Abstract
Background
In 2017, nearly 80% of malaria morbidity and mortality occurred in sub-Saharan African (SSA) countries and India. Rapid diagnostic tests (RDTs), especially those targeting histidine-rich protein 2 (PfHRP2) of Plasmodium falciparum, have become an important diagnostic tool in these malaria-endemic areas. However, the chances of RDT-oriented successful treatment are increasingly jeopardized by the appearance of mutants with deletions in pfhrp2 and pfhrp3 genes. This systematic review and meta-analysis determines the prevalence of field P. falciparum isolates with deletion in pfhrp2 and/or pfhrp3 genes and their proportion among false-negative results in the PfHRP2-based RDTs in SSA and India.
Methods
Eight electronic databases were used for searching potentially relevant publications for the systematic review analysis, wherein the main methodological aspects of included studies were analysed and some missing links in the included studies were identified.
Results
A total of 19 studies were included, 16 from SSA and 3 from India. The pooled prevalence of pfhrp2 deletions was 8 and 5% while 16 and 4% for pfhrp3 gene deletions in Africa and India, respectively. The pooled proportion of pfhrp2 gene deletions found among false negative PfHRP2-based RDTs results was about 27.0 and 69.0% in Africa and India, respectively.
Conclusions
This review study indicates a relatively high proportion of both pfhrp2/3 genes deletions in P. falciparum isolates and among false-negative malaria cases using PfHRP2-based RDT results in SSA and India. Recently the deletions in pfhrp2/3 genes have also been reported from two African countries (Nigeria and Sudan). This review emphasizes the importance of more extensive studies and standardization of studies addressing the pfhrp2/3 gene deletions in malarious areas.
Similar content being viewed by others
Background
According to the World Health Organization (WHO), Plasmodium falciparum was responsible for over 90% of all malaria cases and deaths that occurred worldwide in 2017 [1]. Plasmodium falciparum is highly prevalent in most malaria-endemic areas, especially in sub-Saharan Africa (SSA) and Southeast Asia, where it accounted for 99.7 and 62.8% of malaria cases, respectively [1].
The P. falciparum genome comprises 14 linear chromosomes with a total of 25–30 megabases of nuclear DNA, with two fragments of 35 kb and 6 kb in the apicoplast and mitochondria, respectively [2]. This genome is extremely adenine/thymine-rich (80%) and consists of over 5000 genes encoding diverse proteins [2, 3]. A large number of these proteins are shared between all malarial species; other proteins such as histidine-rich protein 2 (PfHRP2) are specifically expressed by a given species. PfHRP2 is yielded by P. falciparum during its different developmental stages in humans and has been detected at different localization, including the membrane surface of infected erythrocytes and bloodstream [4]. PfHRP2, with other diverse proteins, have enabled the development of immunochromatographic rapid diagnostic tests (RDTs) of P. falciparum blood infections in humans.
Rapid diagnostic tests are becoming increasingly the preferred method for malaria diagnosis in health facilities throughout the world. They accounted for 75% of malaria tests used in health facilities in 2017 against 40% in 2010 globally [1]. The increase in RDT usage is probably due to their easier implementation, handling and interpretation as compared with light microscopy, which has many limitations jeopardizing its utilization in certain areas [5, 6]. The implementation of light microscopy is particularly challenging as it is labour-intensive and requires skilled microscopists in resource-constrained, remote and difficult-to-reach areas with no electricity [1, 5, 6]. As a consequence, malaria diagnosis relies mainly on signs and symptoms presented by patients in these areas [1, 6]. This kind of clinical diagnosis is less sensitive and specific, especially in highly malaria-endemic areas, given other co-endemic infectious diseases may elicit a similar clinic presentation in patients [6]. In contrast, there are evidence-based reports indicating the improved management of malarious patients using RDTs [1, 5, 6]. Another study in Cameroonian children reported that the utilization of a RDT lowered the rate of anti-malarial drugs misuse in them [7].
There are a few other reports of significant rates of false-negative results using PfHRP2-based RDTs, thereby limiting their diagnostic utility in malaria-endemic settings. The main causes of false-negative results include low parasitaemia, poor state of RDT, prozone effect and poor utilization of RDT by user [8, 9]. The absence of PfHRP2 expression due to gene deletions is also a cause of false-negative PfHRP2-based RDTs results [10]. The presence of malaria parasites not expressing PfHRP2 is primarily reported in the Amazon area of Peru [11]. The utilization of PfHRP2-based RDTs is no longer recommended in this area as the proportion of pfhrp2 gene deletions is very high [12]. Other reports indicated that the expression of PfHRP3 may reduce the level of false-negative results as this protein has the potential to cross-react with monoclonal antibodies (MAbs) used by PfHRP2-based RDTs due to its structural similarity with PfHRP2 [9]. Cross-reactions are more likely to occur in high parasitaemia as this protein is lesser expressed than PfHRP2. Thus, real malarial infection cases may be misdiagnosed using PfHRP2-based RDTs and thereby increase the likelihood of survival and transmission of malaria strains [13].
