling CRISPR-Cas9 off-target genome editing. Cell Biol Toxicol. 2019;35(5):403–6.
Schaefer KA, Wu W-H, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR–Cas9 editing in vivo. Nat Methods. 2017;14(6):547–8.
Kang S-H, Lee W-j, An J-H, Lee J-H, Kim Y-H, Kim H, et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat Commun. 2020;11(1):3596.
Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–6.
Barman A, Deb B, Chakraborty S. A glance at genome editing with CRISPR-Cas9 technology. Current genetics. 2020;66(3):447–62.
Uniyal AP, Mansotra K, Yadav SK, Kumar V. An overview of designing and selection of sgRNAs for precise genome editing by the CRISPR-Cas9 system in plants. 3 Biotech. 2019;9(6):223-.
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84.
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6.
Lv J, Wu S, Wei R, Li Y, Jin J, Mu Y, et al. The length of guide RNA and target DNA heteroduplex effects on CRISPR/Cas9 mediated genome editing efficiency in porcine cells. J Vet Sci. 2019;20(3):e23-e.
Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37(3):224–6.
Park J, Lim K, Kim JS, Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33(2):286–8. https://doi.org/10.1093/bioinformatics/btw561. Epub 2016 Aug 24.
Guell M, Yang L, Church GM. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics. 2014;30(20):2968–70.
Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168.
Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, van Steensel B. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol Cell. 2018;70(5):801-13 e6.
Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015;31(22):3676–8.
Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One. 2014;9(9):e108424.
Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171-W4.
Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11(2):122–3.
Blin K, Pedersen LE, Weber T, Lee SY. CRISPy-web: An online resource to design sgRNAs for CRISPR applications. Synth Syst Biotechnol. 2016;1(2):118–21.
Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol Plant. 2017;10(3):530–2.
Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids. 2014;3:e214.
Wong N, Liu W, Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218.
Park J, Bae S, Kim JS. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31(24):4014–6.
Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–5.
Prykhozhij SV, Rajan V, Gaston D, Berman JN. CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One. 2015;10(3):e0119372.
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9.
Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402.
Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, et al. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell. 2016;167(7):1829–38.e9.
Zhu Y, Gao A, Zhan Q, Wang Y, Feng H, Liu S, et al. Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins. Mol Cell. 2019;74(2):296–309.e7.
Wiegand T, Karambelkar S, Bondy-Denomy J, Wiedenheft B. Structures and Strategies of Anti-CRISPR-Mediated Immune Suppression. Annu Rev Microbiol. 2020;74:21–37.
Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493(7432):429–32.
Shin J, Jiang F, Liu J-J, Bray NL, Rauch BJ, Baik SH, et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv. 2017;3(7):e1701620-e.
Davidson AR, Lu WT, Stanley SY, Wang J, Mejdani M, Trost CN, et al. Anti-CRISPRs: Protein Inhibitors of CRISPR-Cas Systems. Annual review of biochemistry. 2020;89:309–32.
Stanley SY, Borges AL, Chen KH, Swaney DL, Krogan NJ, Bondy-Denomy J, et al. Anti-CRISPR-Associated Proteins Are Crucial Repressors of Anti-CRISPR Transcription. Cell. 2019;178(6):1452–64.e13.
Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, et al. Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity. Cell. 2018;174(4):908–16.e12.
Taati Moghadam M, Amirmozafari N, Shariati A, Hallajzadeh M, Mirkalantari S, Khoshbayan A, et al. How Phages Overcome the Challenges of Drug Resistant Bacteria in Clinical Infections. Infect Drug Resist. 2020;13:45–61.
Sinha S, Barbosa K, Cheng K, Leiserson MDM, Jain P, Deshpande A, et al. A systematic genome-wide mapping of oncogenic mutation selection during CRISPR-Cas9 genome editing. Nat Commun. 2021;12(1):6512.
Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8(1):14454.
Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16(1):45–56.
