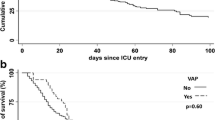
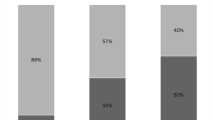
Abstract
Background
Description and comparison of bacterial characteristics of ventilator-associated pneumonia (VAP) between critically ill intensive care unit (ICU) patients with COVID-19-positive, COVID + ; and non-COVID-19, COVID-.
Methods
Retrospective, observational, multicenter study that focused on French patients during the first wave of the pandemic (March–April 2020).
Results
935 patients with identification of at least one bacteriologically proven VAP were included (including 802 COVID +). Among Gram-positive bacteria, S. aureus accounted for more than two-thirds of the bacteria involved, followed by Streptococcaceae and enterococci without difference between clinical groups regarding antibiotic resistance. Among Gram-negative bacteria, Klebsiella spp. was the most frequently observed bacterial genus in both groups, with K. oxytoca overrepresented in the COVID- group (14.3% vs. 5.3%; p < 0.05). Cotrimoxazole-resistant bacteria were over-observed in the COVID + group (18.5% vs. 6.1%; p <0.05), and after stratification for K. pneumoniae (39.6% vs. 0%; p <0.05). In contrast, overrepresentation of aminoglycoside-resistant strains was observed in the COVID- group (20% vs. 13.9%; p < 0.01). Pseudomonas sp. was more frequently isolated from COVID + VAPs (23.9% vs. 16.7%; p <0.01) but in COVID- showed more carbapenem resistance (11.1% vs. 0.8%; p <0.05) and greater resistance to at least two aminoglycosides (11.8% vs. 1.4%; p < 0.05) and to quinolones (53.6% vs. 7.0%; p <0.05). These patients were more frequently infected with multidrug-resistant bacteria than COVID + (40.1% vs. 13.8%; p < 0.01).
Conclusions
The present study demonstrated that the bacterial epidemiology and antibiotic resistance of VAP in COVID + is different from that of COVID- patients. These features call for further study to tailor antibiotic therapies in VAP patients.
Similar content being viewed by others
Introduction
Ventilator-associated pneumonia (VAP) is the second most common nosocomial infection and remains the leading cause of death in critically ill patients [1]. Prior to COVID, the risk of VAP was estimated to be 1.5% per mechanical-ventilation-day, decreasing to < 0.5% daily after 2 weeks of mechanical ventilation, and was associated with a 7 day increase in hospital length of stay and a $40,000 increase in healthcare costs [2].
Since the beginning of the COVID pandemic, significant disparities in prevalence of VAP have been demonstrated. For example, a prevalence rate of 29% in Italy was reported in the first wave, while other countries reported an incidence of 79% (HR of 2.1 to COVID-negative patients) [3, 4]. In this context, it cannot be ruled out that the incidence ratio and the representation of the epidemiology of bacterial superinfections are biased.
Before COVID, the microbial etiology of VAPs varied according to duration of ventilation, length of hospital stays and the local microbial ecology (reflecting local antibiotic prescribing habits). VAPs were caused by Gram-negative bacteria (GNB, mainly Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa), accounting for 50–80% of cases, to a greater extent than Gram-positive bacteria (GPB, Staphylococcus aureus and Streptococcaceae), which accounted for < 40% of cases [5, 6].
The common problem of antimicrobial stewardship for bacterial co-/super-infection in patients with severe SARSCoV2 infection, particularly those requiring intensive care and ventilation, remains a challenge. The French Society of Anesthesia and Resuscitation recommended that these critical care and ventilation issues could be avoided by optimizing the clinical management of patients [7, 8]. The society emphasized the value of limiting the duration of ventilation and of requesting microbial specimens whenever possible in cases of clinical suspicion requiring any type of respiratory specimen for the diagnosis of microbial infections [9, 10]. Optimization of antibiotic therapy in suspected VAP, which is crucial for proper clinical management of the disease, depends on the risk factor for the carriage of multiresistant bacteria (MDR), and cannot be limited to an unwarranted combination of antibiotics [8, 11].
The objective of this study, conducted at the very beginning of the French COVID pandemic, was to compare the bacterial characteristics of VAP in critically ill patients infected with COVID-19, COVID + ; and uninfected, COVID-, admitted to intensive care units (ICU) during the same period.
Methods
Design
This study is a retrospective, observational, multicenter study that focused on French patients during the first French wave of the pandemic. All voluntary-based participating centers were contacted through professional networks (Collège de Bactériologie, Virologie et Hygiène Hospitalière, and Société Française de Microbiologie, SFM). The investigators retrospectively analyzed the clinical records to complete an electronic clinical record file that was blinded by the principal investigators.
