Abstract
Background
To investigate the drug resistant gene profiles and molecular typing of Acinetobacter baumannii isolates collected from clinical specimens in a comprehensive hospital, Jiangsu province.
Methods
This study included 120 patients in a comprehensive hospital with drug-resistant A. baumannii infections on clinical specimens from October 2011 to December 2013. Antibiotic susceptibility test was determined by Vitek 2 Compact system. OXA-51, OXA-23, OXA-24, OXA-58, VIM, IMP, SHV, GES, TEM, AmpC, qacEΔ1-sul1, intI l, CarO, aac(6′)-Ib, and aac(6′)-II were analyzed by PCR. The analysis of molecular typing for 50 multidrug resistant A. baumannii isolates was performed by PFGE.
Results
A total of 64(53%) isolates were multidrug-resistant A.baumannii. The antibiotic susceptibility tests showed that the resistant rates to common antibiotics of mutidrug-resistant A. baumannii were extremely high, most of which over 60%. One hundred and ten isolates harbored OXA-51 (91.7%), 100 for OXA-23(83.3%), 103 for VIM-1(85.8%), 90 for AmpC(75.00%), 50 for aac(6′)-Ib(41.7%), 77 for the loss of CarO (64.2%), 85 for intl1(70.8%), and 64 for qacEΔ1-sul1(53.33%), while OXA-24 was undetected. Fifty multidrug-resistant A. baumannii isolates belong to 14 clones according to the PFGE DNA patterns. Main clone A includes 24 isolates, while clone B and clone C includes 6 and 9 isolates, respectively and others with no common source identified.
Conclusion
There is high morbidity of A. baumannii infections in the hospital, especially in ICU and sputum is the most common sample type.The mainly drug-resistant genes of A. baumannii are OXA-51, OXA-23, and VIM-1 in the hospital. Clonal dissemination provides evidence for the prevalence of multidrug-resistant A. baumannii among clinical isolates. It is suggested that there is an urgent need for effective control and prevention measures.
Similar content being viewed by others
Background
Acinetobacter baumannii (A.baumannii) is a non-fermentative, gram-negative, conditional pathogenic bacterium, which can colonize and survive for prolonged period under a wide range of environmental conditions, such as hospital environment and human skin [1]. Meanwhile, A. baumannii is an emerging opportunistic nosocomial pathogen, with increasingly global prevalence, responsible for a variety of nosocomial infections including nosocomial pneumonia, blood infection, urinary tract infection, surgical wound infection, etc., especially for patients in intensive care unit (ICU) [2,3]. Its great capacity to survive in low-moisture environments and its ability to develop resistance to antimicrobial agents afford A. baumannii the possibility of spreading in hospitals. The increasing threat of antibiotic resistance in microbes impacting on the patient outcomes has been recognized as a challenge for treatment of clinical infection with broad spectrum antibiotics use. According to the report from CHINET, 2012, surveillance data reveal that the resistance rates of Acinetobacter spp. (A. baumannii accounted for 89,6% to imipenem and meropenem were up to 62.8%and 59.4%, respectively [4]. What’s more, multidrug resistant A. baumannii, defined as resistant to at least three different groups of antibiotics, causes numerous nosocomial outbreaks and health care–associated infections around the world [1]. The identification of drug resistance mechanisms in A. baumannii will improve the outcome of infections caused by this organism. The resistance mechanisms of A. baumannii are complex, which include activating or production of enzyme, the integron formation, outer membrane permeability, biofilm formation, drug exocytosis mechanism and so on [5-9].
