Abstract
Background
Acinetobacter baumannii (AB) has emerged as one of the most problematic pathogens affecting critically ill patients. This study aimed to investigate the longitudinal epidemiology of AB causing invasive diseases in children.
Methods
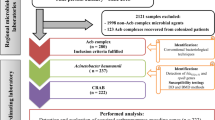
Acinetobacter spp. cultured from sterile body fluids and identified as Acinetobacter calcoaceticus-baumannii (ACB) complexes by automated systems from children aged below 19 years old were prospectively collected during 2001–2020. The discriminative partial sequence of rpoB gene was sequenced to identify the species, and sequence types (STs) were determined. Temporal changes in antimicrobial susceptibilities and STs were analyzed.
Results
In total, 108 non-duplicate ACB isolates were obtained from patients with invasive infections. The median age was 1.4 (interquartile range, 0.1–7.9) years, and 60.2% (n = 65) were male. Acinetobacter baumannii comprised 55.6% (n = 60) of the isolates, and the 30-day mortality was higher in patients with isolated AB than in those with non-baumannii Acinetobacter spp. (46.7% vs. 8.3%, P < 0.001). After 2010, complete genotype replacement was observed from non-CC92 genotypes to only CC92 genotypes. Carbapenem resistance rates were highest in AB CC92 (94.2%), followed by AB non-CC92 (12.5%) and non-baumannii Acinetobacter spp. (2.1%). During 2014–2017, which included clustered cases of invasive ST395, colistin resistance increased to 62.5% (n = 10/16), showing a mortality rate of 88% during this period.
Conclusion
Complete genotype replacement of non-CC92 with CC92 genotypes was observed. AB CC92 was extensively drug-resistant, and pandrug resistance was observed depending on the ST, warranting careful monitoring.
Similar content being viewed by others
Introduction
Acinetobacter baumannii (AB) has emerged as one of the most problematic pathogens worldwide, causing nosocomial invasive infections in critical patients [1]. The prolonged survival of AB in hospital environments facilitates their nosocomial spread, resulting in critical infections of hospitalized patients with breaches in skin integrity or airway protection [2].
The Acinetobacter genus comprises four closely related species: A. nosocomialis, A. pittii, A. calcoaceticus, and AB, referred to as the Acinetobacter calcoaceticus-baumannii (ACB) complex [3]. Species within the ACB complex are difficult to distinguish using biochemical methods and have not been reliably identified by semi-automated commercial identification systems, such as VITEK2, which are widely used in many hospitals [4, 5]. Afterwards, the development of post-processing software for MALDI-TOF/MS identification systems led to successful identification of Acinetobacter spp. [6]. Accurate identification of the species within the ACB complex is required due to the dissimilar clinical significance and clinical outcome of them. A previous retrospective study demonstrated statistically significant less multidrug resistance and lower mortality of non-baumannii Acinetobacter spp. [7]. Thus, distinguishing AB from other species in the ACB complex is relevant for better clinical outcomes.
Carbapenems such as meropenem and imipenem are the drugs of choice for AB infections as they are active against multidrug-resistant (MDR) AB. However, carbapenem-resistant AB (CRAB) have been repeatedly reported globally for recent decades [8,9,10]. In the case of South Korea, the Korean Antimicrobial Resistance Monitoring System (KARMS) and the Korean Nationwide Surveillance of Antimicrobial Resistance (KONSAR) reported an alarming increase in the proportion of CRAB [11,12,13].
Currently, there is a paucity of clinical data, long-term observational studies, and investigations into the clinical characteristics of AB causing invasive infections in children. Therefore, the primary goal of this study was to identify the species of the ACB complex isolated from children with invasive diseases in South Korea and investigate the proportion of infections caused by AB and non-baumannii ACB complexes. Also, this study includes the longitudinal molecular epidemiology of AB at a single center and observes changes in the genotypes causing invasive AB infections in children over a 20-year period.
