Abstract
Background
Bloodstream infections are responsible for thousands of deaths each year. The rapid identification of the microorganisms causing these infections permits correct therapeutic management that will improve the prognosis of the patient. In an attempt to reduce the time spent on this step, microorganism identification devices have been developed, including the VITEK® 2 system, which is currently used in routine clinical microbiology laboratories.
Methods
This study evaluated the accuracy of the VITEK® 2 system in the identification of 400 microorganisms isolated from blood cultures and compared the results to those obtained with conventional phenotypic and genotypic methods. In parallel to the phenotypic identification methods, the DNA of these microorganisms was extracted directly from the blood culture bottles for genotypic identification by the polymerase chain reaction (PCR) and DNA sequencing.
Results
The automated VITEK® 2 system correctly identified 94.7 % (379/400) of the isolates. The YST and GN cards resulted in 100 % correct identifications of yeasts (15/15) and Gram-negative bacilli (165/165), respectively. The GP card correctly identified 92.6 % (199/215) of Gram-positive cocci, while the ANC card was unable to correctly identify any Gram-positive bacilli (0/5).
Conclusions
The performance of the VITEK® 2 system was considered acceptable and statistical analysis showed that the system is a suitable option for routine clinical microbiology laboratories to identify different microorganisms.
Similar content being viewed by others
Background
Sepsis is a global health problem and an estimated 17 million cases of sepsis occur each year in the world [1]. The early initiation of appropriate antibiotic therapy is determinant for the prognosis and survival of patients with bloodstream infections [2]. Patients receiving antibiotic therapy that is adapted based on the susceptibility profile of the infectious agent isolated from blood cultures exhibit lower mortality than those treated initially with inadequate antibiotic therapy [3]. In addition, technological advances that permit the rapid and reliable identification of most pathogens involved in infectious diseases have long been recognized to have clinical benefits, including shorter hospital stays and lower mortality, as well as financial benefits by reducing healthcare costs [4].
The objective of this study was to evaluate the sensitivity of the VITEK® 2 system, a system that automatically performs the processes required for microorganism identification and for the determination of antimicrobial susceptibility using a standard primary inoculum isolated from subcultures of positive blood cultures. Although classical identification methods are still considered the gold standard, these methods are slow, time consuming and prone to subjective interpretations. On the other hand, the VITEK® 2 system reduces the time necessary for identification and permits the standardization of inter- and intra-laboratory results, the storage of results, issuing rapid epidemiological reports, and simultaneous identification and antimicrobial susceptibility testing; however, the system is poorly efficient in identifying certain species of Gram-positive cocci [42].
Studies using direct inoculation of VITEK® 2 cards from blood culture bottles have been conducted in an attempt to further reduce the time of identification of microorganisms that cause bloodstream infections, but the results were acceptable only for Gram-negative bacilli and were inaccurate for Gram-positive cocci [5, 6]. For this reason, the present study used inocula of microorganisms cultured previously on solid media for 24 h.
The difference of this study was the prospective evaluation of the VITEK® 2 system during the routine work of a clinical microbiology laboratory in a university hospital using 400 microorganisms isolated from blood cultures collected during the hospitalization period of patients rather than to conduct a retrospective study of samples stored for years.
Methods
Isolates studied
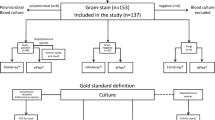
Four hundred microorganisms isolated from positive blood cultures of patients hospitalized in intensive care units of the Botucatu University Hospital between August 2012 and February 2014 were identified. The blood samples were inoculated into blood culture bottles and incubated in the BACTEC™ 9050 apparatus.
Identification of microorganisms in positive blood cultures
Automated phenotypic identification
Samples exhibiting microbial growth were submitted to Gram staining and cultured on solid media directly from the blood culture bottles. After subculture on blood and MacConkey agar, the isolates were inoculated into the following specific identification cards of the automated VITEK® 2 system using the standard protocol: Gram-positive cocci (GP), Gram-positive bacilli (GN), Gram-negative bacilli (ANC), and yeasts (YST). Gram-positive cocci, Gram-positive bacilli and yeast were inoculated into the cards from colonies grown on blood agar and Gram-negative bacilli from colonies grown on MacConkey agar, all diluted in saline (0.9 % NaCl) to a 0.5 McFarland standard.