The present systematic review and meta-analysis is a part of a project aimed at determining and comparing the prevalence of P. falciparum field isolates with deletions in pfhrp2 and pfhrp3 genes, specifically between India and SSA countries. The criteria of this selection was based on three major factors: (i) SSA countries and India contributed nearly 80% of the global malaria burden in 2017; (ii) they are major consumers of malaria RDTs [1]; and, (iii) little is known of the impact of pfhrp2 and pfhrp3 gene deletions on the performances of RDTs, unlike in South American countries where RDTs are no longer advised for diagnosis due to high levels of deletions [11, 14, 15]. Although, there are a few studies that reported the presence of strains with deletions in pfhrp2 and pfhrp3 genes and their impact on results of PfHRP2-based RDTs in India and SSA, there is no systematic review and meta-analysis available on this relevant topic. Additionally, the main methodological aspects of included studies were analysed for any missing links.
Methods
The study was conducted according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (PRISMA) [16]. The PRISMA checklist was used to ensure inclusion of relevant information in the analysis (Additional files 1 and 2).
Search strategy
A comprehensive search of eight electronic databases was done to identify all relevant publications published in the last 10 years on deletions of pfhrp2/3 genes and the impact on results of RDTs, targeting PfHRP2 only due to the absence of RDTs targeting specifically PfHRP3. These databases included Medline, Wiley, EMBASE, Crossref, WHOLIS, ScienceDirect, Popline, and the Cochrane Library. Two search engines (Google and Google Scholar) were also consulted. The search strategy, performed in English and French, is presented in Additional file 3. Titles and abstracts of potentially eligible publications were independently reviewed using the above-mentioned search strategy.
Assessment of the reliability and quality of studies
The methodological quality of studies was assessed using The Joanna Briggs Institute (JBI) Critical Appraisal tools for use in JBI Systematic Reviews Checklist for Prevalence Studies available at http://joannabriggs.org/assets/docs/critical-appraisal-tools/JBI_Critical_Appraisal-Checklist_for_Prevalence_Studies2017.pdf [17]. The tool consists of 9 points ranging from the appreciation of sampling frame to address target population (point 1), to the rate of response (point 9). Each point was scored as 1 for presence and 0 for absence. A score was not given if the point of the appraisal tool was not applicable to the study, or unclear. Given the absence of studies that defined a quality-based clear classification of studies, study with a score of ≥ 5 was considered acceptable for inclusion in the study. A score ponderation was performed for studies if at least 1 point out of 9 was not applicable in these studies. Studies having a quality score ≥ 5 were eligible for the meta-analysis (Additional file 4). Quality of studies was independently evaluated by both authors and any disagreement was resolved through discussion.
Inclusion criteria
Those studies included were considered only if they fulfilled the following prerequisites: (1) addressed the determination of prevalence of P. falciparum field isolates with deletions in pfhrp2/3 genes in SSA and/or India; (2) written in English or French; (3) published and peer-reviewed articles; (4) published between January 2009 and June 2019 in order to provide recent estimates and reduce heterogeneity between studies; (5) had sample size of ≥ 30; and, (6) of acceptable quality (score ≥ 5). Criteria 1–4 were used to include any study in the systematic review while the fifth and sixth criteria were used to include the studies in meta-analysis.
Data management
Data consisted of first author’s name, year of publication, African or Indian state, study population, year of data collection, diagnostic method of malaria, pfhrp gene investigated (pfhrp2 and/or pfhrp3), the genomic sequences genotyped to define deletion in pfhrp2 and/or pfhrp3, the laboratory malarial strains used as control, the number of malarial strains successfully genotyped for pfhrp2/3, the number of strains with deletions in pfhrp2/3 genes among false-negative PfHRP2-based RDT results. Data were keyed into Excel and then exported to the OpenMeta Analyst software for Windows for performing meta-analysis and descriptive analyses [18, 19]. Data were presented as charts or tables where appropriate. The results from meta-analysis were presented graphically using forest plots. I2 statistics was computed to appraise the level of heterogeneity between studies included in the meta-analysis and choose the best statistical model (i.e., binary fixed effect or random effect models) to compute pooled value of prevalence of gene deletion [20]. The variance of individual studies was stabilized using the arcsine transformation prior to pooling estimates of proportion. The I2 statistic appraises the percentage of total variation across studies due to real differences between studies rather than chance, while the heterogeneity was assessed using the Chi-square test based on Cochrane’s Q statistic [21]. Fixed effect model was suitable when I2 was ≤ 25%, while random effect model was suitable when I2 was ≥ 75% [22, 23]. Publication bias was appraised using funnel plot, with meta-analysis being performed separately for India and SSA countries using a sub-group analysis (Additional file 1). The maps were generated using ArcGIS version 10.5 (ESRI, USA) and Adobe Illustrator for Windows (Adobe Inc., USA).