Methods
Study population
This study involved the prospective collection of AB isolates from patients below 19 years old diagnosed and treated for invasive AB infections at a tertiary referral children’s hospital during January 2001 to December 2020. The clinical data of the cases included were retrospectively reviewed from the electronic medical records for the following: patient demographics, underlying conditions, admission duration, cultured specimen, antimicrobial susceptibilities, treatment, and outcome of infection.
Patients that had Acinetobacter species isolated from sterile body fluid cultures, identified as AB (excluding ACB complex) by automated identification systems, and isolates available for storage were included as study participants. A case of invasive AB infection was defined as a patient with at least one infection sign and AB cultured from sterile body fluids (blood, pleural fluid, and ascites). In cases where multiple AB isolates were culture from sterile body fluids from one episode of invasive AB infection, only the first cultured isolate was included in the analyses.
Isolates and DNA extraction
Acinetobacter spp. that were cultured from sterile body fluids of children underwent species identification and antimicrobial susceptibility testing using the following automated systems: from 2001, the MicroScan Walk-Away (Siemens Healthcare Diagnostics, Deerfield, IL, USA), from 2011, the Vitek-2 (bioMerieux, Marcy L’Etoile, France), and from August 2020, an additional M50 (BD Diagnostic Systems, Sparks, MD, USA). These isolates were collected and stored in inositol stocks at -70 °C. Collected isolates were thawed and cultured on MacConkey agar plates overnight in incubators set at 37 °C. The colonies were then collected and underwent chromosomal DNA extraction using a DNA extraction kit (DNeasy Kit; Qiagen GmbH) according to the instructions given by the manufacturer. The extracted DNA were evaluated by NanoDrop (Thermo Fisher Scientific, Inc., Waltham, MA, US) and then stored at -70 °C until use.
Antimicrobial susceptibility testing
The results of antimicrobial susceptibility testing reported by Vitek-2 and MicroScan Walk-Away were collected. The 2016 Clinical and Laboratory Standards Institute (CLSI) guideline was used to determine the cut-off for antibiotic susceptibilities. The definition for MDR Acinetobacter spp. was non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories, and extensively drug resistant (XDR) Acinetobacter spp. was non-susceptibility to ≥ 1 agent in all but ≤ 2 categories, as defined by the international expert proposal for interim standard definitions for acquired resistance.
Colistin susceptibility was tested via broth microdilution (BMD) according to the CLSI-EUCAST guidelines [14]. Minimum inhibitory concentrations (MICs) were determined using untreated MicroWell trays (Thermo Fisher Scientific, Inc., Waltham, MA, US) and 5 × 105 colony-forming units/mL of AB was inoculated. The plates were then incubated for 24 h and read using the Sensititre Manual Viewer (Thermo Fisher Scientific, Inc., Waltham, MA, US). According to the CLSI criteria, colistin MIC ≤ 2 µg/mL was considered susceptible, and ≥ 4 µg/mL was considered resistant [14].
Acinetobacter baumannii genomic species identification
The discriminative partial sequence of rpoB gene was sequenced for genomic species identification using the extracted DNA. Partial rpoB gene underwent polymerase chain reaction (PCR) using the primers Ac696F (TAYCGYAAAGAYTTGAAAGAAG) and Ac1093R (CMACACCYTTGTTMCCRTGA), as reported by Gundi et al. [15]. Upon electrophoresis, the band that appeared in the 350 bp region for each of the isolates were sequenced using Ac696F and Ac1093R as sequencing primers. Using the GenBank (https://www.ncbi.nlm.nih.gov/nuccore/) website, the rpoB sequences of each isolate were entered and the species were identified using the BLAST search tool.
