Abstract
Colorectal cancer (CRC) is one of the most lethal and prevalent solid malignancies worldwide. There is a great need of accelerating the development and diagnosis of CRC. Long noncoding RNAs (lncRNA) as transcribed RNA molecules play an important role in every level of gene expression. Metastasis‐associated lung adenocarcinoma transcript‐1 (MALAT1) is a highly conserved nucleus-restricted lncRNA that regulates genes at the transcriptional and post-transcriptional levels. High expression of MALAT1 is closely related to numerous human cancers. It is generally believed that MALAT1 expression is associated with CRC cell proliferation, tumorigenicity, and metastasis. MALAT1 by targeting multiple signaling pathways and microRNAs (miRNAs) plays a pivotal role in CRC pathogenesis. Therefore, MALAT1 can be a potent gene for cancer prediction and diagnosis. In this review, we will demonstrate signaling pathways associated with MALAT1 in CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) or colorectal adenocarcinoma is a complex and the third cause of malignancies in the world [1, 2]. CRC usually arises from the hyper-proliferative glandular and epithelial cells in the large intestine [3]. Several environmental and genetic factors can stimulate the accumulation of mutations and oncogenes, and inhibit tumor suppressor genes in colon epithelial cells [4]. Currently, surgical resection [5], chemotherapy [6], and radiotherapy [7] are the common types of treatments for CRC [8]. Recently, molecular targeted therapy has emerged as a novel treatment option for targeting CRC cells [9, 10]. Some studies provided evidence that cancer-specific long non-coding RNAs (lncRNAs) can be utilized for anti-CRC drugs [11, 12]. LncRNA (> 200 nucleotides in length) are transcribed RNA molecules that directly or indirectly regulate a variety of biological functions [13]. It has been shown that many lncRNAs are involved in human diseases and cancers through the induction of cell cycle progression, invasion, and metastasis [14]. Metastasis‐associated lung adenocarcinoma transcript‐1 (MALAT1) is a conserved and well-characterized lncRNA that plays an important role in various biological processes through diverse mechanisms [15]. Under hypoxia conditions, MALAT1 plays an important role in inflammation, angiogenesis, and metastasis [16].
The expression of MALAT1 was first detected in patients with non-small cell lung cancer (NSCLC) [17, 18]. The expression of MALAT1 has been upregulated in multiple cancer types include liver [19], cervix [20], breast [21], colorectal [22], renal [23], prostate [24], gastric [25] and other cancers [26, 27]. In tumor cells, MALAT1 by targeting several transcription factors, growth factors, hormones, and epigenetic histone modifications can mediate cancer cell proliferation, tumorigenicity, autophagy, epithelial-mesenchymal transition (EMT), metastasis, and drug resistance phenotypes [28,29,30,31]. Recent studies elucidated the role of MALAT1 in CRC cell growth, migration, invasion, and metastasis [32, 33]. MALAT1 was reported to target several CRC-related pathways such as Wnt/β-catenin, YAP, SOX9, RUNX2, Snail, EGF, PI3K/AKT/mTOR, P53, and VEGF [34, 35]. Besides, MALAT1 by suppressing multiple microRNAs (miRNAs) plays a pivotal role in CRC pathogenesis [36, 37]. miRNAs are epigenetic modulators that target mRNAs and function in various biological and pathological processes [38].
Therefore, MALAT1 can be a potent biomarker for CRC prediction and diagnosis [39, 40]. In this review, we summarized MALAT1-related signaling pathways in CRC.
Biogenesis of MALAT1
MALAT1 (known as nuclear-enriched abundant transcript 2 (NEAT2) or hepcarcin (HCN)) is the most widely investigated lncRNA and RNA polymerase II transcripts that localizes to nuclear speckles [41, 42]. MALAT1 coding gene is located on human chromosome 11q13.1 (> 8000 nucleotides) [28] and functions in alternative splicing [26]. The MALAT1 precursor contains a highly conserved triple-helix element at the 3′ end named MALAT1-associated small cytoplasmic RNA (mascRNA) that protects the 3′ end from degradation and facilitates the localization of MALAT1 [43, 44]. mascRNA with a tRNA-like structure is separated from MALAT precursor by tRNA endonucleases RNase P to generate pre-mature MALAT1 [45, 46]. RNase P can also generate a 61-nt tRNA-like small RNA at the 5’ end of the abundant MALAT1 transcript which is exported to the cytoplasm [47]. Pre-mature MALAT1 with a short poly(A) tail-like moiety is cleaved by tRNA endonucleases RNase Z in the nucleus, prior to addition of the CCA motif. After processing, mascRNA with CCA trinucleotide tail exported to the cytoplasm, while the stable MALAT1 transcript accumulates in the nucleus [48] (Fig. 1).