Phenotypic identification by conventional methods
Phenotypic identification consisted of Gram staining for the observation of morphology and specific staining, followed by a series of biochemical tests specific for each group of microorganisms. Gram-positive cocci were submitted to the catalase test for differentiation between Staphylococcus and Enterococcus. The following biochemical test battery was used for the identification of species of the genus Staphylococcus: coagulase, sugar fermentation (sucrose, maltose, trehalose, xylose, and mannitol), anaerobic growth on semi-solid sodium thioglycolate medium and, if necessary, ornithine and urease production and novobiocin susceptibility [7]. Isolates previously identified as Gram-positive, catalase-negative, bile esculin-positive, NaCl-positive (growth in brain heart infusion broth with 6.5 % NaCl) and pyrrolidonyl-aminopeptidase test-positive cocci were submitted to biochemical tests of fermentation of mannitol, arabinose, arginine and sorbitol, motility, and presence or absence of a pigment on sheep blood agar. Gram-negative bacilli were first tested for glucose fermentation. Glucose-fermenting bacilli were submitted to manual biochemical tests known as EPM/MILi/Citrate, an identification system based on the following tests: production of H2S, urease and l-tryptophan desaminase; motility; indol production; lysine decarboxylase production, and the ability to use citrate as a single carbon source. Non-glucose-fermenting Gram-negative bacilli were identified based on motility, growth at a temperature of 42 °C, and production of DNAse. Yeasts were isolated on Sabouraud agar, replated on CHROMagar, and identified based on the color, texture and shape of their colonies [8].
DNA extraction from the isolates
Extraction of bacterial DNA
Bacterial DNA was extracted directly from the blood sample of the blood culture bottle using the Illustra Kit (GE Healthcare) according to the protocol of the manufacturer, with modifications in the first centrifugation [9] and the addition of 800 µL benzyl alcohol [10]. For sample collection, the lid of the blood culture bottle was first disinfected with cotton soaked in 70 % alcohol. Next, 1.5 mL of the culture was aspirated with a sterile needle and syringe and transferred to sterile microtubes. The microtubes were centrifuged at 850g for 2 min and the supernatant was removed by aspiration with a micropipette and sterile tips and the supernatant stored (directly in a DNA-free microtube) at −20 °C until the time of extraction.
For DNA extraction, the sample was centrifuged at 10,000g for 1 min, the supernatant was discarded, and 500 µL lysozyme was added to the sediment. The mixture was vortexed, 800 µL benzyl alcohol was added, and the mixture was again shaken and centrifuged at 7000g for 5 min. Next, 300 µL of the supernatant was carefully removed and transferred to a new sterile microtube (which was used for extraction). Ten microliter lysozyme (10 mg/mL) was added and the microtube was left to stand at room temperature for 15 min, with vortexing every 5 min. After this period, 10 µL proteinase K (20 mg/mL) was added and the mixture was vortexed. The microtube was incubated for 15 min at 56 °C, with vortexing every 5 min. This mixture was then transferred to an extraction microcolumn and centrifuged at 11,000g for 1 min. The filtrate was discarded and 500 µL washing solution was added to the microcolumn. The column was again centrifuged at 11,000g for 3 min. The supernatant was discarded, the microcolumn was transferred to a new sterile microtube, and 200 µL Milli-Q water previously heated to 70 °C was added. The microcolumn was kept at room temperature for 1 min and centrifuged at 11,000g for 1 min. The columns were discarded and the filtered material was frozen until analysis by the polymerase chain reaction (PCR).
Extraction of yeast DNA
Yeast DNA was extracted according to the protocol proposed by McCullough et al. [11]. The isolates were seeded onto inclined Sabouraud agar and incubated for 36 h at 37 °C. A loopful of this culture was resuspended in a 2-mL tube containing 1 mL 1 M sorbitol, 125 mM EDTA, and 500 mg glass beads. The tube was shaken twice in a Precellys® homogenizer for 45 s and centrifuged at 13,000g for 10 min. The supernatant was discarded and the sediment together with the glass beads was resuspended in 500 µL of a buffer solution containing 50 mM Tris–HCl, 50 mM EDTA and 2 % SDS and incubated for 1 h at 65 °C. After incubation, 500 µL 3 M sodium acetate was added. The mixture was homogenized by inverting the tube and kept on ice for 2 h, followed by centrifugation for 10 min at 25 °C. The supernatant was transferred to a 1.5-mL centrifugation microtube containing 1 mL ice-cold absolute ethanol, homogenized by inversion, and centrifuged for 10 min at 4 °C. The supernatant was discarded and the DNA retained on the tube wall was resuspended in 50 µL autoclaved Milli-Q water and frozen until the time of PCR.