Multilocus sequence typing of Acinetobacter baumannii isolates
All AB isolates from collected from 2001 to 2020 underwent multilocus sequence typing (MLST) by conventional sanger sequencing of the internal fragments of the 7 housekeeping genes using the oxford scheme for AB reported by Bartual et al. [16] The sequences of each loci were then submitted to the AB MLST database (https://pubmlst.org/abaumannii) and allotted allele numbers at each of the 7 loci, ultimately giving each isolate a 7-digit allele profile. This profile was then used to determine the sequence type (ST). The STs that shared 6 identical alleles of the 7 loci were clustered into a clonal complex (CC) using the eBURST (PHYLOViZ, https://phyloviz.readthedocs.io/en/latest/index.html) program.
Statistical analyses
Categorical variables were compared by using Chi-square test or Fisher’s exact test, and continuous variables were compared by Kruskal-Wallis H test. The cox proportional hazards regression analysis was used to find the risk of mortality at 7 and 30 days after the onset of sepsis, and logistic regression analyses was used to show the inverse relation between colistin prescription and 30-day mortality. The P for trend was analyzed using One-Way ANOVA (analysis of variance) linear analyses and the weighted P value were considered in the analyses. All tests were two-tailed and were considered statistically significant when the P-value was < 0.05.
Results
Demographics and clinical characteristics
During the 20-year study period, a total of 108 non-duplicate isolates were cultured and identified as AB using commercial identification systems from sterile fluids of children admitted at Seoul National University Children’s Hospital. Of the 108 patients, 60.2% (n = 65) were male patients, and the median age was 1.4 (interquartile range, 0.1–7.9) years old. In total, 98.1% (n = 106) of the isolates were cultured from children with underlying diseases. The most common underlying diseases were non-malignant chronic diseases (n = 40, 37.0%), malignancies (n = 23, 21.3%), and congenital heart disease (n = 16, 14.8%). The isolates were cultured from the blood in 85.2% (n = 92) of the cases and from ascites fluid in 12.0% (n = 13) (Table 1).
Genomic species identification
All 108 isolates reported as AB via commercial identification systems underwent further species identification via partial sequencing of the rpoB gene. Only 55.6% (n = 60) were identified as true AB species, whereas 44.4% (n = 48) were identified as non-baumannii Acinetobacter spp.
Of the 48 non-baumannii Acinetobacter spp. identified via partial rpoB sequencing, 87.5% (n = 42) of the isolates belonged to the ACB complex: A. nosocomialis (n = 25), A. pittii (n = 13), A. seifertii (n = 3), and A. calcoaceticus (n = 1). The non-ACB Acinetobacter spp. were identified as follows: A. soli (n = 3), A. bereziniae (n = 1), A. iwoffii (n = 1), and A. junii (n = 1) (Fig. 1).
A higher percentage of non-baumannii Acinetobacter spp. were isolated from patients who were in the general wards, as opposed to the intensive care unit (ICU), compared to patients with AB isolated (62.5% vs. 13.3%, respectively) (P < 0.001). The 7-day and 30-day mortality rates were significantly higher in patients with AB isolated (P < 0.001) (Table 1).
Genotype identification of Acinetobacter baumannii isolates
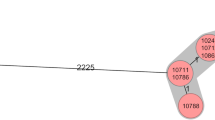
Isolates identified as AB species by partial rpoB sequencing were subjected to MLST. The phylogenetic relationships and diversification among STs are shown in the eBURST forest diagram (Fig. 2). One clonal complex, CC92 (n = 52, 86.9%), and eight singletons: ST17 (n = 3), ST159 (n = 2), ST868 (n = 1), ST1201 (n = 1), and ST1536 (n = 1) were found. Within CC92, there were 15 different STs: ST138 (n = 9), ST395 (n = 9), ST1125 (n = 4), ST75 (n = 4), ST1656 (n = 3), ST190 (n = 2), ST735 (n = 2), ST784 (n = 11), ST357 (n = 2) and one isolate each belonging to ST829, ST92, ST137, ST436, ST469, and ST184.
eBURST analysis of the 60 invasive AB isolates. The size of the colored shaded areas correlates with the number of isolates in the CC and STs. The nodes in the diagram represent an ST, and branches represent their relationship. The distance of the branches represents the relatedness off the STs. The number of isolates and isolated year are represented within parentheses. CC, clonal complex; ST, singleton
Changes in species and genotype distribution during 2001–2020
During the 20-year study period, one cluster of cases caused by A. nosocomialis was observed during 2001–2002 (32.0%, n = 8/25). Subsequently, a consistent number of cases were observed throughout the study period. A. pittii, the second most frequent cause of non-baumannii Acinetobacter spp. infections, was consistently isolated throughout the study period (Fig. 3).
