Abstract
Background
Uropathogenic Escherichia coli (UPEC) is a major cause of urinary tract infection (UTI); however, treatment of UTI has been challenging due to increased antimicrobial resistance (AMR). One of the most important types of AMR is carbapenem resistance (CR). CR bacteria are known as an important threat to global public health today. Class B metallo-beta-lactamases (MBLs) are one of the major factors for resistance against carbapenems. We aimed to investigate the characteristics of UPEC isolates producing MBL.
Methods
A cross-sectional study was conducted from October 2018 to December 2019 in Ahvaz; Iran. UPEC isolates were identified by biochemical and molecular methods. Metallo-beta-lactamase-producing isolates were detected using modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM) tests. MBL genes, phylogenetic group, and virulence genes profile of carbapenem resistant isolates were determined. Conjugation assay and plasmid profiling were conducted to evaluate the ability of transferring of CR to other E. coli isolates. Clonal similarity of isolates were assessed using Enterobacterial intergenic repetitive element sequence (ERIC)-PCR.
Results
Among 406 UPEC isolates, 12 (2.95%) carbapenem-resistant were detected of which 11 were phenotypically MBL-producing strains. Four isolates were resistant to all investigated antimicrobial agents and were considered possible pandrug-resistant (PDR). blaNDM, blaOXA-48, blaIMP-1, and blaIMP-2 genes were found in 9, 5, 1, and 1 isolates, respectively. Among 30 virulence genes investigated, the traT, fyuA followed by fimH, and iutA with the frequency of 8 (66.7%), 8 (66.7%), 7 (58.3%), and 7 (58.3%) were the most identified genes, respectively. Siderophore production was the main virulence trait among carbapenem-resistant UPEC isolates. Except for two, all other isolates showed weak to moderate virulence index. In all recovered isolates, CR was readily transmitted via plasmids to other isolates during conjugation experiments.
Conclusion
MBL and carbapenemase genes, especially blaNDM and blaOXA-48 are spreading rapidly among bacteria, which can be a threat to global public health. Therefore monitoring the emergence and dissemination of new AMR is necessary to continuously refine guidelines for empiric antimicrobial therapy. Understanding the mechanisms of resistance and virulence in this group of bacteria can play an effective role in providing new therapeutic methods.
Similar content being viewed by others
Introduction
Urinary tract infection (UTI) is one of the most common infectious diseases that affects people of all ages. The main etiologic agent causing UTI is a group of Escherichia coli strains that named uropathogenic Escherichia coli (UPEC) [1]. UPEC is a subgroup of extraintestinal pathogenic E. coli (ExPEC) which is differentiated from intestinal E. coli strains. UPEC strains are capable to colonize and invade urogenital tract which eventually leads to infection of urinary tract system. Disease establishment is caused by virulence factors (VFs) of bacteria and host characteristics [2, 3]. Various VFs have been incriminated in UPEC pathogenesis including invasins, adhesins, toxins, iron-acquisition systems, and serum resistance factors [4]. VFs may be present in the chromosome or acquired horizontally through mobile genetic elements such as transposons, plasmids, and pathogenicity islands, thereby leading to great diversity among UPEC strains [5, 6].
Antimicrobial therapy usually is the first strategy in treating a UTI. The choice of antimicrobial compound depends on the patient's health condition and the type of bacteria causing the UTI. However, due to misuse of antibiotics, antimicrobial resistance (AMR) has greatly expanded among bacteria. Many E. coli strains, especially UPEC have become multi-, extensively- or pan-drug resistant (MDR, XDR, or PDR) [7, 8]. This poses a great challenge in the treatment of UTI infections. Therefore, monitoring the emergence and dissemination of new AMR is necessary to continuously refine guidelines for empiric antimicrobial therapy [1, 9]. Resistance against trimethoprim-sulfamethoxazole, fluoroquinolones, and beta-lactams is increasing among UPEC isolates [2, 10, 11]. Carbapenems such as imipenem, meropenem, and doripenem are stable antibiotics against extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases [12] and considered as a last-resort drugs for treating infections by MDR bacteria. However extensive use of carbapenems has led to the emergence of carbapenem-resistant Enterobacteriaceae (CRE) [12]. CRE are known as a global clinical and public health problem because their infections are resistant to most classes of antibiotics including trimethoprim-sulfamethoxazole, fluoroquinolones, and beta-lactams and so-called superbugs. CRE are associated with high mortality due to limited treatment options [12,13,14].
Several mechanisms have been proposed for resistance against carbapenems in Enterobacteriaceae. Production of carbapenemases that are categorized in Ambler classification as follows: class A carbapenemases including Klebsiella pneumonia carbapenemase (KPC), Guiana extended-spectrum β-lactamase (GES), Serratia marcescens enzyme (SME), imipenemase/non-metallocarbapenemase-A (IMI/NMC-A), sulfhydryl variable lactamase (SHV), and Serratia fonticola carbapenemase (SFC-1); class B metallo-beta-lactamase (MBL) including New Delhi metallo-beta-lactamase (NDM), imipenemase (IMP), and Verona integron-encoded metallo-beta-lactamase (VIM), as well as class D oxacillinases (OXA). Moreover, over-expression of ESBLs, AmpC enzymes and efflux pumps combined with porin loss can also lead to carbapenem resistance (CR) [12, 15,16,17,18]. Carbapenemase genes are predominantly located on plasmids and are transmitted to other bacteria or integrated into chromosomes [16].