Genotypic identification of the isolates
Polymerase chain reaction of bacteria
Gram-positive bacteria of the genus Staphylococcus that belonged to the group of coagulase-negative staphylococci (CoNS) were identified by internal transcribed spacer PCR (ITS-PCR) using primers targeting conserved sequences adjacent to the 16S and 23S genes: G1 (5′-GAAGTCGTAACAAGG-3′) and L1 (5′-CAAGGCATCCACCGT-3′) [12, 13].
The remaining isolates of Gram-positive cocci and Gram-negative bacilli were submitted to PCR carried out in 0.2-mL microtubes containing 15.8 µL Milli-Q water, 10 pM of the forward primer and 10 pM of the reverse primer, 100 µM triphosphate desoxyribonucleotides (GE Healthcare), 10 U Taq DNA polymerase (Biotools), 20 mM MgCl2-free buffer (Biotools), 0.75 mM MgCl2 (Biotools), and 3 µL DNA. Primers targeting conserved sequences of each species were used for DNA amplification. The temperature and time parameters and number of amplification cycles reported in the literature and described in Table 1 were used.
The efficiency of the amplifications was monitored by electrophoresis on 2 % agarose gel prepared in 1× TBE buffer (89 mM Tris (pH 7.6), 89 mM boric acid, and 2 mM EDTA) and stained with SYBR® Safe at 90 V for 60 min (Figs. 1, 2). The following international reference strains were used as controls: Acinetobacter baumannii (ATCC 19606), Enterobacter cloacae (ATCC 23355), Enterococcus faecalis (ATCC 29212), Enterococcus faecium (ATCC 6569), Escherichia coli (ATCC 10536), Klebsiella pneumoniae (ATCC 4352), Morganella morganii (ATCC 8019), Proteus mirabilis (ATCC 15290), Pseudomonas aeruginosa (ATCC 15442), Serratia marcescens (ATCC 14756), Staphylococcus aureus (ATCC 25923), and Stenotrophomonas maltophilia (ATCC 13637).
Figures 1 and 2 illustrate the genotypic identification of Enterobacter cloacae and Escherichia coli, respectively.
DNA sequencing of bacteria
The yeast and Gram-positive bacillus isolates were sequenced for identification to species level. Gram-negative bacilli identified as Enterobacter aerogenes by the conventional phenotypic methods and by the automated test were sequenced for species confirmation.
Amplification and purification of bacterial DNA for sequencing
The bacterial isolates identified as Gram-positive bacilli by Gram staining and the Gram-negative bacilli identified as Enterobacter aerogenes by the conventional phenotypic methods and automated test were submitted to simple PCR. The efficiency of the reactions was monitored as described in item 2.4.1. For PCR, protocols described in the literature were followed using universal primers for Gram-positive bacilli [14] and primers of enterobacteria for Enterobacter aerogenes isolates [15]. The temperature and time parameters and number of amplification cycles are shown in Table 1. The amplified DNA fragments were purified using the HiYield Gel/PCR DNA Fragments Extraction Kit (RBC).
Amplification and purification of yeast DNA
The yeast isolates were submitted to ITS-PCR as described by Kurtzman et al. [16]. These reactions were carried out in 0.2-mL microtubes containing a mixture of 6.7 µL Milli-Q water, 10 µM forward primer and 10 µM reverse primer, 12.5 µL GoTaq®Green Master Mix, and 3 µL DNA. The efficiency of the amplifications was monitored by electrophoresis (60 min, 80 V) on 1.5 % agarose gel prepared in 1× TBE buffer and stained with SYBR® Safe. Strains previously identified at the Laboratory of Fungal Biology, Department of Microbiology and Immunology, IBB, were used as controls. The amplified fragments were purified using the Illustra™ ExoProStar™ Kit (GE Healthcare Life Sciences).
DNA sequencing reaction of yeast and bacteria
The sequencing reactions were carried out in a mixture containing 3.25 µL water, 1.75 µL 5× BigDye buffer (Applied Biosystems), 1 µL of each primer (5 pmol/µL), 2 µL of the PCR product, and 2 µL BigDye (Applied Biosystems). The cycle sequencing reaction was initiated at 96 °C for 1 min, followed by 40 cycles of denaturation at 96 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 4 min. The sequencing reaction product was precipitated with 1 µL 125 mM EDTA, 1 µL 3 M sodium acetate, and 25 µL 100 % ethanol. After homogenization, the solution was left to stand for 15 min and then centrifuged at 3000g for 15 min at 4 °C. The supernatant was removed by inverting the tube and 35 µL 70 % ethanol was added. The solution was centrifuged at 1650g for 15 min at 4 °C. After removal of the supernatant by inversion, 10 µL HiDi formamide (Applied Biosystems) was added and the mixture was left to stand for 5 min at 95 °C and for 2 min on ice. The product was run on an 8-capillary ABI 3500 sequencer (50 cm) using POP7 polymer (Applied Biosystems).