A distinct change in AB genotypes causing invasive infections in children was observed. During the early study period from 2001 to 2007, non-CC92 genotypes were predominant, and CC92 genotypes were intermittently isolated. However, after the 2010–2011 outbreak caused by ST138 belonging to CC92, complete genotype replacement was observed, from non-CC92 genotypes to only CC92 genotypes. The predominant strain in 2014–2017 was ST395, which changed to ST784 in 2018–2020 (Fig. 3).
Antimicrobial susceptibility
The antimicrobial susceptibility of non-baumannii Acinetobacter spp. and AB non-CC92 showed higher antimicrobial susceptibility rates compared to AB CC92 (Table 2). The AB CC92 isolates showed high resistance to ceftazidime, cefepime, piperacillin/tazobactam, ciprofloxacin, imipenem, trimethoprim/sulfamethoxazole, and amikacin. Among the susceptible antibiotics, colistin showed the highest susceptibility rate of 40.4%, followed by amikacin with a susceptibility rate of 23.1%.
Imipenem resistance rates were highest in AB CC92 (94.2%) followed by AB non-CC92 (12.5%), and non-baumannii Acinetobacter spp. (2.1%), showing a significantly higher resistance rate of AB CC92 to carbapenems compared to the other two groups (P < 0.001 for both) (Table 2).
Changes in antimicrobial susceptibility and mortality in AB
For AB, changes in the imipenem and colistin resistance rates were investigated during the 20-year study period. Colistin-resistant AB strains were isolated as early as in 2001 (ST1536). The proportion of AB isolates resistant to imipenem was below 50.0% in 2001–2005 and at 50% in 2006–2009. However, after 2010, resistance rates increased to 94.0% in 2010–2013 and 100% in 2014–2017. Furthermore, the proportion of patients with colistin included in the treatment regimen during 2006–2009 was 0%, whereas starting 2010–2013, colistin use increased significantly (P < 0.001).
The colistin resistance rate was 30.0% (n = 3/10) during 2001–2005, and 0% during 2006–2009. In the period following, 2010–2013, an outbreak caused by ST138 was detected, and colistin resistance increased to 50.0% (n = 9/18). During 2014–2017, clustered cases of invasive ST395 infections were observed, and the colistin resistance rate increased further, reaching 62.5% (n = 10/16). However, during the 2018–2020 period which included an outbreak caused by ST784, colistin resistance rate decreased to 12.5% (n = 2/16). The 30-day mortality showed a similar curve, with the highest mortality observed during 2014–2017 at 88%. During 2018–2020, although the carbapenem resistance rate was similar to that in the previous time period, the 30-day mortality rate decreased to 19%. Furthermore, during the last two quinquennium, colistin prescription decreased the 30-day mortality in a statistically significant manner (OR, 15.750; 95% CI, 1.675-148.119; P = 0.016) (Fig. 4).