Carbapenem resistance in A.baumannii is increasingly being observed worldwide [5]. The most important mechanism of carbapenem resistance in A. baumannii is the enzymatic hydrolysis mediated by carbapenem-hydrolyzing β-lactamases, including class A (TEM, SHV, and GES), class B (IMP, VIM, and SIM), class C (AmpC), and class D (OXA-23-like, OXA-24-like, OXA-51-like, and OXA-58-like) [10]. Resistance to aminoglycosides in A. baumannii is mainly mediated by the production of aminoglycosides-modifying enzymes (AMEs). The most frequent AMEs in A. baumannii are AAC(6′)-Ib and AAC(6′)-II [3,11]. What’ more, the loss of outer-membrane protein(OMP) and the acquisition of class 1 integron are also contributed to an increasing incidence of drug resistance [12]. In addition, many commercial products based on ammonium quaternary compounds (QAC) are currently used in considerable quantities as antiseptic agents in hospitals, but due to the intrinsic and chronic resistance to QAC, infections with A. baumannii are growing [13].
To date, there are few reports of combinations of different resistance mechanisms, but there is a correlation of antimicrobial resistance with the enzyme production and porin and integrons. In the present study, from 2011 to 2013, surveillance at a provincial hospital in Jiangsu detected 120 drug-resistant A. baumannii isolates. We aimed to analyze the genetic linkage and drug-resistance gene profiles of these drug-resistant A. baumannii isolates and investigate various mechanisms of drug resistance in isolates.
Materials and methods
Subjects and bacterial isolates
This investigation was conducted at a comprehensive hospital in Nanjing, Jiangsu, China, from Oct 2011 to Dec 2013. Written informed consent was obtained from the participants for the use of samples in this study. This study was approved by the Nanjing Medical University Clinical Research Ethics Committee, Nanjing, China. No patients received antibiotic therapy before samples were collected. All the clinical isolates were routinely collected and stored at −80°C until use. One hundred and twenty A. baumannii strains were isolated from clinical samples. Multidrug resistant A. baumannii was defined as A. baumannii isolates which were resistant to more than 3 classes of antimicrobials.
Antibiotic susceptibility testing
A. baumannii identification and general antimicrobial susceptibilities were performed using Vitek 2 Compact system (bioMérieux, Inc., Marcy-l’Etoile, France). Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), and Staphylococcus aureus (ATCC 29213) were used as quality control strains. Susceptibility results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
PCR of drug resistant genes
Genomic DNA of A.baumannii isolates was extracted using TIANamp Bacteria DNA Kit(Tiangen). PCR was performed using Taq PCR Master Mix(TaKaRa).Primers were synthesized by Jierui, Shanghai. The details of primer sequences were showed in Table 1. Each reaction was performed in a final volume of 50 μl consisting of 25 μl Taq Mix, 1 μl primers, 1 μl DNA template and 22 μl RNase Free H2O. OXA-51 and 16S rRNA was used as the internal control. The cycling conditions were as follows: an initial denaturation step at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 90 s. Then the PCR products were electrophoresed in agarose gel to detect the target band.
Molecular typing by pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed using previously described methods [14]. In brief, the purified bacterial genomic DNA was digested by the restriction enzyme ApaI( TaKaRa), and the fragments were separated in a CHEF Mapper system (Bio-Rad Laboratories, Hercules, CA, USA) with pulses ranging from 5 to 20 seconds at a voltage of 5 V/cm and switch angle of 120° for 19 hours at 14°C. And then gels were stained with ethidium bromide and DNA patterns were acquired by Bio-Rad Vilber Lourmat. The PFGE profiles were interpreted according to Tenover et al. [15]. We used BioNumerics software (Applied Maths, Kortrijk, Belgium) to analyze similarities between digitized PFGE outputs. The Between-groups linkage method was used to analyze hierarchic clustering. The classification criteria is as followed:(1) The completely consistent patterns are defined as the same clone, for example, Clone A. (2) If there are 1 to 3 different band compared with Clone A, define it as subtype (Clone A1…Clone An). (3)If there are over 3 different stripes, define it as another clone, for example, Clone B, Clone C, and Clone D etc.