Analysis of the DNA sequences of yeast and bacteria
The MEGA 5.0 (Molecular Evolutionary Genetics Analysis) and Lasergene programs were used for visualization and alignment of the DNA sequences obtained, which were compared to sequences published and stored in the GenBank database.
Statistical analysis
Agreement between the results obtained with the different identification methods was analyzed statistically by the kappa test using the SPSS 20 program (IBM, Armonk, NY, USA). Genotypic identification was used as the gold standard. The accuracy of the conventional phenotypic identification methods and identification by the VITEK® 2 system was evaluated by calculating sensitivity and specificity according to Fletcher et al. [17].
Results
The results of identification with the automated VITEK® 2 system showed overall agreement of 94.7 % with the results of the genotypic methods (Tables 2, 3). Overall agreement of 98.7 % was observed between the results obtained by phenotypic identification using conventional methods and the results of the genotypic methods.
All yeast isolates (15/15) were correctly identified by the VITEK® 2 system using the YST card. The same result was obtained with the GN card for the identification of Gram-negative bacilli, with 100 % correct identifications of the 165 strains isolated.
The rate of correct identification obtained with the GP card used for the identification of Gram-positive cocci was 92.6 % for the microorganisms isolated (199/215). The agreement between the results of automated identification and those obtained with the other identification methods for Enterococcus spp. was 91.7 % due to the incorrect identification of one Enterococcus faecalis strain by the VITEK® 2 system. The agreement between species of the genus Staphylococcus was 97.5 % (198/203). The VITEK® 2 system correctly identified all Staphylococcus aureus isolates (17/17) and incorrectly identified 15 of the 186 (9.19 %) isolates belonging to the group of CoNS. The highest rate of incorrect identification was observed for Enterococcus faecalis with 12.5 % (7/8), followed by Staphylococcus epidermidis with 12.3 % (10/81), Staphylococcus hominis with 8.6 % (3/35), Staphylococcus capitis with 5.5 % (1/18), and Staphylococcus haemolyticus with 2 % (1/50).
The VITEK® 2 system incorrectly identified the isolates because some biochemical tests failed during the identification process performed by the device, exhibiting divergent characteristics of the species isolated. These errors are shown in Table 4.
Fewer errors occurred when the conventional phenotypic methods were used for identification compared to the automated VITEK® 2 system. Among Gram-positive cocci, the conventional phenotypic methods correctly identified 211/215 (98.1 %) isolates and the few errors observed mainly occurred in the identification of CoNS species, showing divergent results for a given species. The conventional phenotypic methods correctly identified all yeasts (15/15) and 164/165 (99.4 %) Gram-negative bacilli, with the errors described in Table 4.
One hundred percent discrepant results were obtained for the identification of five isolates of Gram-positive bacilli by the VITEK® 2 system (ANC card) and the genotypic identification methods.
Statistical analysis of the identification results revealed a kappa value of 0.945 (p < 0.001), indicating almost perfect agreement according to the criteria of Landis and Koch [18] (Table 5).
Agreement between the conventional phenotypic methods and genotypic identification was higher than that between the genotypic method and automated identification by the VITEK® 2 system (Table 6).
Comparison of the sensitivity of the conventional phenotypic methods and the VITEK® 2 system showed a better performance of the former (Table 7).
Discussion
The need for the rapid and efficient identification of microorganisms isolated from blood cultures has encouraged studies that investigated automated identification systems to reduce the time of identification. Several of these studies have used direct inoculation from blood culture bottles, but the results were not as efficient as those obtained in studies using standard inocula from subcultures of microorganisms grown for 24 h on solid media. The poor performance of the VITEK® 2 system for the identification of microorganisms using direct inoculation from blood culture bottles is probably due to the small number of cells or to contamination with other microorganisms that impair the correct identification of the causative agent of infection [19–22]. The VITEK system has been investigated for more than two decades and improvement of the ID-GPC (Gram-positive cocci) and ID-GNB (Gram-negative bacilli) identification cards to the GP (Gram-positive cocci) and GN (Gram-negative bacilli) cards has made the system more efficient. Wallet et al. [23] compared the old and new identification cards and found that the GP and GN cards correctly identified 235/249 (94.4 %) Gram-positive cocci and 321/331 (97 %) Gram-negative bacilli, while the ID-GPC and ID-GNB correctly identified 218/249 (87.5 %) and 295/331 (89.1 %) isolates, respectively.