Changes in mortality rates and antibiotic resistance in Acinetobacter baumannii species during the study period. The study period was divided into four periods; the first period was from 2001–2009, when the dominant strains were AB non-CC92AB. From 2010 onwards, the period was divided by 4-year intervals. The P for trend is shown on the graph for each analyzed factors. AB, A. baumannii; CC, clonal complex
Discussion
Acinetobacter baumannii has emerged as a major global threat, especially because of its exceptional ability to acquire resistance genes against all classes of antibiotics currently available. There have been high misidentification rates due to its phenotypic similarities with other Acinetobacter spp. However, differentiating between the species of the ACB complex, especially AB, is extremely important because of the poor outcomes and higher mortality rate in critically ill patients with invasive AB infections [17]. In our study, we found that second-generation commercial identification systems correctly identified only 55.6% of AB strains between 2001 and 2020. Before 2007, non-CC92 AB was predominant. However, after 2010, a complete replacement was observed from non-CC92 to CC92 genotypes at a single center. The AB CC92 isolates showed XDR characteristics, with only 5.8% of the isolates being susceptible to carbapenems. As carbapenem resistance increased, the proportion of patients treated with colistin also increased. The mortality rate decreased in 2018–2020 as majority of the infections were caused by colistin-susceptible ST784.
Automated phenotypic identification methods analyze reactions of the bacteria to different chemicals, creating an analytical biochemical profile, which then matches the profile to the “best fit” bacteria. First-generation automated bacterial identification systems included the analytical profile index (API) (bioMérieux, Craponne, France) and Vitek® system (bioMérieux, Craponne, France). Second-generation systems include MicroScan-Walkaway (Siemens, Munchen, Germany) and Vitek 2® (bioMérieux, Craponne, France), which are more accurate than first-generation systems [18]. However, in this study, second-generation commercial identification systems correctly identified only 55.6% (n = 47/93) of the AB isolated at a single center during 2001–2020, showing a misidentification rate of 44.4%. Similarly, a study evaluating the VITEK 2 System to identify AB showed that 68.0% of AB strains were correctly identified [19].
The correct identification of Acinetobacter spp. is crucial because of the poor prognosis of patients with AB infections, especially critically ill patients, compared with other non-baumannii Acinetobacter spp. infections [20,21,22,23]. In this study, there was a significant difference in 7-day mortality between AB and non-baumannii Acinetobacter spp., 40.0% vs. 4.2% (P < 0.001), and 30-day mortality (46.7% vs. 8.3%, P < 0.001). Differentiating between the species can allow clinicians to focus on early and aggressive interventions to enhance the survival and outcome of patients with invasive AB infections.
Data from Korean adults from the Korean Nationwide Surveillance of Antimicrobial Resistance (KONSAR) data in 2005 showed that the resistance rates of Acinetobacter spp. to imipenem and meropenem were 16% and 29%, respectively, and in 2011, the resistance rates increased to 64% and 63%, respectively [11, 12]. By 2015, the resistance rates were even higher, with the proportion of AB resistant to imipenem and meropenem being 85% and 84%, respectively [13]. Both AB and non-baumannii ACB complex Acinetobacter spp. were included in the adult data. However, in children with data including only invasive AB during similar time periods in this study, carbapenem resistance rates were similar during 2001–2009 and increased to > 90% after 2010. In the molecular epidemiology of both invasive and noninvasive ACB complexes isolated from children in Mexico, carbapenem resistance was 47% in 2017, and all XDR ACB complexes were AB, showing that carbapenem resistance is a problem in children as well as adults [24].
Pathogens identified as non-baumannii Acinetobacter spp. and AB non-CC92 isolates showed similar antibiotic susceptibility patterns, with susceptibility to cephalosporins, carbapenems, amikacin, and fluoroquinolones exceeding 85%, although the number of AB-non-CC92 isolates was limited. However, all AB CC92 isolates showed XDR characteristics, with complete resistance to 3rd/4th generation cephalosporins, piperacillin-tazobactam, and fluoroquinolones. Only 5.8% of isolates were susceptible to carbapenems. Susceptibility rates to colistin (40.4%) and amikacin (23.1%) were the highest, although both were below 50%.