In the present study we aimed to investigate the antimicrobial susceptibility pattern and presence of MBL-producing among UPEC isolates. As the information on the virulence characteristics of MBL-producing UPEC isolates is very limited, the virulome, phylogenetic groups, clonal relationship, and mechanism of CR transfer among these isolates were investigated.
Materials and methods
Ethics
The ethics of the study was confirmed by the Ethics Committee of Shahid Chamran University of Ahvaz according to Declaration of Helsinki (EE/98.24.3.26336/scu.ac.ir). Before collecting information, participants or parents (for children cases) were asked to read, accept and sign an informed consent form.
Sample collection and identification
A cross-sectional study was performed from October 2018 to December 2019. The sample size was estimated using a single population proportion formula based on the prevalence of 0.15 [1], 95% confidence interval, and margin error of 5%. With considering a 10% non-response rate, the minimum samples size was 225; however, we collected 427 E. coli isolates for more accuracy.
The suspected E. coli isolates were obtained from hospitals and laboratories in Ahvaz city; Iran. These bacteria were isolated from patients who suffered from UTI and referred to laboratories by physicians to identify the pathogen and performing the antibiogram test. UTI was defined as the presence of at least 105 cfu/mL of pathogenic agent in urine and pyuria (≥ 104 leukocytes/mL of urine). The midstream urine samples of these patients were cultured on Blood Agar and McConkey and after purification, they were examined for antibiotic susceptibility profile. Thus, 427 isolated Escherichia coli isolates were collected and further investigated. E. coli isolates of patients who had recently consumed antimicrobial drugs were excluded from the study. Briefly, to identify and confirm the isolates, they were cultured onto MacConkey (Biolife Italiana; Italy) and Eosin Methylene Blue (EMB, Merck; Germany), and subsequently were incubated at 37 °C for 24 h. Lactose-fermenting colonies on MacConkey, or colonies with metallic sheen on EMB were investigated by conventional biochemical tests including production of lysine decarboxylase, oxidase, Sulfur Indole Motility (SIM), Simmon’s Citrate and Methyl Red/Voges-Proskauer (MR/VP). Finally, purified isolates were analyzed by PCR for the presence of uspA gene, which is the highly specific gene of E. coli. Detection of uspA gene was performed as described previously [19].
Antimicrobial susceptibility pattern
Kirby-Bauer disc diffusion method was performed to evaluate the resistance and susceptibility of isolates to antimicrobial agents recommended by Clinical & Laboratory Standards Institute 2018 (CLSI-2018) [20]. The antimicrobial discs included Nalidixic-acid, Ampicillin, Tetracycline, Streptomycin, Sulfamethoxazole-trimethoprim, Ciprofloxacin, Kanamycin, Gentamycin, Fosfomycin, Imipenem, Meropenem, Cefotaxime, Ceftazidime, Cefazolin, and Nitrofurantoin. The isolates resistance to three or more different antimicrobial families were considered multidrug resistant (MDR).
As meropenem or imipenem resistant UPEC isolates were the focus of present study, the resistant isolates were subjected to further analyses. The minimum inhibitory concentration (MICs) for imipenem or meropenem were determined as recommended by CLSI-2018. MIC breakpoints for both antibiotics were defined ≥ 4 μg/mL. The resistance of these isolates to ceftriaxone, cefoxitin, cefepime, amikacin, ampicillin-sulbactam, aztreonam, piperacillin/tazobactam, and colistin antimicrobials was also measured to determine XDR and PDR isolates. The CLSI recommendation for Acinetobacter spp. was applied for colistin.
Combined-disc (CD) and double disc synergy (DDS) tests
Phenotypic CD and DDS tests were performed to identify MBL-producing isolates. For CD test, two discs of imipenem (10 µg) and imipenem-EDTA (10 µg-1460 µg) were placed on the Muller-Hinton agar (MHA; Biolife Italiana; Italy) inoculated by 0.5-McFarland test isolates. After incubation of the plates at 35 °C for 16–18 h, the inhibition zone around discs were measured. The isolates were considered MBL-producer when the diameter of inhibition zone around the imipenem-EDTA disc increased by ≥ 7 mm compared to imipenem disc alone [21].
For DDS test, two discs of imipenem and EDTA (1460 µg) were placed on MHA plates inoculated with test isolates. The distance between two discs was considered 15 mm. After incubation of the plates at 35 °C for 16–18 h, the zone around discs were measured. Increasing of the inhibition zone or the formation of a phantom zone between the two discs indicated the MBL production [21].
Modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM)
Either mCIM and eCIM tests are extensively used for the epidemiological or infection prevention aims. The mCIM test could identify the bacteria that produce all carbapenemases, while the eCIM test was performed to differentiate MBL-producers from the serine carbapenemases. To perform mCIM, 1 µL loopful of the isolates were emulsified in 2 mL of tryptone soya broth (TSB). Then, one meropenem disc was immersed in the suspension for 4 h at 37 °C. A MHA plate was inoculated by 0.5-McFarland standard E. coli ATCC25922. Meropenem disc was removed from the suspension and excess liquid was expelled. Meropenem disc was placed on the inoculated plate and incubated at 37 °C for 24 h. Inhibition zone diameter of 6–15 mm or appearance of pinpoint colonies within a 16–18 mm zone around imipenem disc indicate the presence of carbapenemase [20].
eCIM test was performed when the mCIM test was positive. This test was done as similar to mCIM, except that after adding test isolate to the TSB, 20 µL of 0.5 M EDTA was added; then meropenem disc was immersed. Meropenem discs of eCIM and mCIM tests were placed on one plate and analyzed simultaneously. An increase of ≥ 5 mm in inhibition zone for eCIM versus mCIM was considered MBL-positive, while no change in zone diameter or an increase of ≤ 4 mm indicated the presence of carbapenemase [20].
Phenotypic differentiation of MLBs and class A KPC carbapenemases
To differentiate MBLs- and class A KPC carbapenemase-producing isolates, phenyl boronic acid (PBA) disc test was applied. The test was performed as a combined-disc of meropenem with and without PBA. Phenyl boronic acid was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 20 mg/mL. Then meropenem disc was inoculated by 20 µL of PBA solution (400 µg PBA/disc). The test was performed as given for the standard disc diffusion method. An increase of ≥ 5 mm in inhibition zone around meropenem-PBA disc versus meropenem was considered class A KPC carbapenemase producer [22, 23].
Detection of resistance genes
Imipenem or meropenem-resistant isolates were analyzed for the presence of MBL-genes including blaVIM-1, blaVIM-2, blaIMP-1, blaIMP-2, blaSPM-1, blaNDM, blaSIM, and blaGIM. The other CR genes of blaKPC, blaOXA-23 and blaOXA-48 were also investigated. Genomic DNA of isolates was extracted using the boiling lysis procedure. PCR reactions were performed as previously described [24,25,26,27,28,29,30,31].
Virulence genotyping and phylogenetic grouping
The isolates were assayed for the presence of 30 virulence traits using five multiplex-PCR panel as previously described [32]. The investigated VGs were including papEF, papA, fimH, papG allele I-III, papG allele I, papG allele II, papG allele III, kspMTIII, gafD, focG, sfa/focDE, nfaE, papC, afa/draBC, sfaS, bmaE, ibeA, traT, cvaC, cdtB, hlyA, cnf1, fyuA, chuA, iutA, K1, K5, kpsMTII, and rfc. Determination of major E. coli phylogroups was performed based on the quadruplex PCR, and complementary tests as described by Clermont et al. [33]. According to the presence of three genes of arpA, chuA and yjaA, and TSPE4.C2 DNA fragment, the isolates assigned to one of A, B1, B2, C, D, E and F phylogenetic groups.
Enterobacterial intergenic repetitive element sequence (ERIC)-PCR
Fingerprinting of imipenem or meropenem resistant isolates was performed using ERIC-PCR based on conditions and primers described previously [34]. To evaluate the relationship between isolates, the presence or absence of bands compared to the standard DNA molecular marker was assessed. The clustering of the isolates was done based on Unweighted Pair Group Method with Arithmetic Mean (UPGMA) analysis using the SAHN NTSYS program version 2.02e. A Dice similarity index was used for the definition of ERIC clusters.
Plasmid profiling and conjugation
Carbapenem resistant isolates were assayed for plasmid content. Plasmids were extracted using alkaline lysis method [35], then electrophoresed on agarose gel (1%). Estimation of plasmid sizes was acquired using a molecular weight marker, made from a lambda/Hind III digest.
The MBLs- or carbapenemase-producing isolates were conjugated with a lactose-negative enteroinvasive E. coli (EIEC) strain that was susceptible to imipenem and meropenem. The donors and recipient bacteria were cultured in nutrient broth for 16 h; then mixed in a ratio of 1:10 (donor: recipient) [35]. After incubation for 48 h at 37 °C, the mixtures were inoculated on MCA containing imipenem or meropenem (4 μg/mL). The plates were incubated overnight at 37 °C. The resistant lactose-negative isolates were analyzed for the presence of inv and resistance genes via PCR reaction.
Results
Antimicrobial susceptibility pattern of UPEC isolates
Out of 427 cultured samples, 406 isolates were phenotypically and molecularly confirmed as UPEC. As defined in Table 1, the most antimicrobial resistance was against ampicillin (82.3%) followed by cefazolin (80%), tetracycline (59.4%), and nalidixic-acid (59.1%). Only 13 (3.2%) isolates were susceptible to all antimicrobials tested and the rest (96.8%) were resistant to at least one or more antimicrobials. MDR profile was found in 377 (92%) isolates. The highest frequency of antimicrobial susceptibility was recorded for meropenem (97.5%), imipenem (97.3%), followed by nitrofurantoin (95.4%), and fosfomycin (92.4%).
In total, 12 isolates were resistant to imipenem, meropenem or both (Table 1) and archived for further analysis. Five isolates were XDR meaning non-susceptible to at least one drug in all but two or fewer antibiotic or antimicrobial family [8], also 4 (1%) isolates were possible PDR meaning were non-susceptible to all investigated antimicrobial categories [8].