Participants
This study included all patients hospitalized in French intensive care units (private, general, military, and university hospitals) who required mechanical ventilation during the first month of the first French wave of the COVID pandemic (March to April 2020). All patients in whom the diagnosis of VAP was established bacteriologically (“identification of a bacterium at a concentration above a threshold depending on the nature of the respiratory specimen in which it was identified”) before introduction of antibiotics were included [12]. The application of these thresholds allowed to differentiate between infection (above the threshold) and colonization (below the threshold). The CDC VAP 2020 diagnostic guidelines, adapted in the present study, required at least one of the following [13]: i/Fever ≥ 38.5 ℃; ii/Leukopenia or leukocytosis (white blood cell count ≤ 4 G/L or ≥ 12G/L); and two of the following: i/Change in sputum character, or newly purulent sputum; ii/Increased airway secretions or need for aspiration; iii/New or worsening cough, tachypnea, or dyspnea; iv/Noisy breathing or abnormal bronchial noise; v/Poor gas exchange possibly requiring more oxygen or the use of a ventilator. In addition, and as mentioned in the above guidelines, it should be noted that to be considered, the patient must also have been on mechanical ventilation for at least two calendar days, and mechanical ventilation must be present either on the day of diagnosis of pneumonia or the day before.
Microbiological diagnosis
All samples were analyzed according to SFM standards, and concentrations of the respective bacteria were interpreted according to the recommended thresholds (105 CFU/mL for aspirates, 104 CFU/mL for bronchoalveolar lavage, 103 CFU/mL for the protected distal sample). Antibiotic susceptibility testing (AST) (disc diffusion or liquid dilution) was interpreted according to the thresholds and recommendations of the European Committee for Antimicrobial Susceptibility Testing (EUCAST).
Objective of the study and evaluation criteria
The primary objective was to evaluate the prevalence of above-the-threshold bacteria responsible for superinfection in mechanical ventilation-requiring COVID +. The secondary objectives were to describe the AST of these bacteria before any antibiotic therapy.
According to the French Society of Hospital Hygiene, MDR included methicillin-resistant S. aureus (MRSA), extended-spectrum-βlactamase/Carbapenemase-producing Enterobacterales (E-ESBL/CPE), ceftazidime-resistant P. aeruginosa, carbapenem-resistant Acinetobacter baumannii (CRAB), and glycopeptide-resistant Enterococcus faecium/faecalis (GRE) [14, 15].
Statistical methods
Descriptive statistics were expressed as percentages for categorical variables and as mean with standard deviation or median with interquartile range for continuous variables. Demographic characteristics, underlying conditions, and clinical characteristics were functions of COVID status using Pearson’s chi-square test or Fisher’s exact test (GraphPad Prism v9.0.0). Results with p < 0.05 were considered as significant.
Ethical procedure
All samples were pseudonymized during completion of the eCRF by local investigators prior to collection and analyses by the national coordinators. This study was authorized by the Commission Nationale de l'Informatique et des Libertés (CNIL, n°920232) and informed consent was waived for this particular study (due to the context of the first wave).
Results
Clinical and demographic characteristics of patients at diagnosis
During the study period, 935 patients with identification of at least one bacteriologically proven VAP from 65 hospital centers (including 802 COVID + ; Table 1) were included. Regarding age distribution, COVID + were significantly older than COVID- (70.0 vs. 60.4 years; p < 0.01), without difference in sex (3.7 men per woman). As expected, the proportion of patients with a risk factor for severe COVID (as defined by French national authorities, Haut Conseil de la Santé Publique, March 14, 2020) was higher in COVID + than in COVID- (72.7% vs. 62.1%; p < 0.05), with more cardiovascular disease (34.9% vs. 29.7%; p < 0.05), diabetes mellitus (16.8% vs. 6.1%; p < 0.01) but less chronic respiratory disease (and/or severe liver disease; 0.1% vs. 4.7%; p < 0.01) in COVID + [16]. Of note, the number of risk factors per patient was similar between groups (1.69 vs. 1.80). VAP was discovered earlier in the clinical history of COVID + with a time lapse between intubation and the first positive specimen of 8.9 days (vs. 12.3; p < 0.05) without differential time lapse between the groups regarding the time between symptomatology and specimen, or symptomatology and intubation. The nature of the respiratory samples, taken before antibiotic therapy, differed between the groups, with a greater need for distal samples (25.8 vs. 14.4; p < 0.01) in COVID +, with no difference in the number of pathogenic bacteria identified. Finally, over the course of the patients’ clinical history, COVID + had longer duration of hospitalization than COVID- (31 days vs. 22 days; p < 0.01) with a lower rate of return home (7.9% vs. 13.0%; p < 0.01) but shorter duration of intubation (15 vs. 21 days; p < 0.01). No difference in case fatality rate could be observed in this cohort (33.8% vs. 38.2%).
Bacterial epidemiology
During the study period, 950 pathogenic bacteria were considered to be involved in VAP, including 802 in COVID + (Table 2). Without difference in proportion between the two groups, GPB represented approximately one quarter of the bacteria identified (24.8% vs. 27.0%) while GNB represented most isolates (75.2% vs. 73.0%).