Characterization of MALAT1. MALAT1 contains a highly conserved triple-helix element at the 3′ end named MALAT1-associated small cytoplasmic RNA (mascRNA) that protects the 3′ end from degradation and facilitates the localization of MALAT1. mascRNA is a tRNA-like structure that is separated from MALAT precursor by tRNA endonucleases RNase P. Then, pre-mature MALAT1 with a short poly(A) tail-like moiety is cleaved by tRNA endonucleases RNase Z. MALAT1 plays functional roles in transcriptional regulation, translation activation, epigenetic regulation, RNA processing, physiological processes, and cancer
MALAT1 interacts with multiple miRNAs and small nuclear RNAs (snRNAs) to regulate various biological processes in human tissues [46]. It has been reported that several nuclear speckles such as RNPS1 (RNA-binding protein with serine-rich domain 1), SRm160 (serine/arginine-rich (SR)-related protein), and IBP160 (intron binding protein) regulate the proper localization of MALAT1 to nuclear speckles [17]. MALAT1 also changes the distribution of SR splicing factor (SRSF), SON1, hnRNPC, and hnRNPH1 as pre-mRNA splicing factors [17, 49]. MALAT1 can target polycomb repressive complex 2 (PRC2) components (enhancer of Zeste 2 (EZH2), SUZ12, and EED), increase trimethylation of histone H3 at lysine 27 (H3K27me3), and decrease target gene or miRNA expression [50]. Down-regulation of MALAT1 influenced the distribution of SR proteins and changed splicing of pre-mRNA [51]. Nowadays, various specific gene manipulation methods using siRNAs, miRNAs, and shRNA mediated the knockdown of MALAT1 have been introduced for diagnostic, prognostic, and therapeutic values of MALAT1 and its downstream targets [17, 52]. Although the exact mechanism of MALAT1 is unclear, its expression is misregulated in numerous human malignancies. MALAT1 as a competing endogenous RNA (ceRNA) can sponge miRNAs to inhibit their expression and stimulate their downstream targets.
MALAT1 was suggested to be a prognostic marker in multiple cancerous tissues. Below, we summarized the overview function of MALAT1 in colorectal cancer.
The role of MALAT1 in colorectal cancer
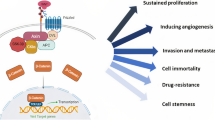
The MALAT1 fragment at the 3' end is known to be associated with CRC cell metastasis [47, 53]. However, the exact mechanisms of MALAT1 in CRC are not fully understood. Previous studies have established that MALAT1 promotes CRC cell proliferation, apoptosis, migration, metastasis, and angiogenesis (Table 1). MALAT1 by targeting several signaling pathways and miRNAs plays a pivotal role in CRC pathogenesis (Fig. 2).
Based on a previous study, MALAT1 as a prognostic risk factor decreased the survival outcomes of patients with CRC [54]. In advanced CRC patients, overexpression of MALAT1 is associated with drug resistance [22]. MALAT1 is able to increase oxymatrine resistance and the invasion ability of CRC cells [55]. MALAT1 by targeting at least 243 genes stimulates tumor development and enhances CRC cell invasion. The expression of PRKA kinase anchor protein 9 (AKAP-9) has been recognized that was increased in CRC tissues with lymph node metastasis [35].
A study reported that tumor-associated dendritic cells (TADCs) promoted migration and EMT in CRC [56]. C–C motif ligand 5 (CCL5) is a chemokine that mimics the impact of TADCs on CRC cells. Therefore, the inhibition of CCL5 expression via neutralizing antibodies or siRNA reduced cancer progression by TADCs. It has been suggested that Snail as the downstream target of MALAT1 participates in TADC-mediated CRC progression [56].
Further studies have found that MALAT1 can target miR‑619‑5p and increase the clinicopathological features of patients with CRC [57]. In CRC, MALAT1 through interaction with EZH2 can inhibit the expression of E-cadherin and induce Oxaliplatin (Ox) resistance. Also, MALAT1 interacts with miR-218 to enhance EMT, metastasis, and FOLFOX resistance [22]. MALAT1 by targeting miR-363-3p can enhance EZH2 expression levels and promote CRC cell proliferation [58].