Phenotypic detection of MBL-producing UPEC isolates
In total, 9 isolates were confirmed by disc diffusion and MIC to be resistant against imipenem and meropenem, however, 2 and 1 isolates were only resistant against imipenem and meropenem, respectively. To detect MBL-producers, CDT, DDST, and mCIM-eCIM were done. We found that 2 and 6 UPEC isolates were positive for CD, or both CD, DDS tests, respectively; while four UPEC isolates were negative in both tests (Table 2; Fig. 1).
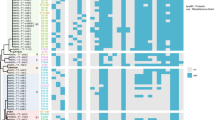
ERIC-PCR clustering and all phenotypic and genotypic characters of carbapenem-resistant UPEC isolates. blaVIM-1, blaVIM-2, blaSIM, blaSPM, blaGIM, blaOXA-23, blaKPC, kpsMTIII, papEF, ibeA, gafD, cdtB, focG, bmaE, sfa/focDE, papG allel I, II, and III, K1, rfc, nfaE, sfaS, and afa/draBC genes were not found; therefore were not shown. Blue and yellow colors indicate resistance and susceptibility, respectively. Green color shows presence of gene or phenotypic character and white color indicates absence of gene or phenotypic character. Abbreviations were as follow: PDR pan-drug resistant, XDR extensively-drug resistant, MDR multi-drug resistant, NA nalidixic-acid, AMP ampicillin; TET tetracycline, STR streptomycin, STX sulfamethoxazole-trimethoprim, CP ciprofloxacin, K kanamycin, GN gentamycin, FOS fosfomycin, IMP imipenem, MEN meropenem, CTX cefotaxime, CAZ ceftazidime, CZ cefazolin, FM nitrofurantoin, CRO ceftriaxone, FOX cefoxitin, FEP cefepime, AN amikacin, SAM ampicillin-sulbactam, AZT aztreonam, PTZ piperacillin/tazobactam, CL colistin, DDST double disc synergy test, CDT combined disc test, PBA phenylbronic acid disc test, mCIM modified carbapenem inactivation method, eCIM EDTA-CIM, MIC minimum inhibitory concentration
Phenotypic tests of mCIM, eCIM were used to detect and differentiate MBLs from serine carbapenemases. Out of 12 resistant UPEC isolates, 11 were positive by mCIM-eCIM tests, while one isolate was negative by eCIM. Details of results are presented in Table 2 and Fig. 1.
Phenotypic detection of KPC-producing UPEC isolates
PBA disc test was applied to detect class A KPC-producing isolates. Two isolates were phenotypically positive by this test and were considered KPC-producing UPEC. However, these isolates were also previously detected as MBL producers (Table 2; Fig. 1).
Resistance gene assays of MBL- and carbapenemase-producing isolates
The distribution of MBLs and carbapenemase genes among UPEC isolates are shown in Table 2. Out of 12 carbapenem resistant isolates, 11 were positive for MBL genes. blaNDM, blaIMP-1, and blaIMP-2 were found in 9, 1, and 1 isolates, respectively. None of the isolates carried blaVIM-1, blaVIM-2, blaSIM, blaSPM, and blaGIM genes. Three main non-MBL carbapenemase genes of blaOXA-48, blaOXA-23, and blaKPC were investigated [36,37,38,39,40,40]. blaOXA-48 was found in five isolates while blaOXA-23, and blaKPC were not detected. The simultaneous carriage of two genes blaNDM and blaOXA-48 was found in four isolates (Table 4; Fig. 1). Gene sequences were registered under the accession numbers of MT321108 (blaNDM), MT321107 (blaNDM), MT321106 (blaOXA-48), and MT293867 (blaIMP) in the GenBank.
Virulome and phylogenetic grouping
The content of VGs for carbapenem resistant UPEC isolates is shown in Table 3. The most detected genes were traT, fyuA followed by fimH, and iutA with the frequency of 8 (66.7%), 8 (66.7%), 7 (58.3%), and 7 (58.3%), respectively. kpsMTIII, papEF, ibeA, gafD, cdtB, focG, bmaE, sfa/focDE, papG allel I, II, and III, K1, rfc, nfaE, sfaS, and afa/draBC genes were not detected among the isolates. Two UPEC isolates had none of the tested virulence genes while they showed resistance against all investigated antibiotics. According to phylogenetic grouping 2, 2, 2, 1, and 1 isolates were B2, C, D, B1, and E phylogroups, respectively. Four isolates were positive for all three genes and DNA fragment and according to Clermont Escherichia coli phylo-typing were considered as “unknown” [33].
ERIC-PCR
The isolates produced PCR products with different sizes from 250 to 2500 bp and various patterns yielded 5–12 bands. Two isolates showed the same ERIC pattern and virulence gene profile; however the AMR patterns were different. Also two isolates showed ~ 95% similarity index. The similarity of the other isolates was < 80%, indicating a different source of isolates. Drawn dendrogram had a matrix correlation of 0.91.
Conjugal transfer and plasmid profiling
Conjugation test showed that imipenem or meropenem resistance could be transmitted to other bacteria; therefore, these isolates were capable to disseminate MBL or carbapenemase genes horizontally. All 12 UPEC donors transferred carbapenem resistance phenotype to EIEC recipient strain and this strain was able to grow on the medium containing the antibiotic carbapenem. Transconjugants were positive for MBL or carbapenemase genes and inv by gene-specific PCR. Plasmid profiling showed different patterns for plasmids ranging 5 to upper 50 kb. All isolates presented different plasmid pattern. Plasmid profiling of donors and recipients cells were not equal after conjugation, indicating that not all plasmids were transferred during conjugation.