Among GPB, in both COVID + and COVID-, staphylococci accounted for more than two-thirds of the bacteria involved (69.9% vs. 67.5%), followed by Streptococcaceae (13.57% vs. 17.50%) including pneumococci (32.4% vs. 57.1% of Streptococcaceae) followed by enterococci (11.6% vs. 15.0% of GPB). Enterococci isolation was more frequently due to Enterococcus faecium in COVID- (3.5% vs. 33.3%; p < 0.05).
Among Enterobacterales, Klebsiella sp. was the most frequently observed bacterial genus in both groups (21.9% vs. 19.4%), with greater representation of K. oxytoca isolated in COVID- (14.3% vs. 5.3%; p < 0.05), followed by E. coli (13.0% vs. 11.6%), Enterobacter sp. (9.1% vs. 9.3%), Serratia marcescens (5.5% vs. 8.3%), Proteus sp. (4.3% vs. 7.4%), Citrobacter koseri (4.3% vs. 2.8%), Hafnia alvei (3.5% vs. 2.8%) and Morganella morganii (3.0% vs. 1.9%).
Among non-Enterobacterales GNB, Pseudomonas sp. were more frequently isolated from VAPs of COVID + (23.9% vs. 16.7%; p < 0.01) without difference in the proportion of respective species. Other bacteria identified were Acinetobacter sp. (3.2% vs. 1.9%), Stenotrophomonas maltophilia (3.5% vs. 3.7%), Achromobacter sp. (0.7% vs. 0.9%), Haemophilus sp. (4.3% vs. 8.3%; mainly H. influenzae 92.3% vs. 100% of Haemophilus sp.) and Moraxella catarrhalis (0.3% vs. 0.5%). Burkholderia sp., Neisseria sp. and Prevotella sp. were observed only in COVID +, in contrast to Sphingomonas sp. observed only in COVID-.
Epidemiology of antibiotic resistance
Among GPB, the AST profile was characterized for all different species (enterococci, Streptococci including S. pneumoniae) without difference between the two clinical groups (Additional file 2) except for Staphylococci (Additional file 2: Table S1). For Staphylococci, no differences were observed for all the antibiotic families tested, including β-lactams, quinolones, glycopeptides, fosfomycin, fusidic acid, rifampicin, and cotrimoxazole except for aminoglycoside phenotype (0.7 vs. 7.4%; p < 0.05). Due to lack of power, none of these differences could be observed after stratification by bacterial species (coagulase-negative Staphylococci, CNS, vs S. aureus).
Among Enterobacterales, no differences between clinical groups were observed with respect to quinolone resistance (17.8% vs. 9.7%) (Table 3). A trend towards overrepresentation of fosfomycin resistance (29.8% vs. 15.0%) and overrepresentation of cotrimoxazole-resistant bacteria was observed in COVID + (18.5% vs. 6.1%; p < 0.05). This was also observed when stratified by species, for K. pneumoniae (39.6% vs. 0%; p < 0.05). Overrepresentation of aminoglycoside-resistant strains was observed in COVID- (13.9% vs. 20%; p < 0.01), due mainly to an association of tobramycin and amikacin resistance (3.5% vs. 11.4%; p < 0.01). Regarding beta-lactams, in COVID-, E. coli was more frequently resistant to amoxicillin and ticarcillin (8.7% vs. 28.6%; p < 0.05), K. pneumoniae was less resistant to cephalosporins (p < 0.05) and the Enterobacter cloacae complex overexpressed more frequently its cephalosporinase (27.4% vs. 60%; p < 0.05). Considering each Enterobacterales, no difference was observed with respect to quinolone or fosfomycin resistance (p > 0.05), whereas with respect to aminoglycoside-resistance, this difference in Enterobacterales was likewise observed in S.marcescens (55.1% vs. 87.5%; p < 0.05).
Among the non-Enterobacterales GNB (Table 4), no difference between groups was observed regarding the antibiotic resistance profile of H. influenzae. Strains of P. aeruginosa also showed different phenotypes according to the clinical group: for beta-lactams, higher resistance to carbapenem (0.8% vs. 11.1%; p < 0.05); higher resistance to at least two aminoglycosides (1.4% vs. 17.7%, mainly gentamicin and tobramycin; p < 0.05), and higher resistance to fluoroquinolones in COVID- (7.1% vs. 53.6%; p < 0.05).
Distribution of multiresistant bacteria by clinical group
Out of the bacteria analyzed, approximately one-fifth (18.4%) could be considered multidrug-resistant according to the previous definition (Table 5). COVID- patients were statistically more frequently infected with multidrug-resistant bacteria than COVID + (40.1% vs. 13.8%; p < 0.01). One third of these multiresistant GNB concerned enterobacteria (n = 55; 33.1%), CPE (7/55; 12.8%) or ESBL (48/55; 87.3%). For these multiresistant GNB, differential distribution could be observed between COVID + and COVID-, mainly associated with an over-representation in COVID + (12.2% vs. 2.8%; p < 0.05). Without difference between clinical groups, MDRs were due to ceftazidime resistant Pseudomonadaceae, CRAB and MRSA. Of note, in the present cohort, no GRE could be observed at diagnosis.