Alsaihati BA, Ho K-L, Watson J, Feng Y, Wang T, Dobbin KK, et al. Canine tumor mutational burden is correlated with TP53 mutation across tumor types and breeds. Nat Commun. 2021;12(1):4670.
Biagioni A, Laurenzana A, Margheri F, Chillà A, Fibbi G, Del Rosso M. Delivery systems of CRISPR/Cas9-based cancer gene therapy. Journal of Biological Engineering. 2018;12(1):33.
Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differentiation. 2018;25(1):154–60.
Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nature protocols. 2018;13(1):195–215.
Lentsch E, Li L, Pfeffer S, Ekici AB, Taher L, Pilarsky C, et al. CRISPR/Cas9-Mediated Knock-Out of Kras(G12D) Mutated Pancreatic Cancer Cell Lines. Int J Mol Sci. 2019;20(22).
Gonatopoulos-Pournatzis T, Aregger M, Brown KR, Farhangmehr S, Braunschweig U, Ward HN, et al. Genetic interaction mapping and exon-resolution functional genomics with a hybrid Cas9-Cas12a platform. Nat Biotechnol. 2020;38(5):638–48.
Aregger M, Xing K, Gonatopoulos-Pournatzis T. Application of CHyMErA Cas9-Cas12a combinatorial genome-editing platform for genetic interaction mapping and gene fragment deletion screening. Nat Protoc. 2021;16(10):4722–65.
Johansson-Åkhe I, Mirabello C, Wallner B. Predicting protein-peptide interaction sites using distant protein complexes as structural templates. Sci Rep. 2019;9(1):4267.
Yip BH. Recent Advances in CRISPR/Cas9 Delivery Strategies. Biomolecules. 2020;10(6):839.
Mout R, Ray M, Lee Y-W, Scaletti F, Rotello VM. In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges. Bioconjug Chem. 2017;28(4):880–4.
van Haasteren J, Li J, Scheideler OJ, Murthy N, Schaffer DV. The delivery challenge: fulfilling the promise of therapeutic genome editing. Nat Biotechnol. 2020;38(7):845–55.
Chen F, Alphonse M, Liu Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. WIREs Nanomedicine Nanobiotechnology. 2020;12(3):e1609.
Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–33.
Martinez-Lage M, Puig-Serra P, Menendez P, Torres-Ruiz R, Rodriguez-Perales S. CRISPR/Cas9 for Cancer Therapy: Hopes and Challenges. Biomedicines. 2018;6(4):105. https://doi.org/10.3390/biomedicines6040105.
Chen F, Alphonse M, Liu Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;12(3):e1609.
Alton E, Armstrong DK, Ashby D, Bayfield KJ, Bilton D, Bloomfield EV, et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir Med. 2015;3(9):684–91.
Kaczmarek JC, Kowalski PS, Anderson DG. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome medicine. 2017;9(1):60.
de Jong OG, Murphy DE, Mäger I, Willms E, Garcia-Guerra A, Gitz-Francois JJ, et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat Commun. 2020;11(1):1113.
Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–3.
Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–74.
Yu Q-h, Wang B, Li N, Tang Y, Yang S, Yang T, et al. CRISPR/Cas9-induced Targeted Mutagenesis and Gene Replacement to Generate Long-shelf Life Tomato Lines. Sci Rep. 2017;7(1):11874.
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91.
Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol. 2016;34(8):807–8.
Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11(3):2452–8.
Alapati D, Zacharias WJ, Hartman HA, Rossidis AC, Stratigis JD, Ahn NJ, et al. In utero gene editing for monogenic lung disease. Sci Transl Med. 2019;11:488.
Ricobaraza A, Gonzalez-Aparicio M, Mora-Jimenez L, Lumbreras S, Hernandez-Alcoceba R. High-Capacity Adenoviral Vectors: Expanding the Scope of Gene Therapy. Int J Mol Sci. 2020;21(10):3643.
Bak RO, Dever DP, Reinisch A, Cruz Hernandez D, Majeti R, Porteus MH. Correction: Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AAV6. Elife. 2018;7.