Discussion
UTI is one of the most important bacterial infectious diseases in humans, which is mainly caused by E. coli. Antibiotics are commonly used to treat UTIs. However, the emergence and dissemination of AMR has posed a major challenge in the treatment of these diseases. Detection of antibiotic resistance type, its transmission mechanism, and the characteristics of resistant bacteria can be very effective in designing treatment guidelines.
Investigation of antimicrobial susceptibility profile of UPEC isolates shows high antimicrobial resistance against most of the antibiotics evaluated in this study. However, imipenem, meropenem, nitrofurantoin, fosfomycin, and to some extent gentamycin and kanamycin showed noticeable activity against UPEC isolate. Developing resistance to these antibiotics can be very costly to the healthcare system. Other studies in Iran regarding the antibiotic resistance of UPEC isolates show almost similar profiles [42,43,44,44]. Some studies in Iran have not found resistance to imipenem or meropenem among E. coli isolates. More than 90% of UPEC isolates were MDR and 2.2% were possible XDR. This statistic indicates a high frequency of antibiotic resistance among these isolates. Four isolates were resistant to all studied antibiotics and therefore were considered possible PDR. The XDR and PDR Enterobacteriaceae are important because the mortality rate is high among patients infected by these bacteria [46,47,47]; although, some researchers disagree [48]. PDR E. coli is very rare [45]; however, in our study, four isolates (1%) were resistant to all the antibiotics studied. Risk factors for infections with XDR and PDR E. coli isolates have not yet been identified and little information is available. Understanding the mechanisms of AMR and virulence characteristics of these pathogens helps researchers to find solutions for the long-standing AMR problem [7].
To detect MBL-producing isolates and differentiate from other carbapenemases CDT, DDST, mCIM-eCIM and PBA test were performed. Previously, Modified Hodge test was recommended as a phenotypic method for detecting carbapenemases; however, this test was excluded from the CLSI (2018) due to the inability to identify some carbapenemase-producing bacteria, including NDM-producing strains [21]. Among the tests performed in the present study, the mCIM-eCIM method was able to more effectively detect the MBL-producing isolates. MBL genes were found in all isolates identified as MBL-producers by this method. One isolate was positive by mCIM but negative by eCIM and also PBA disc test. Eventually it was found that this isolate carries blaOXA-48 and belonged to group D carbapenemases. Although two isolates were positive for PBA disc test, but they lacked blaKPC gene. These isolates were also positive for mCIM-eCIM and both had the blaNDM gene. The lack of detection of blaKPC may be due to the inability of the primers to target this gene or the PBA disc test results were false positive.
PCR detection and sequencing showed that 11 isolates carry MBL genes. The predominant gene was blaNDM that was found in nine UPEC isolates. For the first time blaNDM was detected in a Swedish patient (originally from India) who traveled to New Delhi, India. In this patient, the blaNDM was isolated from Klebsiella pneumoniae, which caused UTI [49]. After that, bacteria carrying blaNDM have been reported from other parts of the world including Iran [51,52,53,53]. In Iran blaNDM is mostly isolated from carbapenem-resistant Klebsiella isolates [54, 55]; however, there have been reports of carbapenem-resistant E. coli isolates carrying this gene [53, 57,58,58]. A study conducted in Ahvaz; Iran from 2014 to 2015 reported that there were no MBL-producing Enterobactericeae isolates carrying blaNDM in this region [59]. However, in 2018, this gene was found in commensal E. coli and Pseudomonas aeruginosa which indicates the dissemination of this type of resistance among bacteria in Ahvaz; Iran [57, 60]. In the present study, 2.2% of UPEC isolates harbored blaNMD; therefore, this gene is rapidly spreading among the strains of this region. Conjugation experiments showed that all blaNDM genes identified in this study are transferred by plasmids and have the ability to be transmitted to other strains. The other detected MBL genes were blaIMP-1 and blaIMP-2. Little is known about the presence of these genes among carbapenem-resistant Escherichia coli isolates in Iran. We found two reports about the presence of blaIMP among E. coli strains in northern Iran and Tehran (capital of Iran) [61, 62]. blaOXA-48, which is class D carbapenemase, was detected in five carbapenem-resistant UPEC isolates. This gene also seems to play an important role in carbapenem resistance among E. coli isolates. blaOXA-48 is endemic in Iran and dissemination of this gene is mainly driven by the composite transposon Tn1999 and its variants [63].
In the present study, the co-existence of blaOXA-48 and blaNDM genes was found in four UPEC isolates. However, it is not clear that these two genes are carried on a single plasmid or placed in the separate plasmids. The presence of these two genes in an isolate has been reported in several studies [15, 17, 63].
Conjugation experiments revealed that all carbapenem resistance genes were able to transfer to other isolates; however, plasmid profiling showed that these genes carried by plasmids with different sizes and probably different sequences. We could not identify the sequence and types of the plasmids which is another limitation of the present study. Complete sequencing of these plasmids can clear up many ambiguities about how they are transmitted, distributed, and their role in the resistance and virulence of pathogenic bacteria. ERIC-PCR clusters indicated that some isolates were clonally similar, but revealed different AMR profiles and different carbapenem- resistant genes that may be due to having different plasmids.