Discussion
This study is one of the major French studies of data from the first French wave of the COVID-19 pandemic. Unlike most publications focusing on this period in France, this study is multicentric, summarizing data from throughout France, including fifty-six hospitals. The results allow us to understand the bacterial presentation of VAP in a totally naïve human population during the first contact of the French population with SARS-CoV-2.
Influenza-associated VAPs are primarily caused by S. pneumoniae, S. aureus, and H. influenzae, whereas SARS-Cov2 demonstrated an association with S. aureus followed by P. aeruginosa and Klebsiella sp. [17]. Our cohort confirmed that VAPs associated with COVID had a different bacterial presentation and etiology than VAPs of other origins [17]. This could have a double interest. First, these bacteria are frequently associated with multidrug resistance/high resistance, which may require careful management of infection control in order to avoid disseminating multidrug-resistant bacteria (plasmid support, especially in CPE and E-ESBL, as in the current cohort) [18]. Second, preventive treatment of VAP, as suggested by national and international guidelines, may be more complicated in COVID + patients. Indeed, these patients more often require combined and/or longer treatment. For example, influenza superinfections are more frequently (compared to COVID +) associated with bacteria that could be controlled by more manageable antibiotic therapy (beta-lactam monotherapy) [19]. It is to note that the present cohort presented with a larger number of micro-organisms frequently implicated in community-acquired pneumonias (such as Haemophilus sp. and Streptococccus pneumoniae) more than in hospital-acquired pneumonia. This point could be considered as different from other descriptions in the literature but remains coherent, with the main species (Staphyloccocus aureus for Gram-positive bacteria and Pseudomonas aeruginosa for Gram-negative bacteria) remaining found to be the most prevalent [20].
In a context of tension in health care institutions and hospitals, especially in intensive care units, it is important to analyze the proportion and spread of MDR bacteria. The present study has shown that a significant proportion (about one fifth) of MDR bacteria were observed during this period. Although this proportion is higher than usual in France, it is different from that observed by Moretti et al. in the Belgian cohort with the same proportions, but with regard to susceptible bacteria [19]. Despite these differences from one nation to another, it is crucial to observe that the presence of these MDR bacteria was higher in COVID-, which could have been due not only to the reinforcement and involvement of infection control specialists during this period, but also to the differences between this population (younger, less exposed to antibiotics, …) and the usual ICU population (both types of patients were recruited during the same period). This could also be explained by the multiple antibiotic therapies applied in patients with chronic respiratory diseases, which select resistant bacteria in all anatomical niches and in the airways, causing self-infection during mechanical ventilation [21]. Finally, it should be noted that even though it occurred 8 days after intubation, similarly to COVID- VAP in the literature, the diagnosis of VAP was earlier in COVID + patients than in COVID- patients, which could be another factor leading to a change in antibiotic susceptibility testing [22, 23]. This study also highlights the fact that some bacteria were mistakenly considered virulent enough to be treated. (Lactobacillus, Alloscavordia, alpha-hemolytic Streptococci, …), which may be associated with the globally high prescription of inappropriate antibiotic treatments during the early months of COVID pandemics [24]. In order to obtain an overview of the bacterial epidemiology of VAP during this very particular infectious period, all bacteria were considered to be involved in VAP (or as a therapeutic target) regardless of their involvement in the disease if they were alone. This decision could be justified by the fact that the clinicians could have modified their antibiotic therapy management to include this AST.
The study has limitations. First, the first French lockdown was a very particular time in 2020. At that time, clinical departments and medical analysis laboratories were faced with a heavy workload, resulting in logistical difficulties. To overcome these difficulties, while this study was conducted over a 1 month period, and considered cross-sectional at the time, some of the data were collected retrospectively during the summer and early fall of 2020. Even if the situation has evolved, this study benefits from the homogeneity of this first wave, during which only the original viral strain circulated in France, thereby limiting the possible impact of variants of concern (such as delta or omicron) [25]. Second, while this study includes many hospitals (academic and general), it cannot be considered comprehensive, insofar as the two largest teaching hospitals did not respond to the request for information. Nevertheless, this bias can be considered negligible, as the geographic distribution of the responding hospitals and their diversity provided a very good representation of the situation (Additional file 1: Fig. S1). Third, even if this study did not provide answers on the clinical and biological management of these patients during this period, the epidemiology described here could be considered robust because of its representativeness in the absence of ongoing antibiotic therapy at the time of sampling. This point is of great interest insofar as the development of antibiotic resistance during the clinical management of patients may have been considerably favored at that time, during which more than three quarters of patients worldwide received antibiotics, even in the absence of proven/suspected bacterial co-susceptibility (only 1 to 8%) [26].