Statistical analysis
Statistical analyses were performed using SPSS18.0. P values <0.05 are considered statistically significant. Statistical significance was assessed via χ2 test of Fisher’s exact test for categorial variables and Student’s test or the Mann–Whitney U test for continuous variables.
Results
The characteristics of drug-resistant A. baumannii infection subjects
During the study period there were 120 strains isolated from adults aged 23 to 98 years old. The average age of the patients was 73.43 ± 13.09 years, and male–female ratio was 1.24:1. The age distribution were shown as followed in Table 2. A. baumannii isolates were mainly distributed in respiratory department, ICU, and emergency department, accounting for 21.7%, 16.7% and 11.7%, respectively. while multidrug-resistant strains, were mainly in ICU and emergency department, accounting for 29.7% and 20.3%, respextively. According to Chi-square test, there were statistical differences in the incidence of multidrug-resistant strains among different wards (χ2 = 195.504, P < 0.001) (Table 3). The strains were mainly isolated from sputum(112, 93.3%), and the others were from blood, urine, endotracheal tube suction, bronchoalveolar and so on.
Antimicrobial susceptibility of A. baumannii isolates
The susceptibility patterns of the 120 A. baumannii strain were shown in Table 4. Among 120 drug resistant strains, 64 were multidrug resistant. Most exhibited resistance across different classes of agents notably imipenem, gentamicin, ampicillin/sulbactam, ceftazidime, and ciprofloxacin.
The detection of drug-resistant genes in these A. baumannii isolates
All 120 isolates were identified as A. baumannii using 16S rRNA gene validation. With regard to the 120 isolates, one hundred and ten strains carried OXA-51 gene characteristic of A. baumannii. The OXA-23 genes were carried by 100 strains. Coexistence of the OXA-51 and the OXA-23 genes was detected in 95 strains. Only one strain harbored OXA-58 gene, and all strains were negative for OXA-24 gene. In multidrug resistant strains, 73.4% of which were positive for SHV gene, as Amber class A of β-lactamases; 95.3% for VIM, one of MBL genes; 82.8% for AmpC β-lactamases; 50.0% for aac(6′)-Ib, the most frequent aminoglycoside-modifying enzymes; 32.8% for CarO, outer membrane protein; 82.8% for class 1 integrase( Intl 1); 56.3% for qac△E1-sul1, which is related to resistance to quaternary ammonium compounds and sulfonamide (Table 5).
PFGE typing of multidrug-resistant A. baumannii isolates
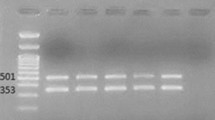
Figure 1 showed that the PFGE results of the multidrug resistant A. baumannii isolates. More than 25 DNA fragments were observed in each isolate. The results of genetic linkage were presented as a dendrogram. PFGE revealed that the isolates had diversity with multivariate clones. Fifty strains were typed into 14 clones. Clone A had 25 isolates (50%), including A1 for 19 , and A2 for 6. Clone B had 5. Clone C had 9,including C1 for 5, C2 for 2, and C3 and C4 for 1,respectively. The others had no common resource. Clonal distribution was shown as followed in Table 6.
Molecular typing of representative multidrug resistant A. baumannii isolates by PFGE. Participating wards, clone type, and source of the samples were also indicated. GE: Geriatrics; R: Respiration; C: Cardiology; E: Emergency; NEU: Neurology; W:wound; GA: Gastroenterol; NEP: Nephrology; HE: Hematology; UL: Urology; S:sputum; CF: cerebrospinal fluid; U:urine.