Little is known about the pathogenicity of carbapenem-resistant E. coli strains. Most virulence and antibiotic resistance genes, especially CR genes, are transmitted to bacteria via plasmids. However, the ability of bacteria to accept different plasmids is limited because plasmids have cost for bacteria and the energy resources are limited. It is not clear whether bacteria move toward greater resistance, greater pathogenicity, or both during their evolution. The finding of Gottig et al. revealed that NDM-1 carriage and expression exerted a fitness cost and does not significantly affect virulence [48]. Investigation of the presence of 30 virulence genes in carbapenem-resistant isolates showed that these isolates had from 0 to 10 virulence genes. No virulence gene were found in the isolates 303 and 47 (Table 4). Although they had different antibiotic resistance profile, ERIC-PCR clustering showed that they are the same clone. The isolate 4, which was considered possible PDR, had ten virulence genes; thus, this isolate had high potential virulence and antibiotic resistance at the same time. The isolate 10 also showed a strong virulence profile; however, the virulome of the rest of them reflected low virulence index.
The genes including traT, fimH, fyuA, and iutA were more prevalent among CR isolates. However, when the frequency of virulence genes was assessed based on their function, the genes involved in iron uptake and siderophore production (fyuA, iutA, and chuA) were highest and 75% of these isolates carried at least one of these genes. fyuA (yersiniabactin siderophore) also plays an important role in biofilm formation in urinary tract system and detaching host-derived copper, it thus protect against intracellular killing [64]. El Ghany et al. characterized 10 carbapenem resistant UPEC isolates. They found that although these isolates have different virulence structure; they nevertheless contained at least one iron system gene [2]. Serum resistance traits (traT and cvaC genes) was the other important virulence factor that was found among these isolates. Toxin, protectin, and adhesion virulence factors were of lower importance. Although 58% of the isolates had the fimH gene, the evaluation of the presence of another 15 adhesion genes revealed that the acquisition or maintenance of these genes was less important for these bacteria. In contrast to our results, Ranjan et al. reported that all five investigated NDM-harboring strains inherited genes belonging to adhesions and reflected moderate virulence [16].
Further studies are needed to determine what virulence factors exist in carbapenem-resistant bacteria after gaining resistance. Perhaps a new treatment can be found to overcome these pathogens by targeting common biochemical pathways for the synthesis of proteins related to the virulence and resistance genes.
Conclusion
Antimicrobial resistance, especially resistance to carbapenems, is rapidly expanding. CR is easily transmitted to other bacteria through HGT. The blaNDM followed by blaOXA-48 were the most common cause of producing metallo-beta-lactamase and carbapenemase among the carbapenem-resistant isolates. Virulome analysis of these isolates revealed the low virulence index in most of them; however, genes involved iron uptake genes especially siderophores were prevalent in the CR isolates. Understanding the spreading and pathogenicity in this group of emerging resistant bacteria can help to improve the therapeutic options and stewardship strategies in different regions.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Sequence data of this project have been deposited in the GenBank of the National Center for Biotechnology Information (NCBI) under the accession number MT321108, MT321107, MT321106, and MT293867.
Abbreviations
- UPEC:
-
Uropathogenic Escherichia coli
- UTI:
-
Urinary tract infection
- ERIC:
-
Enterobacterial intergenic repetitive element sequence
- ExPEC:
-
Extraintestinal pathogenic E. coli
- VF:
-
Virulence Factor
- UPGMA:
-
Unweighted pair group method with arithmetic mean
- TSB:
-
Trypton soya broth
- AMR:
-
Antimicrobial resistance
- MDR:
-
Multi-drug resistant
- XDR:
-
Extensively-drug resistant
- PDR:
-
Pan-drug resistant
- CR:
-
Carbapenem resistance
- MBLs:
-
Metallo-β-lactamases
- KPC:
-
Klebsiella pneumoniae Carbapenemase
- HGT:
-
Horizontal gene transfer
- EMB:
-
Eosin methylene blue
- CLSI:
-
Clinical and Laboratory Standards Institute
- CDT:
-
Combined disc test
- DDST:
-
Double disc synergy test
- mCIM:
-
Modified carbapenem inactivation method
- eCIM:
-
EDTA-CIM
- PBA:
-
Phenylboronic acid
- DMSO:
-
Dimethyl sulfoxide (DMSO)
- EIEC:
-
Enteroinvasive E. coli
- NDM:
-
New Delhi metallo-beta-lactamase
- IMP:
-
Imipenemase
- VIM:
-
Verona integron-encoded metallo-beta-lactamase
- OXA:
-
Oxacillinases
- ESBL:
-
Extended-spectrum β-lactamases
- SIM:
-
Sulfur indole motility
- MR/VP:
-
Methyl red/Voges-Proskauer
References
Ghadiri H, Vaez H, Razavi-Azarkhiavi K, Rezaee R, Haji-Noormohammadi M, Rahimi AA, et al. Prevalence and antibiotic susceptibility patterns of extended-spectrum ß-lactamase and metallo-ß-lactamase-producing uropathogenic Escherichia coli isolates. Lab Med. 2014;45:291–6.
Abd El Ghany M, Sharaf H, Al-Agamy MH, Shibl A, Hill-Cawthorne GA, Hong PY. Genomic characterization of NDM-1 and 5, and OXA-181 carbapenemases in uropathogenic Escherichia coli isolates from Riyadh, Saudi Arabia. PLoS ONE. 2018;13:e0201613.
Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. 2008;46:2529–34.
Khairy RM, Mohamed ES, Abdel Ghany HM, Abdelrahim SS. Phylogenic classification and virulence genes profiles of uropathogenic E. coli and diarrhegenic E. coli strains isolated from community acquired infections. PLoS ONE. 2019;14:e0222441.
Hossain M, Tabassum T, Rahman A, Hossain A, Afroze T, Momen AMI, et al. Genotype-phenotype correlation of β-lactamase-producing uropathogenic Escherichia coli (UPEC) strains from Bangladesh. Sci Rep. 2020;10:14549.
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51.
Wang M, Wang W, Niu Y, Liu T, Li L, Zhang M, et al. A clinical extensively-drug resistant (XDR) Escherichia coli and role of Its β-Lactamase Genes. Front Microbiol. 2020;11:590357.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81.
Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010;13:558–64.
Sanchez GV, Baird AM, Karlowsky JA, Master RN, Bordon JM. Nitrofurantoin retains antimicrobial activity against multidrug-resistant urinary Escherichia coli from US outpatients. J Antimicrob Chemother. 2014;69:3259–62.
Sanchez GV, Master RN, Karlowsky JA, Bordon JM. In vitro antimicrobial resistance of urinary Escherichia coli isolates among U.S. outpatients from, to 2010. Antimicrob Agents Chemother. 2000;2012(56):2181–3.
Singh T, Singh PK, Das S, Wani S, Jawed A, Dar SA. Transcriptome analysis of beta-lactamase genes in diarrheagenic Escherichia coli. Sci Rep. 2019;9:3626.
Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, et al. Phenotypic and genotypic characterization of carbapenem-resistant enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67:S196–205. https://doi.org/10.1093/cid/ciy660.
Cole SD, Peak L, Tyson GH, Reimschuessel R, Ceric O, Rankin SC. New Delhi metallo-β-lactamase-5-producing Escherichia coli in companion animals, United States. Emerg Infect Dis. 2020;26:381–3.
Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17:101.
Ranjan A, Shaik S, Mondal A, Nandanwar N, Hussain A, Semmler T, et al. Molecular epidemiology and genome dynamics of New Delhi metallo-β-lactamase-producing extraintestinal pathogenic Escherichia coli Strains from India. Antimicrob Agents Chemother. 2016;60:6795–805.
Sfeir MM, Hayden JA, Fauntleroy KA, Mazur C, Johnson JK, Simner PJ, et al. EDTA-modified carbapenem inactivation method: a phenotypic method for detecting metallo-β-lactamase-producing enterobacteriaceae. J Clin Microbiol. 2019;57.
Sawa T, Kooguchi K, Moriyama K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care. 2020;13.
Chen J, Griffiths MW. PCR differentiation of Escherichia coli from other gram-negative bacteria using primers derived from the nucleotide sequences flanking the gene encoding the universal stress protein. Lett Appl Microbiol. 1998;27:369–71.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
Picão RC, Andrade SS, Nicoletti AG, Campana EH, Moraes GC, Mendes RE, et al. Metallo-β-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol. 2008;46:2028–37.
Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, et al. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother. 2010;65:1664–71.
Tsakris A, Kristo I, Poulou A, Themeli-Digalaki K, Ikonomidis A, Petropoulou D, et al. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J Clin Microbiol. 2009;47:362–7.
Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, et al. PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41:5407–13.
Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother. 2007;59:321–2.
Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid-and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–7.
Tsakris A, Pournaras S, Woodford N, Palepou M-FI, Babini GS, Douboyas J, et al. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000;38:1290–2.
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–54.
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61.
Donald HM, Scaife W, Amyes SG, Young H-K. Sequence analysis of ARI-1, a Novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–9.
Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22.
Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–72.
Clermont O, Bonacorsi S, Bingen E. The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65.
Helalat H, Rezatofighi SE, Roayaei Ardakani M, Dos Santos LF, Askari BM. Genotypic and phenotypic characterization of enteroaggregative Escherichia coli (EAEC) isolates from diarrheic children: an unresolved diagnostic paradigm exists. Iran J Basic Med Sci. 2020;23:915–21. https://doi.org/10.22038/ijbms.2020.42119.9959.
Mukherjee SK, Mukherjee M. Characterization and bio-typing of multidrug resistance plasmids from Uropathogenic Escherichia coli isolated from clinical setting. Front Microbiol. 2019;10:2913.
Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:1–17.
Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, et al. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother. 2009;63:427–37.
Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infectious Dis. 2009;9:228–36.
Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–6.
Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, et al. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J Clin Microbiol. 2013;51:680–3.
Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16:35–40.
Karam MRA, Habibi M, Bouzari S. Relationships between virulence factors and antimicrobial resistance among Escherichia coli isolated from urinary tract infections and commensal isolates in Tehran. Iran Osong Public Health Res Perspect. 2018;9:217–24.
Shahbazi S, Asadi Karam MR, Habibi M, Talebi A, Bouzari S. Distribution of extended-spectrum β-lactam, quinolone and carbapenem resistance genes, and genetic diversity among uropathogenic Escherichia coli isolates in Tehran. Iran J Global Antimicrob Resist. 2018;14:118–25.