Conclusion
In conclusion, this study provided data on bacterial epidemiology and antibiotic resistance during the first wave of COVID pandemics. These observation calls for further studies focusing on the specific impact of these practice changes and assessing the impact of global pandemic management (including optimization of COVID management, vaccination, and development of viral variants).
Availability of data and materials
Supporting data is available upon request from the corresponding author.
References
Torres A, Niederman MS, Chastre J, Ewig S, Fernandez-Vandellos P, Hanberger H, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European respiratory society (ERS), European society of intensive care medicine (ESICM), European society of clinical microbiology and infectious diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur Respir J. 2017;50:1700582.
Bouadma L, Sonneville R, Garrouste-Orgeas M, Darmon M, Souweine B, Voiriot G, et al. Ventilator-associated events: prevalence, outcome, and relationship with ventilator-associated pneumonia. Crit Care Med. 2015;43:1798–806.
Rouzé A, Martin-Loeches I, Povoa P, Makris D, Artigas A, Bouchereau M, et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: a European multicenter cohort study. Intensive Care Med. 2021;47:188–98.
Giacobbe DR, Battaglini D, Enrile EM, Dentone C, Vena A, Robba C, et al. Incidence and prognosis of ventilator-associated pneumonia in critically ill patients with COVID-19: a multicenter study. J Clin Med. 2021;10:555.
Rhodes NJ, Cruce CE, O’Donnell JN, Wunderink RG, Hauser AR. Resistance trends and treatment options in gram-negative ventilator-associated pneumonia. Curr Infect Dis Rep. 2018;20:3.
Luyt C-E, Hékimian G, Koulenti D, Chastre J. Microbial cause of ICU-acquired pneumonia: hospital-acquired pneumonia versus ventilator-associated pneumonia. Curr Opin Crit Care. 2018;24:332–8.
Leone M, Bouadma L, Bouhemad B, Brissaud O, Dauger S, Gibot S, et al. Pneumonies associées aux soins de réanimation. Anesthésie Réanim. 2018;4:421–41.
Leone M, Bechis C, Baumstarck K, Lefrant J-Y, Albanèse J, Jaber S, et al. De-escalation versus continuation of empirical antimicrobial treatment in severe sepsis: a multicenter non-blinded randomized noninferiority trial. Intensive Care Med. 2014;40:1399–408.
Roquilly A, Cinotti R, Jaber S, VourcH M, Pengam F, Mahe PJ, et al. Implementation of an evidence-based extubation readiness bundle in 499 brain-injured patients a before-after evaluation of a quality improvement project. Am J Respir Crit Care Med. 2013;188:958–66.
Berton DC, Kalil AC, Teixeira PJZ. Quantitative versus qualitative cultures of respiratory secretions for clinical outcomes in patients with ventilator-associated pneumonia. Cochrane Database Syst Rev. 2014. https://doi.org/10.1002/14651858.CD006482.pub4.
Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, et al. Executive summary: management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American thoracic society. Clin Infect Dis. 2016;63:575–82.
Bourlet T, Bouchara J-P, Galinier J-L. Rémic: référentiel en microbiologie médicale. Paris: Société française de microbiologie; 2022.
Modi AR, Kovacs CS. Hospital-acquired and ventilator-associated pneumonia: diagnosis, management, and prevention. CCJM. 2020;87:633–9.
SF2H. Recommandations nationales—Prévention de la transmission croisée par voie respiratoire : air ou gouttelettes. 2013. https://sf2h.net/wp-content/uploads/2013/03/SF2H_recommandations_air-ou-gouttelettes_2013.pdf
SF2H. Recommandations nationales - Prévention de la transmission croisée : précautions complémentaires contact. SF2H; 2009.
HCSP. Avis relatif à la prise en charge des patients à risque de forme sévère de COVID-19. 2020. https://www.infectiologie.com/fr/actualites/covid-19-actualites-mises-a-jour_-n.html#:~:text=Avis%20relatif%20%C3%A0%20la%20prise%20en%20charge%20des%20patients%20%C3%A0%20risque%20de%20forme%20s%C3%A9v%C3%A8re%20de%20COVID%2D19.
Shafran N, Shafran I, Ben-Zvi H, Sofer S, Sheena L, Krause I, et al. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Sci Rep. 2021;11:12703.
Calbo E, Garau J. The changing epidemiology of hospital outbreaks due to ESBL-producing Klebsiella pneumoniae: the CTX-M-15 type consolidation. Future Microbiol. 2015;10:1063–75.
Moretti M, Van Laethem J, Minini A, Pierard D, Malbrain MLNG. Ventilator-associated bacterial pneumonia in coronavirus 2019 disease, a retrospective monocentric cohort study. J Infect Chemother. 2021;27:826–33.