PTBP2 or PTB (polypyrimidine-tract-binding protein) is a proto-oncogene that promotes the growth of CRC cells [59]. SFPQ or PSF is a PTB-associated splicing factor and a tumor suppressor gene that binds to PTBP2 [60]. MALAT1 has been observed that interacts with SFPQ, releases PTBP2 from the SFPQ/PTBP2 complex (SFPQ-detached PTBP2), and accelerates tumor growth and metastasis [61, 62].
Sex-determining region Y (SRY)-box 9 (SOX9) is a transcription factor that participates in CRC oncogenesis and metastasis [63]. MALAT1 by suppressing miR-145 could accelerate SOX9 mediated CRC cell proliferation, migration, and tumorigenesis (MALAT1/miR-145/SOX9 axis) [64].
MALAT1 has been proved that directly stimulates the expression of the mRNA‐decapping enzymes 1a (DCP1A), down-regulates miR-203, and enhances CRC cell proliferation and invasion (MALAT/miR-203/DCP1A axis) [65].
High mobility group box protein 1 (HMGB1) is a nuclear protein that enhances CRC cell development [66]. MALAT1 by targeting miR-129-5p increased the expression of HMGB1 (MALAT1/miR-129-5p/HMGB1 axis) and enhanced the proliferation of CRC cells [67].
Moreover, MALAT1 through the activation of Wnt/β-catenin signaling enhances CRC cell proliferation and reduces apoptosis (caspase-3 and Bax reduced, Bcl-2 increased) [68]. Resveratrol has been shown that down-regulates MALAT1 mediated the Wnt/β-catenin signal pathway and reduces CRC cell invasion and metastasis [69]. Therefore, the knockout of MALAT1 suppressed CRC cell migration and proliferation [54, 68].
MALAT1 by targeting key molecules participating in drug resistance, including breast cancer resistance protein (BCRP), ATP-binding cassette transporters (ABC), and multi-drug resistance proteins (MDR1 and MRP1) can increase the metastasis and invasion of CRC cells. Also, MALAT1 blocks the expression of miR-20b-5p and enhances CRC cell tumorigenesis. Hence, inhibition of MALAT1 reduced cell migration and promoted the sensitivity of CRC cells to 5-FU [70].
Yes-associated protein 1 (YAP1) has been reported that increases proliferation and migration of CRC cells [71]. YAP1 by targeting the MALAT1/miR-126-5p axis can stimulate vascular endothelial growth factor (VEGFA), SLUG, and TWIST as metastasis-associated molecules and control EMT and angiogenesis in CRC cells. miR-126-5p by blocking SLUG, TWIST, and VEGFA has a tumor suppressor role in CRC [72].
RUNX2 (Runt-related transcription factor 2) is a key transcription factor and proto-oncogene that plays an important role in various tumors. miR-15s by suppressing LRP6 expression (Wnt receptor) can block activation of β-catenin signaling. MALAT1 interacts with IRES domain in the 5′UTR of the RUNX2 mRNAs and increases translational levels of RUNX2. MALAT1 also via miR-15s/LRP6/β-catenin signaling positively regulates RUNX2 expression and enhances CRC cell metastasis [73].
MALAT1 was recently investigated that suppressed miR-194-5p and enhanced the migration and invasion of CRC cells [74]. In CRC tissues and cell lines, microtubule-associated protein 1A/1B-light chain 3 (LC3-II/I) reflects autophagosome formation [75]. There is a positive correlation between MALAT1 and LC3-II mRNA levels in CRC cells. MALAT1 by binding to miR-101 can stimulate CRC cell proliferation and LC3-II-induced autophagy, and suppress the expression of Sequestosome-1 (p62/SQSTM1) as an autophagosome cargo protein [76].
miR-101-3p was also reported to play as a tumor suppressor in various neoplasms [77]. A recent study confirmed that MALAT1 targeted miR-101-3p in radio-resistance cells and promoted CRC cell viability [78].
It has been found that high-dose Vitamin C administration has an inhibitory effect on MALAT1 and CRC metastasis [79].
A disintegrin and metalloprotease metallopeptidase domain 17 (ADAM17) is a protease for epidermal growth factor receptor (EGF-R) ligand processing [80]. It has been recently shown that ADAM17 can accelerate the tumorigenesis of CRC [81]. MALAT1 through suppression of miR-324-3p and stimulation of ADAM17 (as a target of miR-324-3p) could reduce the Ox-sensitivity of CRC cells in xenograft tumor mice treated with Ox [82]. Besides, MALAT1 was identified to inhibit the expression of the hsa-miR-194-5p and decrease the progression-free survival in patients with CRC [83].