Wang D, Zhang F, Gao G. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell. 2020;181(1):136–50.
Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–7.
Mingozzi F, High KA. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122(1):23–36.
Grieger JC, Samulski RJ. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol. 2005;79(15):9933–44.
Rosenblum D, Gutkin A, Kedmi R, Ramishetti S, Veiga N, Jacobi AM, Schubert MS, Friedmann-Morvinski D, Cohen ZR, Behlke MA, Lieberman J, Peer D. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv. 2020;6(47):eabc9450. https://doi.org/10.1126/sciadv.abc9450.
Ciancio G, Burke GW, Miller J. Current treatment practice in immunosuppression. Expert Opin Pharmacother. 2000;1(7):1307–30.
Follenzi A, Santambrogio L, Annoni A. Immune responses to lentiviral vectors. Curr Gene Ther. 2007;7(5):306–15.
Carter M, Shieh J. Chapter 11 - Gene Delivery Strategies. In: Carter M, Shieh J, editors. Guide to Research Techniques in Neuroscience (Second Edition). San Diego: Academic Press; 2015. p. 239-52.
Hashimoto M, Takemoto T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci Rep. 2015;5(1):11315.
Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci Rep. 2016;6(1):20611.
Rosenblum D, Gutkin A, Dammes N, Peer D. Progress and challenges towards CRISPR/Cas clinical translation. Adv Drug Deliv Rev. 2020;154-155:176–86.
Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26.
Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–3.
D’Astolfo Diego S, Pagliero Romina J, Pras A, Karthaus Wouter R, Clevers H, Prasad V, et al. Efficient Intracellular Delivery of Native Proteins Cell. 2015;161(3):674–90.
Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–57.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–78.
Shinwari ZK, Tanveer F, Khalil AT. Ethical Issues Regarding CRISPR Mediated Genome Editing. Curr Issues Mol Biol. 2018;26:103–10.
Bailey J. CRISPR-Mediated Gene Editing: Scientific and Ethical Issues. Trends Biotechnol. 2019;37(9):920–1.
Ishii T. Germline genome-editing research and its socioethical implications. Trends Mol Med. 2015;21(8):473–81.
Peng R, Lin G, Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. Febs j. 2016;283(7):1218–31.
Guo X, Li XJ. Targeted genome editing in primate embryos. Cell Res. 2015;25(7):767–8.
Tu Z, Yang W, Yan S, Yin A, Gao J, Liu X, et al. Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci Rep. 2017;7:42081.
Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nature methods. 2017;14(6):547–8.
Zischewski J, Fischer R, Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv. 2017;35(1):95–104.
Brokowski C, Adli M. CRISPR Ethics: Moral Considerations for Applications of a Powerful Tool. J Mol Biol. 2019;431(1):88–101.
Niemiec E, Howard HC. Ethical issues related to research on genome editing in human embryos. Computational Structural Biotechnology Journal. 2020;18:887–96.
Howard HC, van El CG, Forzano F, Radojkovic D, Rial-Sebbag E, de Wert G, et al. One small edit for humans, one giant edit for humankind? Points and questions to consider for a responsible way forward for gene editing in humans. Eur J Hum Genet. 2018;26(1):1–11.
Araki M, Ishii T. International regulatory landscape and integration of corrective genome editing into in vitro fertilization. Reprod Biol Endocrinol. 2014;12:108.
Ledford H. Enzyme tweak boosts precision of CRISPR genome edits. Nature. 2016.
Ledford H. Biologists create more precise molecular scissors for genome editing. Nature. 2015.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
MT and AbB designed and supervised the study. SGF, AB, BMH and MFR wrote the draft and revised it. EJ, AS, BSI, PJJ, and AZ collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent of publication
Not applicable.
Competing interests
The authors declare they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Rasul, M.F., Hussen, B.M., Salihi, A. et al. Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy. Mol Cancer 21, 64 (2022). https://doi.org/10.1186/s12943-021-01487-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-021-01487-4