The present study was conducted over a period of 18 months and evaluated the accuracy of the VITEK® 2 system in identifying 400 microorganisms (Gram-positive cocci, Gram-positive bacilli, Gram-negative bacilli, and yeasts) isolated from blood cultures and inoculated by the standard method onto GP, GN, YST (yeast) and ANC (Gram-positive bacilli) cards. The results were compared to genotypic identification (gold standard) and 94.7 % agreement was observed. Similar rates have been reported by De Cueto et al. [24] (95.0 %, 95/100) and by Nakasone et al. [25] (95.8 %, 454/474) who also used standard inocula. Studies using direct inoculation from blood cultures bottles obtained lower agreement of 91.4 % [5] and 81.0 % [6].
All 15 yeasts isolated during the study period were correctly identified by the VITEK® 2 system. Correct identification of all yeast isolates (56/56) has also been observed by Nakasone et al. [25]. Studies involving a larger number of strains and species found lower agreement, 92.1 % (222/241) reported by Graf et al. [26] and 78.9 % (277/351) by Won et al. [27].
The VITEK® 2 system has shown satisfactory identification rates of Gram-negative bacilli for decades, as also observed in this study in which 100 % correct identifications of the isolates were obtained. Funke et al. [28] and Ling et al. [29] analyzed 845 and 281 isolates, with correct identification rates of 84.7 % (716/845) and 95 % (267/281), respectively. Nakasone et al. [25], Gherardi et al. [5] and Prod’hom et al. [6] analyzed 181, 91 and 95 Gram-negative bacilli and obtained correct identifications of 96.7 % (175/181), 100 % (91/91) and 98.8 % (92/95), respectively, with the VITEK® 2 system. Studies using direct inoculation from blood culture bottles reported lower rates of correct identification of 82 [30] and 93 % [31]. De Cueto et al. [24] compared direct inoculation from blood cultures with inoculation from subcultures in 50 isolates. The result was 100 % correct identifications for the standard method and 62 % (31/5) correct identifications for direct inoculation. Similar results have been reported by Kerremans et al. [32] who analyzed 161 isolates; 90 % (145/161) of the isolates were correctly identified by subculture and 80 % (129/161) by direct inoculation from blood cultures.
The identification of Gram-positive cocci by the automated VITEK® 2 system showed 92.6 % agreement with genotypic identification, which is compatible with the rate reported by Ligozzi et al. [33] who obtained 91.5 % (351/381) correct identifications. De Cueto et al. [24] obtained 100 % (50/50) correct identifications of the isolates. Funke and Funke-Kissling [34], Nakasone et al. [25] and Chatzigeorgiou et al. [35] reported higher rates of correct identification than those obtained in this study of 94.5 % (344/364), 96.1 % (226/235) and 97.9 % (144/147), respectively. Gherardi et al. [5] and Prod’hom et al. [6] obtained correct identifications of 75 % (36/48) and 74 % (133/180), while Lupetti et al. [36] found a rate of 89 % (49/55). These rates are lower than those observed in this study and are outside the acceptable parameter of 90 % correct identifications; however, these studies used direct inoculation from blood culture bottles.
With respect to genera of Gram-positive cocci, a difference in the efficiency of the VITEK® 2 system was observed. Agreement was 91.7 % (11/12) for the genus Enterococcus, similar to the rates reported by Nakasone et al. [25] and Jin et al. [37]. Lower efficiencies of 77.8, 83.1, 87.5, 72 and 77.8 % correct identifications of strains of this genus have been observed by Ligozzi et al. [33], d’Azevedo et al. [38], Moore et al. [39] and Paim et al. [42], respectively. In the case of Staphylococcus, a higher rate of incorrect identifications was observed for CoNS isolates (91.9 %), in agreement with Ligozzi et al. [33] (86 %), Funke and Funke-Kissling [34] (93.7 %), Kim et al. [40] (87.5 %), Delmas et al. [41] (78.8 %), and Paim et al. [42] (72.9 %). These unsatisfactory results are due to the fact that automated identification systems are unable to perform a fully reliable differentiation between different CoNS species because of the variable expression of phenotypic characteristics in these microorganisms [43]. The slow metabolism of certain species leads to ambiguous results in their identification, a fact observed by Ligozzi et al. [33]. All Staphylococcus aureus isolates were correctly identified (17/17), as also reported in the studies of Delmas et al. [41] (6/6), Chatzigeorgiou et al. [35] (52/52), Paim et al. [42] (11/11), and Funke and Funke-Kissling [34] (45/45). Ligozzi et al. [33], who evaluated a larger number of isolates, found 99 % agreement (99/100). These rates of correct identification demonstrate a satisfactory performance of the VITEK® 2 system for the identification of Staphylococcus aureus.