Results
Selection of studies included in the meta-analysis
A total of 871 studies were retrieved in hand searching and electronic databases targeted as presented in the PRISMA flowchart (Fig. 1; Additional files 2 and 3). Six-hundred and seventy-four of 871 were screened after removing duplicates. The studies from regions other than SSA and India were excluded from this systematic review and meta-analysis. Reviews, conference papers, modelling studies, comments, letters to the editor and unrelated studies were also excluded. Twenty-five studies were found eligible for analysis, of which 6 were excluded despite having addressed the prevalence of deletions in pfhrp2/3 genes and/or their proportion among false negative PfHRP2-based RDT results in SSA (Additional file 5). Nineteen studies, 16 in SSA and 3 in India, were finally included in the systematic review. Three studies were excluded from the meta-analysis of the prevalence of deletions in pfhrp2/3 genes because of small sample size (i.e., fewer than 30). Despite this threshold being arbitrary, it is generally accepted as sufficient to do realistic calculation and statistical analysis in practice [24, 25]. Eight studies were excluded from the analysis on the proportion of P. falciparum isolates with deletions in pfhrp2/3 genes among false negative PfHRP2-based RDT results because of small sample size and the topic not being addressed. Sixteen and 10 studies were included in the meta-analysis to determine the prevalence of deletions in pfhrp2/3 genes and their proportion among false negative PfHRP2-based RDT results, respectively [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44] (Fig. 1, Additional file 2).
Characteristics of studies included in the meta-analysis
The 16 studies from 13 SSA countries: Mali, Senegal, Ghana, Kenya, Mozambique, Rwanda, Zambia, Eritrea, Eswatini (Swaziland), Nigeria, Sudan, Madagascar and Democratic Republic of Congo, were conducted locally with one exception which was conducted at national level (Table 1) [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44]. Three studies found deletions in pfhrp2 in Uganda, Gambia and Tanzania [45,46,47], but these were excluded from the systematic review (Additional file 5).
From the three Indian studies only one included samples from several regions, whereas the other two studies did analysis in samples collected locally [42,43,44]. The blood samples in the studies were collected from symptomatic and/or asymptomatic patients, including adults, children or both between 1996 and 2018 in SSA and between 2010 and 2016 in India (Table 2). The minimum sample size of P. falciparum isolates for amplifying pfhrp2 gene was 26 in a study reported from Sudan [40]. The highest sample size was 2329 in a national study conducted in Democratic Republic of Congo [48]. Regarding pfhrp3 gene, sample size ranged from 48 to 1529 samples in India [42, 43]. In the present study, the meta-analysis included 4286 and 1953 samples for pfhrp2 deletions, and 1115 and 1953 samples for pfhrp3 deletions evaluated in SSA and India, respectively.
Prevalence of pfhrp2 gene deletions
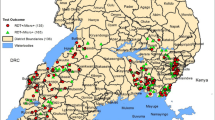
The geographic distribution of pfhrp2 genetic deletions in Africa and India is presented in Fig. 2. Deletions in pfhrp2 gene is reported in 16 African countries (Fig. 2a). Out of the right regions studied in India only six regions reported deletions from India (Fig. 2b).
Geographical distribution of areas in a sub-Saharan Africa and b India. The deletions in pfHRP2 gene observed in Plasmodium falciparum field isolates. The maps were generated using ArcGIS version 10.5 (ESRI, USA) and Adobe Illustrator for Windows (Adobe Inc., USA). A few studies were excluded from this review (Additional file 5 for reasons), however their results have been taken into account to generate this map. *Deletions have been newly reported
In India, the lowest and highest values for prevalence of pfhrp2 gene deletions reported were 2.4 and 9.9% (see Fig. 3) [43, 44]. In Africa, these values were 0 and 62% in Nigeria and Eritrea, respectively [35, 37]. The pooled prevalence of pfhrp2 deletions in field isolates and the values of prevalence were 7.0% (95% CI 4.0–9.0%) and 5% (95% CI 0–11%) in Africa and India, respectively (Figs. 2 and 3, Additional file 6).
Prevalence of pfhrp3 gene deletions
The studies conducted in India also analysed the prevalence of deletions in pfhrp3 gene while in Africa only half of the total reviewed studies focused on this aspect of pfhrp3 gene. Malaria parasites with deletion in pfhrp3 gene were reported from 13 SSA countries while these deletions were found in only five regions of India (Fig. 4). Regarding pfhrp3 deletion prevalence in Africa, highest and lowest values were 82 and 1%, respectively [30, 35]. Pooled prevalence of parasites with deletions in pfhrp3 gene was 16% (95% CI 10–23%) and 4% (95% CI 1–7%) in Africa and India, respectively (Fig. 3, Additional files 6 and 7).
Geographical distribution of areas in sub-Saharan Africa (a) and India (b) The deletions in PfHRP3 gene observed in Plasmodium falciparum field isolates. The maps were generated using ArcGIS version 10.5 (ESRI, USA) and Adobe Illustrator for Windows (Adobe Inc., USA). A few studies were excluded from this review (Additional file 3 for reasons), however their results have been taken into account to generate this map. *Deletions have been newly reported
Prevalence of pfhrp2 and pfhrp3 gene deletions
Seven and three studies from SSA and India were included to calculate the pooled estimates of the prevalence of deletions in both pfhrp2 and pfhrp3 genes. The pooled estimate was 3% (95% CI 1–4%, I2 = 87.22%; P = 0.00) and 3% (95% CI 0–6%, I2 = 98.48%; P = 0.00) in SSA and India, respectively (Additional files 6 and 7).