RAB14 is a small GTPase member of the RAS oncogene family that enhances CRC cell proliferation [84]. MALAT1 as a ceRNA can target miR-508-5p and RAB14 (as a target of miR-508-5p) promote CRC progression [85].
Based on previous studies, endoplasmic reticulum (ER) stress through the unfolded protein response (UPR) pathway is contributed to CRC metastasis [86, 87]. It has been found that the protein kinase R (PKR)‑like ER kinase (PERK), inositol‑requiring enzyme 1 (IRE1), and transcription factor 6 (ATF6) activate signaling pathways involved in the UPR [88]. ER stress by suppressing cyclin D1 (cell cycle machinery) and inducing apoptosis plays an important role in CRC metastasis [89]. Thapsigargin (TG) is an ER stress inducer that stimulates cell migration [90]. TG‑induced MALAT1 is associated with the expression of the PERK and IRE1 pathways. Moreover, in CRC tissue samples, MALAT1 is positively regulated with the X‑box‑binding protein 1 (XBP1) and ATF4 binding sites. Therefore, the IRE1/XBP1 and PERK/ATF4 signaling pathways are involved in MALAT1-induced CRC progression [91].
Exosomes also play critical roles in the progression of CRC [92, 93]. A previous study showed that highly metastatic CRC-derived exosomes could accelerate the fucosyltransferase 4 (FUT4) levels (a key enzyme of fucosylation), invasion, and metastasis in primary CRC cells. They indicated that MALAT1 by targeting miR-26a/26b promoted FUT4-associated fucosylation, stimulated the PI3K/AKT/mTOR pathway, and increased CRC cell proliferation and tumorigenesis (MALAT1/miR-26a/26b/FUT4 axis) [94].
A study identified that MALAT1 can interact with lincRNA-ROR, lncRNA-p21, p53 and increase the tumorigenesis of CRC cells [95].
The RNA-binding protein QUAKING (QK) is involved in apoptosis and the RNA stability of MALAT1. Recently, DANCR (lncRNA) was found to mediate the interaction between QK and MALAT1 (DANCR/QK/MALAT1 axis), increase the anti-apoptotic function of MALAT1, and reduce Doxorubicin (Dox)-induced apoptosis in CRC cells [96].
Therefore, compared to traditional methods, MALAT1 can be a novel biomarker for the early diagnosis and prognosis of CRC.
Conclusion
In this review, we highlighted the recently reported mechanism of MALAT1 in CRC. MALAT1 targets several signaling pathways such as Wnt/β-catenin, YAP, SOX9, RUNX2, Snail, EGF, PI3K/AKT/mTOR, and VEGF. Besides, MALAT1 has been found to modify miRNAs-associated drug sensitivity in CRC. Although these studies showed that MALAT1 plays a pivotal role in CRC tumorigenesis, the exact mechanisms whereby MALAT1 stimulates CRC development or invasion remains largely unclear. Taken together, the MALAT1-mediated treatment can be a critical therapeutic target for chemotherapy and radiotherapy sensitization.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ABC:
-
ATP-binding cassette transporters
- ADAM17:
-
A disintegrin and metalloprotease metallopeptidase domain 17
- AKAP-9:
-
PRKA kinase anchor protein 9
- ATF6:
-
Transcription factor 6
- BCRP:
-
Breast cancer resistance protein
- CCL5:
-
C–C motif ligand 5
- ceRNA:
-
Competing endogenous RNA
- CRC:
-
Colorectal cancer
- DCP1A:
-
Decapping enzymes 1a
- EMT:
-
Epithelial-mesenchymal transition
- ER:
-
Endoplasmic reticulum
- EZH2:
-
Enhancer of Zeste 2
- FUT4:
-
Fucosyltransferase 4
- HCN:
-
Hepcarcin
- HMGB1:
-
High mobility group box protein 1
- H3K27me3:
-
Trimethylation of histone H3 at lysine 27
- IBP160:
-
Intron binding protein
- IRE1:
-
Inositol‑requiring enzyme 1
- LC3-II/I:
-
Microtubule-associated protein 1A/1B-light chain 3
- LncRNA:
-
Long non-coding RNAs
- MALAT1:
-
Metastasis‐associated lung adenocarcinoma transcript‐1
- mascRNA:
-
MALAT1-associated small cytoplasmic RNA
- MDR:
-
Multi-drug resistance proteins
- miRNAs:
-
MicroRNAs
- NSCLC:
-
Non-small cell lung cancer
- NEAT2:
-
Nuclear-enriched abundant transcript 2
- Ox:
-
Oxaliplatin
- PERK:
-
Protein kinase R (PKR)‑like ER kinase
- PFS:
-
Progression-free survival
- PRC2:
-
Polycomb repressive complex 2
- PTB:
-
Polypyrimidine-tract-binding protein
- RNPS1:
-
RNA-binding protein with serine-rich domain 1
- RUNX2:
-
Runt-related transcription factor 2
- SQSTM1:
-
Sequestosome-1
- SRm160:
-
Serine/arginine-rich (SR)-related protein
- SRSF:
-
SR splicing factor
- TADCs:
-
Tumor-associated dendritic cells
- UPR:
-
Unfolded protein response
- VEGFA:
-
Vascular endothelial growth factor
- YAP1:
-
Yes-associated protein 1
References
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89–103.