Chong WH, Saha BK, Ramani A, Chopra A. State-of-the-art review of secondary pulmonary infections in patients with COVID-19 pneumonia. Infection. 2021;49:591–605.
Wicky P-H, Niedermann MS, Timsit J-F. Ventilator-associated pneumonia in the era of COVID-19 pandemic: how common and what is the impact? Crit Care. 2021;25:153.
Cook D, Walter S, Freitag A, Guyatt G, Devitt H, Meade M, et al. Adjudicating ventilator-associated pneumonia in a randomized trial of critically ill patients. J Crit Care. 1998;13:159–63.
Denis J-B, Lehingue S, Pauly V, Cassir N, Gainnier M, Léone M, et al. Multidrug-resistant Pseudomonas aeruginosa and mortality in mechanically ventilated ICU patients. Am J Infect Control. 2019;47:1059–64.
Calderón-Parra J, Muiño-Miguez A, Bendala-Estrada AD, Ramos-Martínez A, Muñez-Rubio E, Fernández Carracedo E, et al. Inappropriate antibiotic use in the COVID-19 era: factors associated with inappropriate prescribing and secondary complications analysis of the registry SEMI-COVID. PLoS ONE. 2021;16:e0251340.
Gaymard A, Bosetti P, Feri A, Destras G, Enouf V, Andronico A, et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Euro Surveill. 2021. https://doi.org/10.2807/1560-7917.ES.2021.26.9.2100133.
Zhang G, Hu C, Luo L, Fang F, Chen Y, Li J, et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan China. J Clin Virol. 2020;127:104364.
Acknowledgements
The authors would like to thank Alexandre Brun for his help in analyzing the data as a pharmacy student. We thank J. Arsham, American translator, for his proofreading and revision of the original English manuscript. Parts of these results were previously presented at the French national congress (eRICAI2020; December 15, 2020).
COVAP study group: Sahar Abdallah; Hopital Simon Veil, Eaubonne, France, Corentine Alauzet; CHU Nancy, Service de Microbiologie, Vandoeuvre-les-Nancy, France, Tom Alix; CHU Nancy, Service de Microbiologie, Vandoeuvre-les-Nancy, France, Kahina Allouche; AP-HP Hopital Bichat, Paris, France, Marlène Amara; Service de Biologie- Unité de microbiologie, Centre Hospitalier de Versailles André Mignot, Versailles, France, Florence Anglade; CHU Clermont-Ferrand, Département de Maladies Infectieuses et Tropicales, Clermont-Ferrand, France, Nadia Anguel; AP-HP Kremlin Bicetre, Service de Médecine Intensive Réanimation, Bicetre, France, Laurence Armand-Lefevre; AP-HP Hopital Bichat, Service de Bactériologie, Paris, France, Francois Barbier; CH Orléans, Service de la Réanimation Médicale, Orléans, France, Clémence Beauruelle; CHU Brest, Unité de Bactériologie, Brest, France, Pascale Bemer; CHU Nantes, Department of Bacteriology, Nantes, France, Hanaa Benmansour; AP-HP Lariboisiere, Laboratoire de Bactériologie, Paris, France, Béatrice Bercot; AP-HP Saint Louis, Service de bactériologie, Paris, France, Ludovic Bergon; CHI Castres-Mazamet, Service de Biologie Médicale, Castres, France, Dominique Bertei; CH de la Miséricorde, Ajaccio, France, Marc Berthon; CH Nevers, Service de Réanimation Polyvalente, Nevers, France, Pascal Beuret; CH , Service de Réanimation et Soins Continus, Roanne, France, Léa Bientz; CHU Bordeaux, Laboratoire de Bactériologie, Bordeaux, France, Laura Billon; CH Albi, Laboratoire, Albi, France, Aurore Bousquet; Hopital d'Instruction des Armees Begin, Département de Biologie Médicale, Saint-Mandé, France, Amélie Brousse; CH Agen-Nérac, Agen, France, Lauranne Broutin; CHU Poitiers, Infectious Agents Department. Bacteriology and Infection Control Laboratory, 2 rue de la Milétrie, 86021, Poitiers, France, Fabrice Bruneel; Centre Hospitalier de Versailles André Mignot, Service de réanimation, Versailles, France, Anne Cady; CHBA Vannes, Laboratoire de Biologie, Vannes, France, Francois Camelena; AP-HP Saint Louis, Service de bactériologie, Paris, France, Amélie Carrer-Causeret; CH Gonesse, Laboratoire de Biologie Médicale, Gonesse, France, Yvan Caspar; CHU Grenoble Alpes, Laboratoire de Bactériologie, Grenoble, France, Lotfi Chemali; AP-HP Hopital Bichat, Service de Bactériologie, Paris, France, Anne Christine Jaouen; CH de la Cote Basque, Laboratoire de Biologie médicale, Boyonne, France, Théophile Cocherie; Service de