Potential role of pfhrp2 gene deletions in the diagnostic performances of PfHRP2-based RDTs
The proportion of isolates with pfhrp2 deletions among the false negative cases using PfHRP2-based RDT ranged from 1.4 to 100% in SSA while in India the proportion was 65.5 to 100% (Table 3). The pooled estimates were 27.0% (95% CI 0–55.0%) and 69.0% (95% CI 61.0–78.0%) in SSA and India, respectively (Additional files 6 and 7).
Discussion
The overall prevalence of malaria parasites with deletions of pfhrp2 gene is relatively high (i.e., ≥ 5%) both in SSA and India. The prevalence of pfhrp2 gene deletions is lower in South American countries with the exception of Peru (prevalence > 40%) where the utilization of PfHRP2-based RDT is no longer recommended given the high risk for false negative results [9,10,11]. The pfhrp2 gene is located on sub-telomeric region of chromosome 8 of P. falciparum. The high prevalence of pfhrp2 gene deletions may be due to fact that sub-telomeric genes are known to be highly polymorphic and susceptible to genetic changes such as deletions during recombination events [47, 49].
In areas where P. falciparum is highly prevalent, as in SSA and India, the circulation of mutants with deletions in pfhrp2 gene might compromise the PfHRP2-based RDT management of patients attending health facilities. High rates of deletions present in pfhrp2 gene in Africa and India among the cases of false negative PfHRP2-based RDT results were found. Although different studies included in this meta-analysis have not clearly proved the causal role of these deletions in false negative cases, the consequences of the circulation of isolates with deletions in pfhrp2 gene are important in terms of public health. A number of P. falciparum mono-infections would be missed and thereby increase the risk of severe malaria due to delay in treatment [9, 50, 51]. This misdiagnosis uselessly exposes malarious individuals to drugs used for treating co-endemic viral and bacterial infectious diseases which have symptomatology similar to that of malaria [52]. Another risk is of misdiagnosis in areas where P. falciparum is not the only circulating main malarial species. In India, P. falciparum and Plasmodium vivax are the main species involved in its malaria burden [1, 51, 53]. Co-infections are frequently reported among malarious patients. Some authors reported a high proportion of mixed infections with both these malarial species in the country [54, 55]. As a consequence, a fraction of mixed infection cases would be diagnosed as P. vivax mono-infection by RDTs able to detect both P. falciparum and P. vivax. Thus, the risk of severe falciparum malaria would increase as the RDTs will have failed to detect P. falciparum. This fact is particularly important in malaria-endemic areas where both the circulation of chloroquine-resistant P. falciparum isolates and chloroquine is preferentially used for treating vivax malaria, as this is the case in India [56,57,58,59,60]. In most endemic countries, chloroquine is first-line treatment of vivax malaria [1]. Keeping in mind providing quality management in malarious patients, it would be helpful to use RDTs that target other P. falciparum antigens as PfHRP2 in areas whose diagnosis strategy relies mainly on RDTs and where suspicions of circulation of strains with deletions in pfhrp2 gene are reported. Some studies reported better specificity of malarial antigen-combining RDTs compared with their counterparts targeting PfHRP2 only [61, 62]. WHO recommended a switch to RDTs that do not rely exclusively on PfHRP2 for detecting P. falciparum if the 95% CI lower value of the reported prevalence is above 5% upon a nationwide study [63].
It should be noted that the prevalence of deletions in pfhrp2 gene found in each individual study could be higher and generally the malaria infections are polyclonal in SSA and Indian endemic regions [64,65,66]. It is likely that individuals are infected with both malaria parasites with deletions in pfhrp2 gene and without deletions wherein, the presence of malaria parasites with no pfhrp2 deletions can overshadow that of their counterparts with deletions in pfhrp2 gene. As a consequence, blood samples of this type would be positive with PfHRP2-based RDTs.
It was also reported a high level of loss of pfhrp3 gene in field isolates in SSA and India. The protein encoded by this gene has capacity to cross-react with monoclonal antibodies of RDTs targeting PfHRP2 and thus reduce the level of false-negative results [9, 10]. However, this likely occurs at high parasitaemia as its expression is much lower than that of PfHRP2 [9].
This review outlines the report of deletions in pfhrp2/3 genes in two supplementary African countries, namely Nigeria and Sudan which are highly malaria endemic, especially Nigeria which accounts for 19% of total malaria-related deaths occurring worldwide [1]. Conversely, this study pinpoints the absence of reports on pfhrp2 and pfhrp3 gene deletions in other countries of SSA, such as Burkina Faso, Sierra Leone and Niger, which account for 6, 5 and 4% of total malaria-related deaths occurring worldwide [1]. Although the cause of death due to PfHRP2-based RDTs related misdiagnosis remains unknown, data are missing from other highly malaria-endemic African countries: Cameroon, Ethiopia and Chad for instance. These countries share borders with countries where pfhrp2/3 deletions were reported (Ethiopia borders Eritrea, Cameroon borders Nigeria). The same logic may be applied to Indian areas bordering those where gene deletions cases have been reported.