Sayhan S, Kahraman DS. Pathologic features of colorectal carcinomas. In: Colon polyps and colorectal cancer, Springer, 2020, pp. 455–480.
Sekar V, Role of cancer stem cells in colitis-associated colorectal cancer, In: Diagnostic and treatment methods for ulcerative colitis and colitis-associated cancer, IGI Global, 2021, pp. 201–219.
Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJH, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065–15065.
Colov EP, Degett TH, Raskov H, Gögenur I. The impact of the gut microbiota on prognosis after surgery for colorectal cancer—a systematic review and meta-analysis. APMIS. 2020;128:162–76.
Mo X, Huang X, Feng Y, Wei C, Liu H, Ru H, Qin H, Lai H, Wu G, Xie W. Immune infiltration and immune gene signature predict the response to fluoropyrimidine-based chemotherapy in colorectal cancer patients. OncoImmunology. 2020;9:1832347.
Häfner MF, Debus J. Radiotherapy for colorectal cancer: current standards and future perspectives. Visc Med. 2016;32:172–7.
Ghani S, Bahrami S, Rafiee B, Eyvazi S, Yarian F, Ahangarzadeh S, Khalili S, Shahzamani K, Jafarisani M, Bandehpour M, Kazemi B. Recent developments in antibody derivatives against colorectal cancer: a review. Life Sci. 2021;265:118791.
Barani M, Bilal M, Rahdar A, Arshad R, Kumar A, Hamishekar H, Kyzas GZ. Nanodiagnosis and nanotreatment of colorectal cancer: an overview. J Nanopart Res. 2021;23:1–25.
Ali O, Tolaymat M, Hu S, Xie G, Raufman J-P. Overcoming obstacles to targeting muscarinic receptor signaling in colorectal cancer. Int J Mol Sci. 2021;22:716.
Yang Y, Yan X, Li X, Ma Y, Goel A. Long non-coding RNAs in colorectal cancer: novel oncogenic mechanisms and promising clinical applications. Cancer Lett. 2021;504:67–80.
Yang Y, Feng M, Bai L, Zhang M, Zhou K, Liao W, Lei W, Zhang N, Huang J, Li Q. The effects of autophagy-related genes and lncRNAs in therapy and prognosis of colorectal cancer. Front Oncol. 2021;11:582040.
Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinform. 2016;14:42–54.
Do H, Kim W. Roles of oncogenic long non-coding RNAs in cancer development. Genomics Inform. 2018;16:e18–e18.
Sun L, Zhang P, Lu W. lncRNA MALAT1 regulates mouse granulosa cell apoptosis and 17β-estradiol synthesis via regulating miR-205/CREB1 Axis. BioMed Res Int. 2021;2021:6671814.
Jiang X, Wang J, Deng X, Xiong F, Zhang S, Gong Z, Li X, Cao K, Deng H, He Y, Liao Q, Xiang B, Zhou M, Guo C, Zeng Z, Li G, Li X, Xiong W. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res. 2020;39:204.
Arun G, Aggarwal D, Spector DL. MALAT1 long non-coding RNA: functional implications. Non-coding RNA. 2020;6:22.
Wei Y, Niu B. Role of MALAT1 as a prognostic factor for survival in various cancers: a systematic review of the literature with meta-analysis. Dis Markers. 2015;2015:164635.
Ji D-G, Guan L-Y, Luo X, Ma F, Yang B, Liu H-Y. Inhibition of MALAT1 sensitizes liver cancer cells to 5-flurouracil by regulating apoptosis through IKKα/NF-κB pathway. Biochem Biophys Res Commun. 2018;501:33–40.
Sun R, Qin C, Jiang B, Fang S, Pan X, Peng L, Liu Z, Li W, Li Y, Li G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol Biosyst. 2016;12:952–62.