Discussion
Acinetobacter baumannii, recently as an increasingly common pathogen, is closely associated with hospital acquired infection [16]. A. baumannii has the characteristics of strong viability and rapid development of drug resistance ability and has raised an important challenge to our therapeutic approach [17]. In this paper, we characterized the occurrence of drug resistant of A. baumannii isolates in a hospital in Nanjing. From 2011 to 2013, a total of 120 drug-resistant isolates were collected. The source of 120 strains was mainly from older patients. There were 107 patients aged more than 60 years old (89.2%). The main ward for drug resistant A. baumannii isolate is ICU and respiratory department. The main source of A. baumannii isolates was respiratory specimen. These were consistent with data presented by others [18]. In other words, respiratory tract infection is the common manifestation of drug resistant A. baumannii infection.
Overall, the resistance rates were high for most antimicrobial agents. The resistance rate to imipenem, ampicillin/sulbactam, ceftazidime, cefepime, aztreonam, and ciprofloxacin were more than 90% in multidrug-resistant A. baumannii. Carbapenems have been the choice in treating infections caused by A. baumannii [8]. However, the number of carbapenem-resistant A. baumannii strains has increased recently [19]. The acquisition of carbapenem resisrance in A. baumannii can be mainly due to the production of class D carbapenem hydrolyzing enzymes OXA-β-lactamases(OBLs)and class B metallo β-lactamases (MBLs) [20]. As shown in our study, sixty-four and 58 of the 64 multidrug-resistant strains studied here carried respectively OXA-51 and OXA-23 genes. Moreover, 58 isolates were positive for both OXA-51 and OXA-23 genes. our data support those of other studies demonstrated that OXA-51 may be used as a marker to identify A. baumannii [21]. The OXA-23 genes have been documented in strains associated with outbreaks of carbapenem resistant A. baumannii in Asia, Europe and South America [22]. Only one strain is OXA-58 positive and all strains were negative for OXA-24. OXA-58 belongs to OXA-58 cluster, and OXA-24 belongs to OXA-40 cluster, which has been both reported in Europe and the United States.
And 103 of 120(85.8%) isolated are VIM-1 positive. Our finding indicates that VIM -producing A. baumannii is more prevalent and that MBL-producing A. baumannii is increasing [5]. The percentage of wards with MBL-producing isolates might have been higher if a larger number of carbapenem-non susceptible isolates had been collected for this study.
Screening for genes encoding for AMEs demonstrated that 50 isolates contained the acetyltransferase gene aac(6′)-Ib. Few articles were published describing the presence of the aminoglycoside-encoding genes in strains isolated in china. In Guangzhou, Yang et al. have reported that 89.0% of 73 amikacin-resistant A. baumannii. were aac(6′)-Ib positive. And it is interesting that the aac(6′)-Ib gene is often present in integrons, transposons, and plasmids [23].
In addition to this enzymatic resistance, the loss of membrane permeability, due to alterations in specific porin, is an intrinsic carbapenem resistance mechanism in A. baumannii. A 29 kDa OMP, designated as CarO, if disrupted by insertion sequence, changes in the primary structure or decreased expression, would have a dramatic impact on the entry of antibiotic in the cell, thud contributing to resistance to this antibiotic [24]. But this is not limited to carbenem-resistant strains of A. baumannii. We showed the loss expression of carO in 77 strains. Further research on the outer membrane permeability is necessary.