The unrestricted use of RDTs targeting PfHRP2 might create pressure for a selective sweep of PfHRP2-negative strains [67, 68]. A recent study outlined a slight to high risk of selection of mutants with deletions in pfhrp2 gene where the treatment of malaria would be based on PfHRP2-based RDTs alone [69]. This finding reinforces the utilization of RDTs targeting several plasmodial antigens simultaneously instead of RDTs targeting PfHRP2 alone as discussed above. The genetics of pfhrp2/3 gene deletions in malaria parasites, their adaptive cost in mutants, along with the influence of epidemiological parameters are elusive and need to be studied in detail [10]. The elucidation of these main issues will allow better understanding of the biological significance of pfhrp2/3 gene deletions in endemic areas.
It would be interesting to define standard methodology in order to better follow up the patterns of mutants with deletions in pfhrp2 gene in a given area and make comparisons between studies. In this regard, a methodology has been proposed by some authors and the WHO in order to standardize the results [61, 70].
The results of this study should be interpreted with caution in the context of its limitations. First, a high heterogeneity was recorded in the meta-analysis and this is inescapable in meta-analyses of prevalence and observational studies [71]. The high heterogeneity reported in the present study might be explained by the discrepancies in epidemiological patterns in each region, characteristics of study population, and methodology for detecting gene deletions. Indeed, it was noted variability between studies in this methodology especially on the amplified genomic regions, the utilization of laboratory-maintained strains (HB3, 3D7, Dd2) and the strategy used for distinction between PCR-negative samples for pfhrp due to either low parasitaemia or real absence of the gene. Hence, the absence of such a distinction might lead to overestimation of P. falciparum isolates with deletions in pfhrp2 and pfhrp3 genes. Second, the small sample size in some studies did not allow the evaluation of possible sources of a high variation between studies.
Conclusion
This study outlined a relatively high proportion of pfhrp2 and pfhrp3 gene deletions as well as their important role in diagnostic performance of PfHRP2-based RDTs in SSA and India. It also pointed out the need for further studies with standardized framework in order to have a clearer picture of the extent of mutants with deletions in pfhrp2/3 genes and follow their patterns over time and space. The rates of deletions in pfhrp2/3 genes are not high enough, compared to those reported in Latin America, to reconsider the usefulness of RDTs. RDTs remain the reliable tool for diagnosis of malaria in SSA and India.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files.
Abbreviations
- A:
-
adenine
- Ag:
-
antigen
- CI:
-
confidence interval
- DNA:
-
deoxyribonucleic acid
- ICMR:
-
Indian Council of Medical Research
- EMBASE:
-
Excerpta Medica Database
- GLURP:
-
glutamate-rich protein
- JBI:
-
The Joanna Briggs Institute
- LDH:
-
lactate dehydrogenase
- MAbs:
-
monoclonal antibodies
- MSP:
-
merozoite surface protein
- NA:
-
not applicable
- NIMR:
-
National Institute of Malaria Research
- pfcrt :
-
P. falciparum chloroquine resistance transporter
- pfmdr 1:
-
P. falciparum multidrug resistance 1
- PfHRP:
-
histidine-rich protein of P. falciparum
- pfk13 :
-
P. falciparum Kelch 13 gene
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- qPCR:
-
quantitative polymerase chain reaction
- RDTs:
-
rapid diagnostic tests
- RNA:
-
ribonucleic acid
- SD:
-
standard diagnostics
- SNP:
-
single nucleotide polymorphism
- T:
-
thymine
- WHO:
-
World Health Organization
References
WHO. World malaria report 2018. Geneva: World Health Organization; 2018. http://www.who.int. Accessed 28 Dec 2018.
Meyer CG, May J, Arez AP, Gil JP, Do Rosário VE. Genetic diversity of Plasmodium falciparum: asexual stages. Trop Med Int Health. 2002;7:395–408.
Gardner MJ, Hall N, Fung E, White O, Berriman M, Richard W, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2013;419:498–511.
Pal P, Daniels BP, Oskman A, Diamond MS, Klein RS, Goldberg DE. Plasmodium falciparum histidine-rich protein II compromises brain endothelial barriers and may promote cerebral malaria pathogenesis. mBio. 2016;7:e00617-16.
Ali IM, Bigoga JD, Forsah DA, Cho-Ngwa F, Tchinda V, Moor VA, et al. Field evaluation of the 22 rapid diagnostic tests for community management of malaria with artemisinin combination therapy in Cameroon. Malar J. 2016;15:31.
Kamgain L, Assam-Assam J-P, Kojom Foko LP, Fouamno H. Prevalence of malaria infection and reliability of ACCUCARE one step malaria test® for diagnosing malaria in people living with human immunodeficiency virus infection in Cameroon. Int J Trop Dis Health. 2017;21:1–10.
Sayang C, Soula G, Tahar R, Basco LK, Gazin P, Moyou-Somo R, et al. Use of a histidine-rich protein 2-based rapid diagnostic test for malaria by health personnel during routine consultation of febrile outpatients in a peripheral health facility in Yaoundé, Cameroon. Am J Trop Med Hyg. 2008;81:343–7.