Qiao F-H, Tu M, Liu H-Y. Role of MALAT1 in gynecological cancers: pathologic and therapeutic aspects. Oncol Lett. 2021;21:1–8.
Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739–51.
Zhang H, Li W, Gu W, Yan Y, Yao X, Zheng J. MALAT1 accelerates the development and progression of renal cell carcinoma by decreasing the expression of miR-203 and promoting the expression of BIRC5. Cell Prolif. 2019;52:e12640.
Lu X, Chen D, Yang F, Xing N. Quercetin inhibits epithelial-to-mesenchymal transition (EMT) process and promotes apoptosis in prostate cancer via downregulating lncRNA MALAT1. Cancer Manag Res. 2020;12:1741.
Dai Q, Zhang T, Li C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag Res. 2020;12:1929.
Fu S, Wang Y, Li H, Chen L, Liu Q. Regulatory networks of LncRNA MALAT-1 in cancer. Cancer Manag Res. 2020;12:10181.
Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63:977–83.
Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, Tassone P. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11:1–19.
Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, Tassone P. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11:63–63.
Zhang X-Z, Liu H, Chen S-R. Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers. 2020;12:1245.
Zhao K, Jin S, Wei B, Cao S, Xiong Z. Association study of genetic variation of lncRNA MALAT1 with carcinogenesis of colorectal cancer. Cancer Manag Res. 2018;10:6257–61.
Chen Q, Zhu C, Jin Y. The oncogenic and tumor suppressive functions of the long noncoding RNA MALAT1: an emerging controversy. Front Genet. 2020;11:93.
Syllaios A, Moris D, Karachaliou GS, Sakellariou S, Karavokyros I, Gazouli M, Schizas D. Pathways and role of MALAT1 in esophageal and gastric cancer. Oncol Lett. 2021;21:1–7.
Li Z-X, Zhu Q-N, Zhang H-B, Hu Y, Wang G, Zhu Y-S. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–68.
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–74.
Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20:5758.
Su K, Wang N, Shao Q, Liu H, Zhao B, Ma S. The role of a ceRNA regulatory network based on lncRNA MALAT1 site in cancer progression. Biomed Pharmacother. 2021;137:111389.
Farzaneh M, Alishahi M, Derakhshan Z, Sarani NH, Attari F, Khoshnam SE. The expression and functional roles of miRNAs in embryonic and lineage-specific stem cells. Curr Stem Cell Res Ther. 2019;14:278–89.
Chen M, Wu D, Tu S, Yang C, Chen D, Xu Y. A novel biosensor for the ultrasensitive detection of the lncRNA biomarker MALAT1 in non-small cell lung cancer. Sci Rep. 2021;11:3666.
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–48.
Zhang X, Hamblin MH, Yin K-J. The long noncoding RNA Malat 1: its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14.
Wilusz JE, Spector DL. An unexpected ending: noncanonical 3’ end processing mechanisms. RNA. 2010;16:259–66.
Donlic A, Zafferani M, Padroni G, Puri M, Hargrove AE. Regulation of MALAT1 triple helix stability and in vitro degradation by diphenylfurans. Nucleic Acids Res. 2020;48:7653–64.
Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188502.
Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci. 2012;109:19202–7.
McCown PJ, Wang MC, Jaeger L, Brown JA. Secondary structural model of human MALAT1 reveals multiple structure–function relationships. Int J Mol Sci. 2019;20:5610.
Wilusz JE, Freier SM, Spector DL. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32.
Song Z, Lin J, Li Z, Huang C. The nuclear functions of long noncoding RNAs come into focus. Noncoding RNA Res. 2021;6:70–9.
Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175–81.
Qu D, Sun W-W, Li L, Ma L, Sun L, Jin X, Li T, Hou W, Wang J-H. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47:3013–27.
Lizarbe MA, Calle-Espinosa J, Fernández-Lizarbe E, Fernández-Lizarbe S, Robles MÁ, Olmo N, Turnay J. Colorectal cancer: from the genetic model to posttranscriptional regulation by noncoding RNAs. Biomed Res Int. 2017;2017:7354260.
Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: a long non-coding RNA highly associated with human cancers. Oncol Lett. 2018;16:19–26.
Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–75.
Zheng H-T, Shi D-B, Wang Y-W, Li X-X, Xu Y, Tripathi P, Gu W-L, Cai G-X, Cai S-J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–81.
Xiong Y, Wang J, Zhu H, Liu L, Jiang Y. Chronic oxymatrine treatment induces resistance and epithelial-mesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol Rep. 2018;39:967–76.