Quaternary ammonium compounds(QAC) are used as antiseptics for the skin in hospitals. Until now many Gram-negative bacteria resistant to QAC have been reported. Resistance mechanisms are coded by qacE and qacΔE1, whose products are transmembrane proteins [25]. The qacE and qac EΔ1 genes are commonly found in Gram-negative bacteria, which are located in conserved sequences of integron class 1. The qacEΔ1 gene is a mutation of the qacE gene, which acts as a multidrug transfer gene [26]. Sulfonamides act as a structural analogue of ρ-amino-benzoic acid and bind dihydropteroate synthase (DHPS), a catalytic enzyme in the folic acid biosynthesis pathway, resulting in the inhibition of dihydrofolic acid formation [27]. Resistance is conferred from the acquisition of an alternative DHPS gene (sul). One of the three known alternative DHPS genes, sul1, is usually located on the conserved region of integron class 1 [28]. The class 1 integrons are one of the most frequent elements in the acquisition, abundance, maintenance and spread of antimicrobial resistance gene cassettes among gram-negative bacteria isolated from clinical samples [29]. An integron possesses a gene for an integrase (intI), that permits the expression of gene cassettes incorporated in the variable region [30]. And 3′-conserved region at the end of the variable region contains the qacEΔ1 gene, followed by the sul1 gene. The class 1 integrons confer a benefit to the bacteria due to their ability to acquire gene cassettes that could provide advantages for survival in hostile environments [31]. We evaluated genetic elements, sul1 and qacEΔ1 genes and genetic elements associated to lateral genetic transfer, intI1, genes, which comprise the genetic platforms of class 1 integrons. Genomic detection of intI1 and qacΔE1-sul1 showed that 85 (70.8%) and 64 (53.3%) were positive, out of 120 A. baumannii isolates, respectively. Resistance genes related to QAC and sulfonamides antibiotics are both carried by class 1 integrons, so it increases concerns about gene expression that is resistant to QAC and sulfonamides, along with the increasing resistance to antibiotics by class 1 integrons. However, it is difficult to identify the sources of class 1 integrons from “environmental” or “clinical” one.
Pulsed-field gel electrophoresis (PFGE )which is based on the length polymorphism of bacterial chromosome DNA restriction fragments, is still considered the gold standard for typing outbreak-related isolates of A. baumannii [32]. It can be used to separate large digested DNA fragments, and then determine bacterial genotyping through the comparison of DNA band patterns. In many health care institutions, multidrug-resistant A. baumannii infection demonstrates complex epidemiologic profiles and coexistence of multiple strain type. PFGE results showed that among the multidrug-resistant A. baumanii strains in the hospital, DNA fingerprinting by pulsed-field gel electrophoresis showed fourteen clusters. Twenty-five of the 50 multidrug-resistant A. baumannii strains belonged to clone A, and type A1 was the most predominant (19 of 50 strains) in every ward. Based on the data presented, the interhospital transmission of multidrug-resistant A. baumanii isolates was apparent. These data also suggested that cross transmission between patients may contributed to the rise in the rates of multidrug resistance. Type A2, B, C1, C2, C3, C4 and other minor epidemic strains spread in different wards, which indicated that resistance towards antibiotic become more common in the hospital. In addition, type A1, B, C3 and C4 strains was found in ICU, type A1, A2, J and L in respiratory wards, type A1, A2, B, C1, C2, and G in neurology wards, and type A2, C1, D, and F in geriatrics ward, respectively, which suggested that the mix of multiple types of drug resistant strains make clinical treatment more difficult. Therefore, active surveillance is needed to detect and prevent the dissemination of such isolates.
The main limitation of this study is that it was confined to a single centre and it would be valuable to extend the origin of the strains. Another limitation is the small sample size which led to a lack of power to determine the individual effects of each broad spectrum antibiotics.
In conclusion, the emergence of broad-spectrum antibiotic resistance profiles in A. baumannii clinical isolates is worrying in the hospital. The mechanism of multidrug resistance in A. baumannii has not yet been fully understood. Multiple mechanisms are likely to work in synergism to produce this phenotype. our results highlight that enhanced surveillance and health policies for the detection and control of these MDR pathogens are urgently needed to avoid the emergence and spreading of such organism.
Reference
Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–63.
Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–51.
Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82.
Wang F, Zhu D, Hu F, Jiang X, Hu Z, Li Q, et al. CHINET 2013 surveillance of bacterial resistance in China. Chin J Infect Chemother. 2014;14(5):369–78.
Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826–36.
Falagas ME, Koletsi PK, Bliziotis IA. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Microbiol. 2006;55:1619–29.
Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med. 2008;358:1271–81.
Livermore DM. The impact of carbapenemases on antimicrobial development and therapy. Curr Opin Investig Drugs. 2002;3:218–24.