Luchavez J, Baker J, Alcantara S, Belizario V, Cheng Q, McCarthy JS, et al. Laboratory demonstration of a prozone-like effect in HRP2-detecting malaria rapid diagnostic tests: implications for clinical management. Malar J. 2011;10:286.
Gendrot M, Fawaz R, Dormoi J, Madamet M, Pradines B. Genetic diversity and deletion of Plasmodium falciparum histidine-rich protein 2 and 3: a threat to diagnosis of P. falciparum malaria. Clin Microbiol Infect. 2019;25:580–5.
WHO. P. falciparum hrp2/3 gene deletions: conclusions and recommendations of a Technical Consultation. In: Malaria policy advisory committee meeting 14–16 September 2016, Geneva, Switzerland Background document for Session 7. 2016. https://www.who.int/malaria/mpac/mpac-sept2016-hrp2-consultation-short-report-session7.pdf?ua=1. Accessed 28 Mar 2019.
Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: Implications for malaria rapid diagnostic tests. PLoS ONE. 2010;5:e8091.
Maltha J, Gamboa D, Bendezu J, Sanchez L, Cnops L, Gillet P, et al. Rapid diagnostic tests for malaria diagnosis in the Peruvian Amazon: impact of pfhrp2 gene deletions and cross-reactions. PLoS ONE. 2012;7:e43094.
Nima MK, Hougard T, Hossain ME, Kibria MG, Mohon AN, Johora FT, et al. Case report: a case of Plasmodium falciparum hrp2 and hrp3 gene mutation in Bangladesh. Am J Trop Med Hyg. 2017;97:1155–8.
Murillo Solano C, Akinyi Okoth S, Abdallah JF, Pava Z, Dorado E, Incardona S, et al. Deletion of Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and histidine-rich protein 3 (pfhrp3) genes in Colombian parasites. PLoS ONE. 2015;10:e0131576.
Fontecha G, Mejía RE, Banegas E, Ade MP, Mendoza L, Ortiz B, et al. Deletions of pfhrp2 and pfhrp3 genes of Plasmodium falciparum from Honduras, Guatemala and Nicaragua. Malar J. 2018;17:320.
Moher D, Liberati A, Tetzlaff J, Altman DG, Altman D, Antes G, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Chin Integr Med. 2009;6:e1000097 (Chinese edition).
Munn Z, MClinSc SM, Lisy K, Riitano D, Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int J Evid Based Healthc. 2015;13:147–53.
Meta-Analyst O. Open meta-analyst-the tool | evidence synthesis in health. 2018. https://www.brown.edu/academics/public-health/research/evidence-synthesis-in-health/. Accessed 21 June 2019.
Wallace BC, Schmid CH, Lau J, Trikalinos TA. Meta-analyst: software for meta-analysis of binary, continuous and diagnostic data. BMC Med Res Methodol. 2009;9:80.
Zlowodzki M, Poolman RW, Kerkhoffs GM, Tornetta P III, Bhandari M. How to interpret a meta-analysis and judge its value as a guide for clinical practice. Acta Orthop. 2007;78:598–609.
Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29.
Reid K. Interpreting and understanding meta-analysis graphs: a practical guide. Austr Fam Phys. 2006;35:635–8.
Sedgwick P. Meta-analyses: Heterogeneity and subgroup analysis. BMJ. 2013;346:9–11.
Greenwood JA, Sandomire MM. Sample size required for estimating the standard deviation as a per cent of its true value. J Am Stat Assoc. 1950;45:257–60.
Kar SS, Ramalingam A. Is 30 the magic number? Issues in sample size. Natl J Commun Med. 2013;4:175–9.
Koita OA, Doumbo OK, Ouattara A, Tall LK, Konaré A, Diakité M, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–8.
Wurtz N, Fall B, Bui K, Pascual A, Fall M, Camara C, et al. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar J. 2013;12:34.
Amoah LE, Abankwa J, Oppong A. Plasmodium falciparum histidine rich protein-2 diversity and the implications for PfHRP2: based malaria rapid diagnostic tests in Ghana. Malar J. 2016;15:101.
Beshir KB, Sepúlveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep. 2017;7:14718.
Gupta H, Matambisso G, Galatas B, Cisteró P, Nhamussua L, Simone W, et al. Molecular surveillance of pfhrp2 and pfhrp3 deletions in Plasmodium falciparum isolates from Mozambique. Malar J. 2017;16:416.
Kozycki CT, Umulisa N, Rulisa S, Mwikarago EI, Musabyimana JP, Habimana JP, et al. False-negative malaria rapid diagnostic tests in Rwanda: impact of Plasmodium falciparum isolates lacking hrp2 and declining malaria transmission. Malar J. 2017;16:123.
Menegon M, L’Episcopia M, Nurahmed AM, Talha AA, Nour BYM, Severini C. Identification of Plasmodium falciparum isolates lacking histidine-rich protein 2 and 3 in Eritrea. Infect Gen Evol. 2017;55:131–4.
Parr JB, Verity R, Doctor SM, Janko M, Carey-Ewend K, Turman BJ, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis. 2017;216:36–44.