The failure of the ANC card to identify Gram-positive bacilli can be explained by the variability in the genera and species of these microorganisms and the consequent difficulty in developing cards that contain variable biochemical tests necessary for correct identification. These microorganisms were not identified by conventional phenotypic methods since this identification is infeasible in routine clinical microbiology because it requires numerous expensive and time-consuming biochemical tests.
The better performance of the conventional methods for CoNS identification was responsible for the higher sensitivity of these methods compared to the VITEK® 2 system. This finding can be explained by the fact that the conventional methods used consisted of specific tests for each CoNS species and by the incubation period of 72 h, which is necessary for this correct identification since some species have a slower metabolism on some substrates.
Conclusions
The kappa values indicate reliability of the results obtained with the VITEK® 2 system. Analysis of specificity showed a performance higher than 90 % as required for commercial systems in clinical microbiology, demonstrating that this system is suitable for the identification of microorganisms isolated in routine clinical microbiology laboratories.
References
Instituto Latino Americano para Estudos da Sepse. Sepse: um problema de saúde pública. Brasília: CFM; 2015.
Diament D, Salomão R, Rigatto O, Gomes B, Silva E, Carvalho NB, Machado FR. Guidelines for the treatment of severe sepsis and septic shock—management of the infectious agent—diagnosis. Rev Bras Ter Intensiva. 2011;23(2):134–44.
Weinstein MP, Murphy JR, Reller LB, Lichtenstein KA. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations, with special reference to factors influencing prognosis. Rev Infect Dis. 1983;5(1):54–70.
Barenfanger J, Drake C, Kacich G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol. 1999;37(5):1415–8.
Gherardi G, Angeletti S, Panitti M, Pompilio A, Di Bonaventura G, Crea F, Avola A, Fico L, Palazzo C, Sapia GF, Visaggio D, Dicuonzo G. Comparative evaluation of the VITEK-2 Compact and Phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn Microbiol Infect Dis. 2012;72(1):20–31.
Prod’hom G, Durussel C, Greub G. A simple blood-culture bacterial pellet preparation for faster accurate direct bacterial identification and antibiotic susceptibility testing with the VITEK 2 system. J Med Microbiol. 2013;62(5):773–7.
Cunha MLRS, Sinzato YK, Silveira LVA. Comparison of methods for the identification of coagulase negative staphylococci. Mem Inst Oswaldo Cruz. 2004;99(8):855–60.
Winn WC Jr, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, Woods GL. Koneman. Diagnóstico microbiológico: texto e altlas colorido. 6th ed. Rio de Janeiro: Editora Guanabara Koogan; 2010.
Hogg GM, McKenna JP, Ong G. Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert blood culture bottles using real-time polymerase chain reaction: evaluation and comparison of 4 DNA extraction methods. Diagn Microbiol Infect Dis. 2008;61(4):446–52.
Fredericks DN, Relman DA. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36(10):2810–6
McCullough MJ, Di Salvo AF, Clemons KV, Park P, Stevens DA. Molecular epidemiology of Blastomyces dermatitidis. Clin Infect Dis. 2000;30(2):328–35.
Barry T, Colleran G, Glennon M, Dunican LK, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1(1):51–6.
Couto I, Pereira S, Miragaia M, Sanches IS, Lencastre H. Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR. J Clin Microbiol. 2001;39(9):3099–103.
Khamis A, Raoult D, La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004;42(9):3925–31.
Gouveia C, Asensi MD, Zahner V, Rangel EF, Oliveira SM. Study on the bacterial midgut microbiota associated to different Brazilian populations of Lutzomyia longipalpis (Lutz & Neiva) (Diptera: Psychodidae). Neotrop Entomol. 2008;37(5):597–601.
Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 59 end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35(5):1216–23.
Fletcher RH, Fletcher SW, Wagner EH. Diagnóstico. In: Fletcher RH, Fletcher SW, Wagner EH, editors. Epidemiologia Clínica. 3rd ed. Porto Alegre: Artes Médicas; 1991. p. 68–107.
Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–74.
Fontanals D, Salceda F, Hernández J, Sanfeliu I, Torra M. Evaluation of wider system for direct identification and antimicrobial susceptibility testing of gram-negative bacilli from positive blood culture bottles. Eur J Clin Microbiol Infect Dis. 2002;21(9):693–5.
Waites KB, Brookings ES, Moser SA, Zimmer BL. Direct bacterial identification from positive BacT/Alert blood cultures using MicroScan overnight and rapid panels. Diagn Microbiol Infect Dis. 1998;32(1):21–6.