Alarming levels of antibiotic resistance have been observed in AB worldwide. Resistance to carbapenems has been reported to reach 80.7% in Brazil, and only strains susceptible to polymyxins have been reported in Europe[25, 26]. In this study, since 2010, the increase in carbapenem resistance consequently led to increasing use of colistin in children with CRAB. However, between 2014 and 2017, a high mortality rate of up to 88% was observed in patients with invasive AB infections (Fig. 4). The high mortality can be attributed to clustered cases caused by both carbapenem and colistin-resistant pan-drug-resistant ST395. To date, there have been no previously published reports describing colistin resistance in ST395, and our study shows the need to monitor this pan-drug-resistant strain. During the consecutive period, 2018–2020, the mortality rate decreased to 19%. During this period, an outbreak of cases was caused by ST784, which was carbapenem-resistant and colistin-sensitive. However, factors associated with mortality are very complicated, as shown by the colistin resistance rate during 2001–2005, which was much higher than in 2018–2020, but with lower mortality rate. The high colistin resistance rate in 2001–2005 may have been driven by different resistance mechanisms than the colistin resistance after 2010, and further studies are needed to explain changes in mortality associated with antibiotics resistance.
The main mechanism underlying the poor outcome of children with invasive AB is their exceptional ability to acquire MDR-genes rapidly. As observed globally, there was a distinct change in the genotypes of AB causing invasive infections in children in this study. Before 2007, non-CC92 AB was predominant. However, after 2010, a complete replacement was observed from non-CC92 to CC92 genotypes at a single center. With genotype replacement, a significant increasing trend in carbapenem resistance and mortality was observed. This change has been reported globally, especially in China, where CC92 CRAB is increasing [27, 28]. Data on adults also show similar findings. A study including 19 different hospitals in South Korea reported a wide dissemination of ceftazidime resistant CC92 in South Korea [29]. During 2013–2017, wide distribution of CC92 and high prevalence of acquired carbapenemase genes among CRAB was reported in the USA [30]. Isolates collected during 2014–2015 from patients within a university hospital in southern Iran also reported the spread of closely related XDR genotypes of CC92 [31]. The epidemic dissemination of CC92 and near synchronous emergence worldwide in many countries are attributed to the successful and rapid acquisition of antimicrobial resistance genes [32, 33].
Furthermore, the pattern of circulating dominant STs within CC92 show two major patterns: in the case of ST1125, cases are observed throughout the entire study period. However, for majority of the STs, we see clustered cases or outbreaks during a certain timeframe within the study period. This latter pattern may be attributed to infection control measures applied to eradicate the ST causing clustered cases, hence elimination of the ST. However, due to the characteristics of AB, we see different STs continually emerging.
This study has several limitations. This was a single-center study; therefore, caution must be exercised in generalizing the data to other hospitals. However, our center has the largest pediatric ICU in the country, with 20 ICU beds, and is representative. Our data also show that the global trend of increase in CRAB CC92 strains applies even to hospital-acquired infections in children from a single center. Furthermore, this is the first study to investigate the 20-year long-term longitudinal molecular epidemiology of AB isolated from children with invasive infections at a single center and is therefore deemed to be valuable for monitoring and treating invasive AB infections in children. Further studies exploring the changes in resistance genes and virulence factors of these strains will provide insight into the changes in molecular epidemiology.
Conclusions
To conclude, not only distinguishing AB from non-baumannii Acinetobacter spp. is important, but also identifying CC92 AB from non-CC92 AB is imperative because of their XDR characteristics. Furthermore, along with the global phenomenon, CC92 has become the dominant circulating strain in children. Many efforts are needed to prevent outbreaks in critical patients, including methods such as decreasing overall antibiotic use and relieving selective pressure, strict contact precautions, hand hygiene of medical staff to prevent transmission, and environmental disinfection to decrease sources of possible infection.
Data availability
The datasets used and/or analyzed during the current study are available in part from the corresponding author on reasonable request.