Ranadive N, Kunene S, Darteh S, Ntshalintshali N, Nhlabathi N, Dlamini N, et al. Limitations of rapid diagnostic testing in patients with suspected malaria: a diagnostic accuracy evaluation from Swaziland, a low-endemicity country aiming for malaria elimination. Clin Infect Dis. 2017;64:1221–7.
Berhane A, Anderson K, Mihreteab S, Gresty K, Rogier E, Mohamed S, et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis. 2018;24:462–70.
Nderu D, Kimani F, Thiong’o K, Akinyi M, Karanja E, Meyer CG, et al. PfHRP2–PfHRP3 diversity among Kenyan isolates and comparative evaluation of PfHRP2/pLDH malaria RDT with microscopy and nested PCR methodologies. Parasitol Int. 2018;67:793–9.
Willie N, Mehlotra RK, Howes RE, Rakotomanga TA, Ramboarina S, Ratsimbasoa AC, et al. Insights into the performance of SD Bioline malaria Ag Pf/Pan rapid diagnostic test and Plasmodium falciparum histidine-rich protein 2 gene variation in Madagascar. Am J Trop Med Hyg. 2018;98:1683–91.
Funwei R, Nderu D, Nguetse CN, Thomas BN, Falade CO, Velavan TP, et al. Molecular surveillance of pfhrp2 and pfhrp3 genes deletion in Plasmodium falciparum isolates and the implications for rapid diagnostic tests in Nigeria. Acta Trop. 2019;196:121–5.
Kobayashi T, Sikalima J, Parr J, Chaponda M, Stevenson J, Thuma P, et al. The search for Plasmodium falciparum histidine-rich protein 2/3 deletions in Zambia and implications for Plasmodium falciparum histidine-rich protein 2-based rapid diagnostic tests. Am J Trop Med Hyg. 2019;100:842–5.
Mussa A, Talib M, Mohamed Z, Hajissa K. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. BMC Res Notes. 2019;12:334.
Thomson R, Beshir KB, Cunningham J, Baiden F, Bharmal J, Bruxvoort KJ, et al. pfhrp2 and pfhrp3 gene deletions that affect malaria rapid diagnostic tests for Plasmodium falciparum: analysis of archived blood samples from three African countries. J Infect Dis. 2019;220:1444–52.
Kumar N, Pande V, Bhatt RM, Shah NK, Mishra N, Srivastava B, et al. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 2013;125:119–21.
Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS ONE. 2016;11:e0157949.
Pati P, Dhangadamajhi G, Bal M, Ranjit M. High proportions of pfhrp2 gene deletion and performance of HRP2-based rapid diagnostic test in Plasmodium falciparum field isolates of Odisha. Malar J. 2018;17:394.
Baker J, Mccarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis. 2005;192:870–7.
Deme AB, Park DJ, Bei AK, Sarr O, Badiane AS, Omar Gueye PEH, et al. Analysis of pfhrp2 genetic diversity in Senegal and implications for use of rapid diagnostic tests. Malar J. 2014;13:34.
Baker J, Ho M-F, Pelecanos A, Gatton M, Chen N, Abdullah S. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J. 2010;9:129.
Parr JB, Anderson O, Juliano JJ, Meshnick SR. Streamlined, PCR-based testing for pfhrp2- and pfhrp3-negative Plasmodium falciparum. Malar J. 2018;17:137.
Volkman SK, Hartl DL, Wirth DF, Nielsen KM, Choi M, Batalov S, et al. Excess polymorphisms in genes for membrane proteins in Plasmodium falciparum. Science. 2002;298:216–8.
Trouvay M, Palazon G, Berger F, Volney B, Blanchet D, Faway E, et al. High performance of histidine-rich protein 2 based rapid diagnostic tests in French Guiana are explained by the absence of pfhrp2 gene deletion in P. falciparum. PLoS ONE. 2013;8:e74269.
Singh V, Kojom LP. Deletions in the Plasmodium falciparum histidine-rich protein 2 gene: an emerging threat to the elimination of malaria in India. J Vector Borne Dis. 2019;56:85–6.
Wogu MN, Nduka FO. Evaluating malaria prevalence using clinical diagnosis compared with microscopy and rapid diagnostic tests in a tertiary healthcare facility in Rivers State, Nigeria. J Trop Med. 2018;2018:3954717.
Singh V, Mishra N, Awasthi G, Dash AP, Das A. Why is it important to study malaria epidemiology in India? Trends Parasitol. 2009;25:452–7.
Gupta B, Gupta P, Sharma A, Singh V, Dash AP, Das A. High proportion of mixed-species Plasmodium infections in India revealed by PCR diagnostic assay. Trop Med Int Health. 2010;15:819–24.
Siwal N, Singh US, Dash M, Kar S, Rani S, Rawal C, et al. Malaria diagnosis by PCR revealed differential distribution of mono and mixed species infections by Plasmodium falciparum and P. vivax in India. PLoS ONE. 2018;13:e0193046.
Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:982–91.
Das S, Tripathy S, Chattopadhayay S, Das B, Kar Mahapatra S, Hati AK, et al. Progressive increase in point mutations associates chloroquine resistance: even after withdrawal of chloroquine use in India. Int J Parasitol Drugs Drug Resist. 2017;7:251–61.