Alipour M, Hajiesmaili R, Talebjannat M, Yahyapour Y. Identification and antimicrobial resistance of Enterococcus spp. isolated from the river and coastal waters in northern Iran. Scientific World Journal. 2014; 2014:287458.
Waites KB, Brookings ES, Moser SA, Zimmer BL. Direct susceptibility testing with positive BacT/Alert blood cultures by using MicroScan overnight and rapid panels. J Clin Microbiol. 1998;36(7):2052–6.
Wallet F, Loïez C, Renaux E, Lemaitre N, Courcol RJ. Performances of VITEK 2 colorimetric cards for identification of Gram-positive and Gram-negative bacteria. J Clin Microbiol. 2005;43(9):4402–6.
De Cueto M, Ceballos E, Martinez-Martinez L, Perea EJ, Pascual A. Use of positive blood cultures for direct identification and susceptibility testing with the VITEK 2 system. J Clin Microbiol. 2004;42(8):3734–8.
Nakasone I, Kinjo T, Yamane N, Kisanuki K, Shiohira CM. Laboratory-based evaluation of the colorimetric VITEK-2 Compact system for species identification and of the Advanced Expert System for detection of antimicrobial resistances: VITEK-2 Compact system identification and antimicrobial susceptibility testing. Diagn Microbiol Infect Dis. 2007;58(2):191–8.
Graf B, Adam T, Zill E, Göbel UB. Evaluation of the VITEK 2 system for rapid identification of yeast and yeast-like organisms. J Clin Microbiol. 2000;38(5):1782–5.
Won EJ, Shin JH, Kim MN, Choi MJ, Joo MY, Kee SJ, Shin MG, Suh SP, Ryang DW. Evaluation of the BD Phoenix system for identification of a wide spectrum of clinically important yeast species: a comparison with VITEK 2-YST. Diagn Microbiol Infect Dis. 2014;79(4):477–80.
Funke G, Monnet D, de Bernardis C, von Graevenitz A, Freney J. Evaluation of the VITEK 2 system for rapid identification of medically relevant Gram-negative rods. J Clin Microbiol. 1998;36(7):1948–52.
Ling TKW, Tam PC, Liu ZK, Cheng AFB. Evaluation of VITEK 2 rapid identification and susceptibility testing system against Gram-negative clinical isolates. J Clin Microbiol. 2001;39(8):2964–6.
Ling TK, Liu ZK, Cheng AF. Evaluation of the VITEK 2 system for rapid direct identification and susceptibility testing of Gram-negative bacilli from positive blood cultures. J Clin Microbiol. 2003;41(10):4705–7.
Bruins MJ, Bloembergen P, Ruijs GJ, Wolfhagen MJ. Identification and susceptibility testing of Enterobacteriaceae and Pseudomonas aeruginosa by direct inoculation from positive BACTEC blood culture bottles into VITEK 2. J Clin Microbiol. 2004;42(1):7–11.
Kerremans JJ, Goessens WH, Verbrugh HA, Vos MC. Accuracy of identification and susceptibility results by direct inoculation of VITEK 2 cards from positive BACTEC cultures. Eur J Clin Microbiol Infect Dis. 2004;23(12):892–8.
Ligozzi M, Bernini C, Bonora MG, De Fatima M, Zuliani J, Fontana R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant Gram-positive cocci. J Clin Microbiol. 2002;40(5):1681–6.
Funke G, Funke-Kissling P. Performance of the new VITEK 2 GP card for identification of medically relevant Gram-positive cocci in a routine clinical laboratory. J Clin Microbiol. 2005;43(1):84–8.
Chatzigeorgiou KS, Siafakas N, Petinaki E, Argyropoulou A, Tarpatzi A, Bobola M, Paniara O, Velegraki A, Zerva L. Identification of staphylococci by Phoenix: validation of a new protocol and comparison with VITEK 2. Diagn Microbiol Infect Dis. 2010;68(4):375–81.
Lupetti A, Barnini S, Castagna B, Capria AL, Nibbering PH. Rapid identification and antimicrobial susceptibility profiling of Gram-positive cocci in blood cultures with the VITEK 2 system. Eur J Clin Microbiol Infect Dis. 2010;29(1):89–95.
Jin WY, Jang SJ, Lee MJ, Park G, Kim MJ, Kook JK, Kim DM, Moon DS, Park YJ. Evaluation of VITEK 2, MicroScan, and Phoenix for identification of clinical isolates and reference strains. Diagn Microbiol Infect Dis. 2011;70(4):442–7.
D’Azevedo PA, Cantarelli VV, Inamine E, Superti S, Dias CAG. Evaluation of an automated system for the identification of enterococci. J Bras Patol Med Lab. 2004;40(4):237–9.