Abbreviations
- AB:
-
Acinetobacter baumannii
- ACB:
-
Acinetobacter calcoaceticus-baumannii
- BMD:
-
Broth microdilution
- CC:
-
Clonal complex
- CLSI:
-
Clinical and Laboratory Standards Institute
- CRAB:
-
carbapenem-resistant AB
- IQR:
-
Interquartile range
- KARMS:
-
Korean Antimicrobial Resistance Monitoring System
- KONSAR:
-
Korean Nationwide Surveillance of Antimicrobial Resistance
- MDR:
-
multidrug resistant
- MIC:
-
Minimum inhibitory concentration
- ST:
-
Sequence type
- XDR:
-
Extensively drug resistant
References
Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathogens and disease. 2014;71(3):292–301.
Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102.
Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, et al. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol. 2011;162(4):393–404.
Gerner-Smidt P, Tjernberg I, Ursing J. Reliability of phenotypic tests for identification of Acinetobacter species. 1991;29(2):277–82.
Kim S, Kim MH, Lee WI, Kang SY, Jeon YL. Misidentification of Acinetobacter baumannii as Alcaligenes faecalis by VITEK 2 system; Case Report. Lab Med. 2017;49(1):e14–e7.
Marí-Almirall M, Cosgaya C, Higgins PG, Van Assche A, Telli M, Huys G et al. MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: inclusion of the novel A. seifertii and A. dijkshoorniae species. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2017;23(3):210.e1-.e9.
Park KH, Shin JH, Lee SY, Kim SH, Jang MO, Kang SJ, et al. The clinical characteristics, carbapenem resistance, and outcome of Acinetobacter bacteremia according to genospecies. PLoS ONE. 2014;8(6):e65026.
Zhao Y, Hu K, Zhang J, Guo Y, Fan X, Wang Y, et al. Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in ICU of the eastern Heilongjiang Province, China. BMC Infect Dis. 2019;19(1):452.
Perez S, Innes GK, Walters MS, Mehr J, Arias J, Greeley R, et al. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii infection and colonization in an Acute Care Hospital during a Surge in COVID-19 admissions - New Jersey, February-July 2020. MMWR Morbidity and mortality weekly report. 2020;69(48):1827–31.
Ben-Chetrit E, Wiener-Well Y, Lesho E, Kopuit P, Broyer C, Bier L et al. An intervention to control an ICU outbreak of carbapenem-resistant Acinetobacter baumannii: long-term impact for the ICU and hospital. Critical care (London, England). 2018;22(1):319.
Yong D, Shin HB, Kim YK, Cho J, Lee WG, Ha GY, et al. Increase in the prevalence of Carbapenem-Resistant Acinetobacter isolates and Ampicillin-Resistant non-typhoidal Salmonella species in Korea: a KONSAR Study conducted in 2011. Infect Chemother. 2014;46(2):84–93.
Lee K, Lee MA, Lee CH, Lee J, Roh KH, Kim S, et al. Increase of ceftazidime- and fluoroquinolone-resistant Klebsiella pneumoniae and imipenem-resistant Acinetobacter spp. in Korea: analysis of KONSAR study data from 2005 and 2007. Yonsei Med J. 2010;51(6):901–11.
Kim YA, Park YS. Epidemiology and treatment of antimicrobialresistant gram-negative bacteria in Korea. Korean J Intern Med. 2018;33(2):247–55.
European Committee on Antimicrobial Susceptibility Testing %J EUCAST. : Växjö S. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016.
Gundi V, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009;155(Pt 7):2333–41.
Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4382–90.
Yang YS, Lee YT, Tsai WC, Kuo SC, Sun JR, Yang CH, et al. Comparison between bacteremia caused by carbapenem resistant Acinetobacter baumannii and Acinetobacter nosocomialis. BMC Infect Dis. 2013;13:311.
Martins KB, Ferreira AM, Mondelli AL, Rocchetti TT. Lr de SdCM. Evaluation of MALDI-TOF VITEK(®)MS and VITEK(®) 2 system for the identification of Staphylococcus saprophyticus. Future Microbiol. 2018;13:1603–9.