Mussi MA, Relling VM, Limansky AS, Viale AM. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett. 2007;581:5573–8.
Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;16;289(1036):321–31.
Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. 2004;53(Pt 12):1233–40.
Fonseca EL, Scheidegger E, Freitas FS, Cipriano R, Vicente AC. Carbapenem-resistant Acinetobacter baumannii from Brazil: role of carO alleles expression and blaOXA-23 gene. BMC Microbiol. 2013;13:245.
Mahzounieh M, Khoshnood S, Ebrahimi A, Habibian S, Yaghoubian M. Detection of Antiseptic-Resistance Genes in Pseudomonas and Acinetobacter spp. Isolated From Burn Patients. Jundishapur. J Nat Pharm Prod. 2014;9(2):e15402.
Kaufmann ME, Pitt TL. Pulsed-field gel electrophoresis of bacterial DNA. In: Methods in practical laboratory bacteriology. Boca Raton, FL: CRC Press; 1994. p. 83–92.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.
Munoz-Pric LS, Robert AW. Acinetobacter Infection. N Engl Med. 2008;258(26):1271–81.
Perez F, Hujer AM, Metal HK. Global challenge of multidrug-resistant Acinetobacter baurnannii. Antimicrob Agents Chemother. 2007;51:3471–84.
Rello J. Acinetobacter baumannii infections in the ICU: customization is the key. Chest. 1999;115(5):1226–9.
Livermore DM, Woodford N. Carbapenemases: a problem in waiting. Curr Opin Microbiol. 2000;3(5):489–95.
Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. Emergence and widespread dissemination of OXA-23,-24/40 and −58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations:report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63(1):55–9.
Merkier AK, Centrón D. blaOXA-51-type beta-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int J Antimicrob Agents. 2006;28(2):110–3.
Mugnier PD, Poirel L, Naas T. Nordmann P.v Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16:35–40.
Yinmei Y, Zhaohui L, Wei hong Z, Huifen Y, Huiling C. Identification of aac(6′)-Ib and aac(6)-Ib-cr gene in Acinetobacter baumannii. Public Health and. Clin Med. 2010;6(3):191–3.
Limansky AS, Mussi MA, Viale AM. Loss of a 29, Kilodalton outer Membrane Protein in Acinetobacter baumann if Is Associated with imipenem resistance. J Clin Microbiol. 2002;40(12):4776–8.
Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60(4):575–608.
Paulsen IT, Littlejohn TG, Radstrom P, Sundstrom L, Skold O, Swedberg G, et al. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37(4):761–8.
Skold O. Sulfonamide resistance: mechanisms and trends. Drug Resist. 2000;3:155–60.
Skold O. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1976;9:49–54.
Hall RM, Collis CM. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updates. 1998;1:109–19.
Orman BE, Piñeiro SA, Arduino S, Galas M, Melano R, Caffer MI, et al. Evolution of multiresistance in nontyphoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob Agents Chemother. 2002;46:3963–70.
Fluit AC, Schmitz FJ. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur J Clin Microbiol Infect Dis. 1999;18(11):761–70.
Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. The Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol. 1997;1(6):426–39.
Acknowledgments
The work was supported by the Natural Science Foundation of China (No. 81102182) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SYZ and QW participated on designing the study and wrote the manuscript, DYJ, PCX, YKZ performed parts of the experiments and data analysis. HFS and HLC contributed to samples collection. All authors have read and approved the final manuscript.
Authors’ information
Suying Zhao and Dongyang Jiang are co-first authors on this work.
Su-ying Zhao and Dong-yang Jiang contributed equally to this work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhao, Sy., Jiang, Dy., Xu, Pc. et al. An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China. Ann Clin Microbiol Antimicrob 14, 7 (2015). https://doi.org/10.1186/s12941-015-0066-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-015-0066-4