Kan J-Y, Wu D-C, Yu F-J, Wu C-Y, Ho Y-W, Chiu Y-J, Jian S-F, Hung J-Y, Wang J-Y, Kuo P-L. Chemokine (C-C Motif) ligand 5 is involved in tumor-associated dendritic cell-mediated colon cancer progression through non-coding RNA MALAT-1. J Cell Physiol. 2015;230:1883–94.
Qiu G, Zhang XB, Zhang SQ, Liu PL, Wu W, Zhang JY, Dai SR. Dysregulation of MALAT1 and miR-619-5p as a prognostic indicator in advanced colorectal carcinoma. Oncol Lett. 2016;12:5036–42.
Xie JJ, Li WH, Li X, Ye W, Shao CF. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33:331–43.
Hou P, Chen F, Yong H, Lin T, Li J, Pan Y, Jiang T, Li M, Chen Y, Song J, Zheng J, Bai J. PTBP3 contributes to colorectal cancer growth and metastasis via translational activation of HIF-1α. J Exp Clin Cancer Res. 2019;38:301.
He Q, Long J, Yin Y, Li Y, Lei X, Li Z, Zhu W. Emerging roles of lncRNAs in the formation and progression of colorectal cancer. Front Oncol. 2020;9:1542.
Amirkhah R, Naderi-Meshkin H, Shah JS, Dunne PD, Schmitz U. The intricate interplay between epigenetic events, alternative splicing and noncoding RNA deregulation in colorectal cancer. Cells. 2019;8:929.
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–48.
Prévostel C, Blache P. The dose-dependent effect of SOX9 and its incidence in colorectal cancer. Eur J Cancer. 2017;86:150–7.
Xu Y, Zhang X, Hu X, Zhou W, Zhang P, Zhang J, Yang S, Liu Y. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med. 2018;24:52.
Wu C, Zhu X, Tao K, Liu W, Ruan T, Wan W, Zhang C, Zhang W. MALAT1 promotes the colorectal cancer malignancy by increasing DCP1A expression and miR203 downregulation. Mol Carcinog. 2018;57:1421–31.
Süren D, Yıldırım M, Demirpençe Ö, Kaya V, Alikanoğlu AS, Bülbüller N, Yıldız M, Sezer C. The role of high mobility group box 1 (HMGB1) in colorectal cancer. Med Sci Monit. 2014;20:530–7.
Wu Q, Meng WY, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233:6750–7.
Zhang J, Li Q, Xue B, He R. MALAT1 inhibits the Wnt/β-catenin signaling pathway in colon cancer cells and affects cell proliferation and apoptosis. Bosn J Basic Med Sci. 2020;20:357–64.
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L, Sun J, Cai J, Qin J, Ren J, Li Q. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β-catenin signal pathway. PLoS ONE. 2013;8:e78700.
Tang D, Yang Z, Long F, Luo L, Yang B, Zhu R, Sang X, Cao G. Inhibition of MALAT1 reduces tumor growth and metastasis and promotes drug sensitivity in colorectal cancer. Cell Signal. 2019;57:21–8.
Dehghanian F, Azhir Z, Akbari A, Hojati Z. New insights into the roles of yes-associated protein (YAP1) in colorectal cancer development and progression. Ann Colorectal Res. 2019;7:1–7.
Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, Li G, Wang G, Song J, Li Z, Zhang Z, Yuan W, Li X. YAP1-induced MALAT1 promotes epithelial–mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627–44.
Ji Q, Cai G, Liu X, Zhang Y, Wang Y, Zhou L, Sui H, Li Q. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019;10:378.
Wu S, Sun H, Wang Y, Yang X, Meng Q, Yang H, Zhu H, Tang W, Li X, Aschner M. MALAT1 rs664589 polymorphism inhibits binding to miR-194-5p, contributing to colorectal cancer risk, growth, and metastasis. Can Res. 2019;79:5432–41.
Liu M, Zhao G, Zhang D, An W, Lai H, Li X, Cao S, Lin X. Active fraction of clove induces apoptosis via PI3K/Akt/mTOR-mediated autophagy in human colorectal cancer HCT-116 cells. Int J Oncol. 2018;53:1363–73.
Si Y, Yang Z, Ge Q, Yu L, Yao M, Sun X, Ren Z, Ding C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol Biol Lett. 2019;24:50.
Wang C-Z, Deng F, Li H, Wang D-D, Zhang W, Ding L, Tang J-H. MiR-101: a potential therapeutic target of cancers. Am J Transl Res. 2018;10:3310–21.