Joyanes P, del Ma CC, Martínez-Martínez L, Perea EJ. Evaluation of the VITEK 2 System for the Identification and Susceptibility Testing of Three Species of Nonfermenting Gram-Negative Rods Frequently Isolated from Clinical Samples. 2001;39(9):3247–53.
Liu YM, Lee YT, Kuo SC, Chen TL, Liu CP, Liu CE. Comparison between bacteremia caused by Acinetobacter pittii and Acinetobacter nosocomialis. Journal of microbiology, immunology, and infection = Wei mian yu gan ran za zhi. 2017;50(1):62–7.
Xiao D, Wang L, Zhang D, Xiang D, Liu Q, Xing X. Prognosis of patients with Acinetobacter baumannii infection in the intensive care unit: a retrospective analysis. Experimental and therapeutic medicine. 2017;13(4):1630–3.
da Silva KE, Maciel WG, Croda J, Cayô R, Ramos AC, de Sales RO, et al. A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a brazilian intensive care unit. PLoS ONE. 2018;13(12):e0209367.
Kuo SC, Lee YT, Yang SP, Chiang MC, Lin YT, Tseng FC, et al. Evaluation of the effect of appropriate antimicrobial therapy on mortality associated with Acinetobacter nosocomialis bacteraemia. Clin Microbiol infection: official publication Eur Soc Clin Microbiol Infect Dis. 2013;19(7):634–9.
Mancilla-Rojano J, Ochoa SA, Reyes-Grajeda JP, Flores V, Medina-Contreras O, Espinosa-Mazariego K, et al. Molecular Epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex isolated from children at the Hospital Infantil de México. Federico Gómez. 2020;11:2404.
Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev anti-infective therapy. 2013;11(3):297–308.
Jones RN, Flonta M, Gurler N, Cepparulo M, Mendes RE, Castanheira M. Resistance surveillance program report for selected European nations (2011). Diagnostic microbiology and infectious disease. 2014;78(4):429 – 36.
Ruan Z, Chen Y, Jiang Y, Zhou H, Zhou Z, Fu Y, et al. Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int J Antimicrob Agents. 2013;42(4):322–8.
Chen Y, Gao J, Zhang H, Ying C. Spread of the bla(OXA-23)-Containing Tn2008 in Carbapenem-Resistant Acinetobacter baumannii isolates grouped in CC92 from China. Front Microbiol. 2017;8:163.
Lee Y, Bae IK, Kim J, Jeong SH, Lee K. Dissemination of ceftazidime-resistant Acinetobacter baumannii clonal complex 92 in Korea. J Appl Microbiol. 2012;112(6):1207–11.
McKay SL, Vlachos N, Daniels JB, Albrecht VS, Stevens VA, Rasheed JK et al. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii in the United States, 2013–2017. Microbial drug resistance (Larchmont, NY). 2022;28(6):645–53.
Saffari F, Monsen T, Karmostaji A, Azimabad FB, Widerström M. Significant spread of extensively drug-resistant Acinetobacter baumannii genotypes of clonal complex 92 among intensive care unit patients in a university hospital in southern Iran. J Med Microbiol. 2017;66(11):1656–62.
Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(2):233–8.
Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii.Microbial genomics. 2019;5(10).
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1C1B5085781).
Author information
Authors and Affiliations
Contributions
HK and KY analyzed and interpreted the patient data, performed rpoB gene sequencing and MLST, and wrote the initial and final draft. EC was a major contributor in data curation, validation, interpreting the data and reviewing the final draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics declarations
The institutional review board of Seoul National University Hospital approved this study (H-1812-080-995). Informed consent was waived.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Financial support
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1C1B5085781).
Disclosures
None.
Conflict of interest
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kang, H.M., Yun, K.W. & Choi, E.H. Molecular epidemiology of Acinetobacter baumannii complex causing invasive infections in Korean children during 2001–2020. Ann Clin Microbiol Antimicrob 22, 32 (2023). https://doi.org/10.1186/s12941-023-00581-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-023-00581-3