Lo E, Hemming-Schroeder E, Yewhalaw D, Nguyen J, Kebede E, Zemene E, et al. Transmission dynamics of co-endemic Plasmodium vivax and P. falciparum in Ethiopia and prevalence of antimalarial resistant genotypes. PLoS Negl Trop Dis. 2017;11:e0005806.
Patel P, Bharti PK, Bansal D, Ali NA, Raman RK, Mohapatra PK, et al. Prevalence of mutations linked to antimalarial resistance in Plasmodium falciparum from Chhattisgarh, Central India: a malaria elimination point of view. Sci Rep. 2017;7:16690.
Mohapatra BN, Mohanty CBK. Indian guidelines and protocols: Malaria. Chapt 2-Section 1. 2010.
Adu-Gyasi D, Asante KP, Amoako S, Amoako N, Ankrah L, Dosoo D, et al. Assessing the performance of only HRP2 and HRP2 with pLDH based rapid diagnostic tests for the diagnosis of malaria in middle Ghana, Africa. PLoS Negl Trop Dis. 2018;13:e0203524.
Quakyi IA, Adjei GO, Sullivan DJ, Laar A, Stephens JK, Owusu R, et al. Diagnostic capacity, and predictive values of rapid diagnostic tests for accurate diagnosis of Plasmodium falciparum in febrile children in Asante-Akim, Ghana. Malar J. 2018;17:468.
WHO. P. falciparum hrp2/3 gene deletions. Conclusions and recommendations of a Technical Consultation. Geneva: World Health Organization; 2016. http://www.who.int. Accessed 15 Feb 2019.
Decuypere S, Elinck E, Van Overmeir C, Talisuna AO, Alessandro UD, Dujardin J. Pathogen genotyping in polyclonal infections: application of a fluorogenic polymerase–chain-reaction assay in malaria. J Infect Dis. 2003;188:1245–9.
Das A, Anvikar AR, Cator LJ, Dhiman RC, Eapen A, Mishra N, et al. Malaria in India: the center for the study of complex malaria in India. Acta Trop. 2013;121:267–73.
Zhong D, Lo E, Wang X, Yewhalaw D, Zhou G, Atieli HE, et al. Multiplicity and molecular epidemiology of Plasmodium vivax and Plasmodium falciparum infections in East Africa. Malar J. 2018;17:185.
Gatton ML, Dunn J, Chaudhry A, Ciketic S, Cunningham J, Cheng Q. Implications of parasites lacking Plasmodium falciparum histidine-rich protein 2 on Malaria morbidity and control when rapid diagnostic tests are used for diagnosis. J Infect Dis. 2017;215:1156–66.
Verma AK, Bharti PK, Das A. HRP-2 deletion: a hole in the ship of malaria elimination. Lancet Infect Dis. 2018;18:826–7.
Watson OJ, Slater HC, Verity R, Parr JB, Mwandagalirwa MK, Tshefu A, et al. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. eLife. 2017;6:e25008.
Cheng Q, Gatton ML, Barnwell J, Chiodini P, Mccarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;13:283.
Noubiap JJ, Balti EV, Bigna JJ, Echouffo-Tcheugui JB, Kengne AP. Dyslipidaemia in Africa—comment on a recent systematic review. Lancet Glob Health. 2018;7:e308–9.
Acknowledgements
The authors are grateful to the Department of Biotechnology (DBT), India and The World Academy of Science (TWAS), Italy which provided a fellowship (2017 DBT-TWAS Postgraduate Fellowship Programme: Grant N° 3240300010) to the first author for his PhD studies at the ICMR-National Institute of Malaria Research, India. The authors are grateful to Dr Stephane Koum, Ph.D (Department of Earth Sciences, University of Douala, Cameroon) for generating maps using ArcGIS version 10.5.
This paper bears the NIMR publication committee Approval no. RIC-16/2019.
Funding
None.
Author information
Authors and Affiliations
Contributions
LPK, VS conceived and designed the study. LPK and VS carried out the screen of the literature and data extraction. LPK analysed the results with help of VS. LPK drafted the manuscript and VS revised the manuscript. VS supervised the work at all stages. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This research was based on information/data extracted from published studies and no ethical approval was acquired.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
PRISMA checklist.
Additional file 2.
PRISMA flow diagram.
Additional file 3.
Search terms and strategy used for articles in sub-Saharan African countries and India.
Additional file 4.
Result on quality assessment of studies included in the meta-analysis.
Additional file 5.
List of publications having addressed the deletions in pfhrp2/3 genes in Africa which have been excluded from the meta-analysis.
Additional file 6.
Findings on meta-analysis of the prevalence of deletions in pfhrp2 and/or pfhrp3 gene in sub-Saharan African countries and India.
Additional file 7.
Funnel plots of the prevalence of deletions in pfhrp2 and/or pfhrp3 gene in sub-Saharan African countries and India.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kojom, L.P., Singh, V. Prevalence of Plasmodium falciparum field isolates with deletions in histidine-rich protein 2 and 3 genes in context with sub-Saharan Africa and India: a systematic review and meta-analysis. Malar J 19, 46 (2020). https://doi.org/10.1186/s12936-019-3090-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-019-3090-6