Biologie- Unité de microbiologie, Centre Hospitalier de Versailles André Mignot, Versailles, France, Aurélie Cointe; AP-HP Robert Debré, Service de Microbiologie, Paris, France, Stephane Corvec; CHU Nantes, Department of Bacteriology, Nantes, France, Laura Courtellemont; CH Orléans, Laboratoire de Microbiologie, Orléans, France, Gaelle Cuzon; AP-HP Kremlin Bicetre, Service de Bactériologie Hygiène, Bicetre, France, Anne Dao; CH Beziers, Service de Biologie Médicale, Beziers, France, Agathe Delbove; CHBA Vannes, Service de Réanimation polyvalente, Vannes, France, Camille D’Humieres; AP-HP Hopital Bichat, Service de Bactériologie, Paris, France, Laura Djamdjian; CH Gonesse, Laboratoire de Biologie Médicale, Gonesse, France, Alexandra Doloy; AP-HP Cochin, Service de Bactériologie, Paris, France, Joséphine Dorin; CH Antibes-Juan-les-Pins, Laboratoire de Biologie, Antibes, France, Yann Dumont; CHU Montpellier, Laboratoire de Bactériologie, Montpellier, France, Bruno Dumoulard; CH Cambrai, Cambrai, France, Faten El Sayed; AP-HP Ambroise Paré, Département de Microbiologie, Boulogne-Billancourt, France, Marie-Sarah Fangous; AP-HP Kremlin Bicetre, Service de Bactériologie Hygiène, Bicetre, France, Laurent Favier; CH Beziers, Réanimation, Beziers, France, Alexis Ferre; Centre Hospitalier de Versailles André Mignot, Service de réanimation, Versailles, France, Nicolas Fortineau; AP-HP Kremlin Bicetre, Service de Bactériologie Hygiène, Bicetre, France, Juliette Francois; CH Antibes-Juan-les-Pins, Réanimation polyvalente, Antibes, France, Clémence Gachet; CH La Rochelle, La Rochelle, France, Mahmoud Gargouri; AP-HP Pitie Salpetriere, Laboratoire de Bactériologie , Paris, France, Denis Garot; CHU Tours, Médecine Intensive et Réanimation, Tours, France, Nabil Gastli; AP-HP Cochin, Service de Bactériologie, Paris, France, Elena Gauvin; CH Niort, Service de Réanimation, Niort, France, Isabelle Geneau; CH Niort, Service de Réanimation, Niort, France, Guillaume Geslain; AP-HP Robert Debré, Réanimation Pédiatrique, Paris, France, Antoine Goury; CHU Reims, Médecine Intensive et Réanimation Médicale, Reims, France, Romaric Grenot; CH Niort, Service de Réanimation, Niort, France, Antoine Grillon; CHU Nantes, Department of Bacteriology, Nantes, France, Thomas Guillard; CHU Reims, Laboratoire de Bactériologie-Virologie-Hygiène Hospitalière-Parasitologie-Mycologie, Reims, France, Aurélie Guillouzouic; CHU Nantes, Department of Bacteriology, Nantes, France, Jerome Guinard; CH Orléans, Laboratoire de Microbiologie, Orléans, France, Jennifer Guiraud; CHU Bordeaux, Laboratoire de Bactériologie, Bordeaux, France, Esther Gyde; Centre Hospitalier Francois Quesnay, Mantes-la-jolie, France, Christophe Henry; CH Albi, Réanimation polyvalente, Albi, France, Katy Jeannot; CHU Besancon, Laboratoire de Bactériologie, Besancon, France, Marie Kempf; CHU Angers, Laboratoire de Bactériologie - Département de Biologie des Agents Infectieux, Angers, France, Achille Kouatchet; CHU Angers, Laboratoire de Bactériologie - Département de Biologie des Agents Infectieux, Angers, France, Luce Landraud; AP-HP Louis Mourier, Microbiologie, Colombes, France, Philippe Lanotte; CHU Tours, Service de Bactériologie Virologie, Tours, France, Sebastien Larreche; Hopital d'Instruction des Armees Begin, Département de Biologie Médicale, Saint-Mandé, France, Brice Le Gallou; CH Orléans, Laboratoire de Microbiologie, Orléans, France, Elodie Le Breton; CH Quimper, Laboratoire de Biologie Médicale de l'Union Hospitalière de Cornouaille, Quimper, France, Pierre-Etienne Leblanc; AP-HP Kremlin Bicetre, Département d'Anesthésie Réanimation, Bicetre, France, Hervé Lecuyer; AP-HP Necker Enfants Malades, Laboratoire de Bactériologie, Paris, France, Ludovic Lemee; CHU Rouen, Laboratoire de Bactériologie, Rouen, France, Pauline Lessard; CH Quimper, Service de Réanimation, Quimper, France, David Leyssene; CH de la Cote Basque, Laboratoire de Biologie médicale, Boyonne, France, Pierre Lureau; CH Niort, Laboratoire de Biologie Médicale, Niort, France, Anne-Elisabeth Manteaux; CHU Nancy, Service de Microbiologie, Vandoeuvre-les-Nancy, France, Michael Mervent; CH Niort, Service de