Moore DF, Zhowandai MH, Ferguson DM, McGee C, Mott JB, Stewart JC. Comparison of 16S rRNA sequencing with conventional and commercial phenotypic techniques for identification of enterococci from the marine environment. J Appl Microbiol. 2006;100(6):1272–81.
Kim M, Heo SR, Choi SH, Kwon H, Park JS, Seong MW, Lee DH, Park KU, Song J, Kim EC. Comparison of the MicroScan, VITEK 2, and Crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulase-negative Staphylococci. BMC Microbiol. 2008;23(8):233–9.
Delmas J, Chacornac JP, Robin F, Giammarinaro P, Talon R, Bonnet R. Evaluation of the VITEK 2 system with a variety of Staphylococcus species. J Clin Microbiol. 2008;46(1):311–3.
Paim TGS, Cantarelli VV, D’Azevedo PA. Performance of the VITEK 2 system software version 5.03 in the bacterial identification and antimicrobial susceptibility test: evaluation study of clinical and reference strains of Gram-positive cocci. Rev Soc Bras Med Trop. 2013;47(3):377–81.
Weinstein MP, Mirrett S, Pelt LV, McKinnon M, Zimmer L, Kloos W, Reller LB. Clinical importance of identifying coagulase-negative Staphylococci isolated from blood cultures: evaluation of MicroScan rapid and dried overnight gram-positive panels versus a conventional reference method. J Clin Microbiol. 1998;36(7):2089–92.
Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974–6.
Turton JF, Hyde R, Martin K, Shah J. Genes encoding OXA-134-like enzymes are found in Acinetobacter lwoffii and A. schindleri and can be used for identification. J Clin Microbiol. 2012;50(3):1019–22.
Stumpf AN, Roggenkamp A, Hoffmann H. Specificity of enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic polymerase chain reaction for the detection of clonality the Enterobacter cloacae. Diagn Microbiol Infect Dis. 2005;53(1):9–16.
Williams JM, Trope M, Caplan DJ, Shugars DC. Detection and quantitation of E. faecalis by real-time PCR (qPCR), reverse transcription-PCR (RT-PCR) and cultivation during endodontic treatment. J Endod. 2006;32(8):715–21.
Doumith M, Day MJ, Hope R, Wain J, Woodford N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol. 2012;50(9):3108–10.
Chander Y, Ramakrishnan MA, Jindal N, Hanson K, Goyal SM. Differentiation of Klebsiella pneumoniae and K. oxytoca by multiplex polymerase chain reaction. Int J Appl Res Vet Med. 2011;9(2):138–42.
Kim SH, An H, Visessanguan W, Benjakul S, Morrissey MT, Su YC, Pitta TP. Molecular detection of a histamine former, Morganella morganii, in Albacore, Mackrel, Sardine, and a processing plant. JFS. 2003;68(2):453–7.
Belas R, Schneider R, Melch M. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol. 1998;180(23):6126–39.
Jeong ES, Lee KS, Heo SH, Seo JH, Choi YK. Triplex PCR for the simultaneous detection of Pseudomonas aeruginosa, Helicobacter hepaticus and Salmonella typhimurium. Exp Anim. 2011;60(1):65–70.
Polson SW, Higgins JL, Woodley CM. PCR-based assay for detection of four coral pathogens. In: Proceedings of the 11th international coral reef symposium, Ft. Lauderdale. Florida. 2008. p. 247-51.
Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;6(3):618–23.
Stenotrophomonas maltophilia MLST Databases. http://pubmlst.org/smaltophilia/info/primers.shtml. Accessed 6 Apr 2016.
Authors’ contributions
ACMM conceived and designed the study, performed the tests, analyzed and interpreted the data, and wrote the manuscript. CMCBF substantially contributed to the conception of the study and performed the statistical analyses. AMF collected the blood cultures and performed the automated identification. RSC participated in the data collection and analysis. EB identified the yeast isolates. MLRSC participated in the conception of the study, data analysis, and writing and revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for financial support (Grant 2012/1566-8).
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
The data set supporting the results of this article is included within the article.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Faculdade de Medicina de Botucatu, Botucatu, São Paulo State, Brazil (Protocol 347/2012-CEP).
Funding
Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP)—Grant 2012/1566-8.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Monteiro, A.C.M., Fortaleza, C.M.C.B., Ferreira, A.M. et al. Comparison of methods for the identification of microorganisms isolated from blood cultures. Ann Clin Microbiol Antimicrob 15, 45 (2016). https://doi.org/10.1186/s12941-016-0158-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-016-0158-9