Malekzadegan Y, Khashei R, Sedigh H, Jahanabadi Z. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infectious Diseases 2018;18.
Haghighatpanah M, Mojtahedi A. Characterization of antibiotic resistance and virulence factors of Escherichia coli strains isolated from Iranian inpatients with urinary tract infections. Infect Drug Resist. 2019;12:2747–54.
Souli M, Galani I, Giamarellou H. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro surveillance 2008;13.
Cagnacci S, Gualco L, Roveta S, Mannelli S, Borgianni L, Docquier JD, et al. Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem-hydrolysing VIM-1 metallo-beta-lactamase: first Italian outbreak. J Antimicrob Chemother. 2008;61:296–300. https://doi.org/10.1093/jac/dkm471.
Souli M, Kontopidou FV, Papadomichelakis E, Galani I, Armaganidis A, Giamarellou H. Clinical experience of serious infections caused by Enterobacteriaceae producing VIM-1 metallo-beta-lactamase in a Greek University Hospital. Clin Infect Dis. 2008;46:847–54.
Göttig S, Riedel-Christ S, Saleh A, Kempf VA, Hamprecht A. Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int J Antimicrob Agents. 2016;47:430–5.
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–54.
Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli. Canada Emerg Infect Dis. 2011;17:103–6.
Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602.
Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother. 2010;65:2253–4.
Solgi H, Giske CG, Badmasti F, Aghamohammad S, Havaei SA, Sabeti S, et al. Emergence of carbapenem resistant Escherichia coli isolates producing blaNDM and blaOXA-48-like carried on IncA/C and IncL/M plasmids at two Iranian university hospitals. Infect Genet Evol. 2017;55:318–23.
Fazeli H, Norouzi-Barough M, Ahadi A, Shokri D, Solgi H. Detection of New Delhi metallo-beta-lactamase-1 (NDM-1) in carbapenem-resistant Klebsiella pneumoniae isolated from a university hospital in Iran. Hippokratia. 2015;19:205.
Giamarellou H. Multidrug resistance in Gram-negative bacteria that produce extended-spectrum β-lactamases (ESBLs). Clin Microbiol Infect. 2005;11:1–16.
Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, Nasiri S, Owlia P, Nikbin VS, et al. First report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb Drug Resist. 2013;19:30–6.
Mahmoodi F, Rezatofighi SE, Akhoond MR. Antimicrobial resistance and metallo-beta-lactamase producing among commensal Escherichia coli isolates from healthy children of Khuzestan and Fars provinces. Iran BMC Microbiol. 2020;20:366.
Jamali S, Tavakoly T, Mojtahedi A, Shenagari M. The phylogenetic relatedness of blaNDM-1 harboring extended-spectrum β-lactamase producing uropathogenic Escherichia coli and Klebsiella pneumoniae in the North of Iran. Infect Drug Resist. 2020;13:651.
Koraei M, Moosavian M, Saki M. Investigation of New Delhi metallo-beta-lactamase 1 (NDM-1) in clinical Enterobacteriaceae isolates in Southwest Iran. J Res Med Dent Sci. 2018;6:1–5.
Mombini S, Rezatofighi SE, Kiyani L, Motamedi H. Diversity and metallo-β-lactamase-producing genes in Pseudomonas aeruginosa strains isolated from filters of household water treatment systems. J Environ Manage. 2019;231:413–8.
Deldar Abad Paskeh M, Mehdipour Moghaddam MJ, Salehi Z. Prevalence of plasmid-encoded carbapenemases in multi-drug resistant Escherichia coli from patients with urinary tract infection in northern Iran. Iranian J Basic Med Sci. 2020;23:586–93. https://doi.org/10.22038/ijbms.2020.34563.8199.
Nojoomi F, Ghasemian A. Resistance and virulence factor determinants of carbapenem-resistant Escherichia coli clinical isolates in three hospitals in Tehran. Iran Infect Epidemiol Microbiol. 2017;3:107–11.
Pitout JD, Peirano G, Kock MM, Strydom K-A, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev 2019;33. https://doi.org/10.1128/CMR.00102-19.
Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol. 2012;8:731–6.
Acknowledgements
The authors are very thankful to Shahid Chamran University of Ahvaz for the facilities to accomplish the present research project (Grant Number: SCU.SB98.658). This study was related to MSc thesis of Fatemeh Zanganeh-Matin and Fahimeh Mahmoodi; however, Fatemeh Zanganeh-Matin has played a greater role in this work. We also thank Dr. Mahdi Askari (Ferdowsi University of Mashhad) for helping us to perform the revised Clermont phylogrouping.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of the experiment. SER designed and supervised the research study. FZM and FM carried out the experiments. MRA and MRA participated in the design of the study. Data analysis was performed by MRA. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by Ethics Committee of Shahid Chamran University of Ahvaz (No: EE/98.24.3.26336/scu.ac.ir). Before collecting information, participants or parents (for children cases) were asked to read, accept and sign an informed consent form.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zangane Matin, F., Rezatofighi, S.E., Roayaei Ardakani, M. et al. Virulence characterization and clonal analysis of uropathogenic Escherichia coli metallo-beta-lactamase-producing isolates. Ann Clin Microbiol Antimicrob 20, 50 (2021). https://doi.org/10.1186/s12941-021-00457-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-021-00457-4