Réanimation, Niort, France, Maite Micaelo; CH Argenteuil, Argenteuil, France, Anthony Michaud; CHU Poitiers, Infectious Agents Department. Bacteriology and Infection Control Laboratory, 2 rue de la Milétrie, 86021, Poitiers, France, Olivier Moquet; CH Nevers, Centre de biologie du Nivernais, Nevers, France, Anaelle Muggeo; CHU Reims, Laboratoire de Bactériologie-Virologie-Hygiène Hospitalière-Parasitologie-Mycologie, Reims, France, Evelina Ochin; Hopital Simon Veil, Eaubonne, France, Patrick Ochocki; CH Niort, Laboratoire de Biologie Médicale, Niort, France, Abdelali Ouchikhe; CH Niort, Service de Réanimation, Niort, France, Maxime Paluch; CH Valenciennes, Valenciennes, France, Marie Pancher-Lory; CH Flers, Flers, France, Alix Pantel; CHU Nimes, Laboratoire de Microbiology, Nimes, France, Adeline Pastuszka; CHU Tours, Service de Bactériologie Virologie, Tours, France, Ophélie Perruche; CHU Clermont-Ferrand, Département de Bactériologie, Clermont-Ferrand, France, Olivia Peuchant; CHU Bordeaux, Laboratoire de Bactériologie, Bordeaux, France, Caroline Piau; CHU Rennes Pontchaillou, Laboratoire de Bactériologie, Rennes, France, Chloé Plouzeau-Jayle; CHU Poitiers, Infectious Agents Department. Bacteriology and Infection Control Laboratory, 2 rue de la Milétrie, 86021, Poitiers, France, Kevin Quesnel; CH de la Cote Basque, Service de Réanimation, Bayonne, France, Lucie Richard; CHU Tours, Service de Bactériologie Virologie, Tours, France, Emeline Riverain; Centre Hospitalier Francois Quesnay, Mantes-la-jolie, France, Alexandre Robert; CH Nice, Nice, France, Anne-Laure Roux; APHP Louis Mourier, Médecine Intensive Réanimation - DMI ESPRIT, Colombes, France, Pierre Saint-Sardos; CHU Clermont-Ferrand, Département de Bactériologie, Clermont-Ferrand, France, Laurent Serpin; CH de la Miséricorde, Ajaccio, France, Daniel Silva; Hopital Delafontaine, Service de Médecine Intensive Réanimation, Saint-Denis, France, Valérie Sivadon-Tardy; APHP Louis Mourier, Médecine Intensive Réanimation - DMI ESPRIT, Colombes, France, Karim Toumert; CH Gonesse, Service de Réanimation Polyvalente, Gonesse, France, Céline Tournus; Hopital Delafontaine, Laboratoire de Microbiologie , Saint-Denis, France, Pauline Touroult-Jupin; CH Cholet, Laboratoire, Cholet, France, Antoine Tran Quy; CHU Clermont-Ferrand, Département de Bactériologie, Clermont-Ferrand, France, Anne Vachee; CH Roubaix, Laboratoire de Biologie, Roubaix, France, Christian Vanjak; Hopital Simon Veil, Eaubonne, France, Véronique Vernet-Lefevre; CHU Reims, Laboratoire de Bactériologie-Virologie-Hygiène Hospitalière-Parasitologie-Mycologie, Reims, France, Camille Vinclair; CH de la Cote Basque, Service de Réanimation, Bayonne, France, Jérémie Violette; Groupement de Coopération Sanitaire de Saintonge, Laboratoire Interhospitalier, Saintes, France, Violaine Walewski; AP-HP Avicenne, Bobigny, France.
Funding
No public or private funds were specifically allocated to this study.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization, MP and CB; methodology, MP; software, MP; validation, MP, JC and CB; formal analysis, MP; investigation, MP (and all COVAP Study Group members); resources, MP and CB; data curation, MP; writing—original draft preparation, MP; writing—review and editing, MP, JC, and CB; supervision, MP and CB; project administration, MP and CB. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Because of the context during the first wave of COVID, individual consent was waived. The study was declared to the competent French legal authorities.
Competing interests
The authors declare no competing interests regarding this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Geographical origin of the data analysed in this study.
Additional file 2:
Table S1. AST profile per bacteria.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pichon, M., Cremniter, J., Burucoa, C. et al. French national epidemiology of bacterial superinfections in ventilator-associated pneumonia in patients infected with COVID-19: the COVAP study. Ann Clin Microbiol Antimicrob 22, 50 (2023). https://doi.org/10.1186/s12941-023-00603-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-023-00603-0