Guo J, Ding Y, Yang H, Guo H, Zhou X, Chen X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101–3p sponging. Exp Mol Pathol. 2020;115:104448.
Chen J, Qin F, Li Y, Mo S, Deng K, Huang Y, Liang W. High-dose vitamin C tends to kill colorectal cancer with high MALAT1 expression. J Oncol. 2020;2020:2621308.
Soto-Gamez A, Chen D, Nabuurs AGE, Quax WJ, Demaria M, Boersma YL. A bispecific inhibitor of the EGFR/ADAM17 axis decreases cell proliferation and migration of EGFR-dependent cancer cells. Cancers. 2020;12:411.
Schumacher N, Rose-John S. ADAM17 activity and IL-6 trans-signaling in inflammation and cancer. Cancers. 2019;11:1736.
Fan C, Yuan Q, Liu G, Zhang Y, Yan M, Sun Q, Zhu C. Long non-coding RNA MALAT1 regulates oxaliplatin-resistance via miR-324-3p/ADAM17 axis in colorectal cancer cells. Cancer Cell Int. 2020;20:1–12.
Yang Q, Zheng W, Shen Z, Huang G, Yang G. MicroRNA binding site polymorphisms of the long-chain noncoding RNA MALAT1 are associated with risk and prognosis of colorectal cancer in Chinese Han population. Genet Test Mol Biomarkers. 2020;24:239–48.
Gopal Krishnan PD, Golden E, Woodward EA, Pavlos NJ, Blancafort P. Rab GTPases: emerging oncogenes and tumor suppressive regulators for the editing of survival pathways in cancer. Cancers. 2020;12:259.
Zhang C, Yao K, Zhang J, Wang C, Wang C, Qin C. Long noncoding RNA MALAT1 promotes colorectal cancer progression by acting as a ceRNA of miR-508-5p to regulate RAB14 expression. Biomed Res Int. 2020;2020:4157606.
Huang J, Pan H, Wang J, Wang T, Huo X, Ma Y, Lu Z, Sun B, Jiang H. Unfolded protein response in colorectal cancer. Cell Biosci. 2021;11:26.
Zhao Q, Bi Y, Guo J, Liu Y, Zhong J, Liu Y, Pan L, Guo Y, Tan Y, Yu X. Effect of pristimerin on apoptosis through activation of ROS/endoplasmic reticulum (ER) stress-mediated noxa in colorectal cancer. Phytomedicine. 2021;80:153399.
Adams CJ, Kopp MC, Larburu N, Nowak PR, Ali MMU. Structure and molecular mechanism of ER stress signaling by the unfolded protein response signal activator IRE1. Front Mol Biosci. 2019;6:11.
Choi SS, Lee SK, Kim JK, Park H-K, Lee E, Jang J, Lee YH, Khim KW, Hyun J-M, Eom H-J, Lee S, Kang BH, Chae YC, Myung K, Myung S-J, Park CY, Choi JH. Flightless-1 inhibits ER stress-induced apoptosis in colorectal cancer cells by regulating Ca2+ homeostasis. Exp Mol Med. 2020;52:940–50.
Jaskulska A, Janecka AE, Gach-Janczak K. Thapsigargin—from traditional medicine to anticancer drug. Int J Mol Sci. 2021;22:4.
Jiang X, Li D, Wang G, Liu J, Su X, Yu W, Wang Y, Zhai C, Liu Y, Zhao Z. Thapsigargin promotes colorectal cancer cell migration through upregulation of lncRNA MALAT1. Oncol Rep. 2020;43:1245–55.
Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, Yuan J, Sun J, Dai L, Yang C, Li Z, Wang J, Zhong T. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020;476:13–22.
Zhou J, Li X-L, Chen Z-R, Chng W-J. Tumor-derived exosomes in colorectal cancer progression and their clinical applications. Oncotarget. 2017;8:100781–90.
Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y, Li S, Qi Y, Huang Y, Jia L. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer Res. 2020;39:1–15.
Chaleshi V, Irani S, Alebouyeh M, Mirfakhraie R, Asadzadeh Aghdaei H. Association of lncRNA-p53 regulatory network (lincRNA-p21, lincRNA-ROR and MALAT1) and p53 with the clinicopathological features of colorectal primary lesions and tumors. Oncol Lett. 2020;19:3937–49.
Xiong M, Wu M, Dan P, Huang W, Chen Z, Ke H, Chen Z, Song W, Zhao Y, Xiang AP, Zhong X. LncRNA DANCR represses doxorubicin-induced apoptosis through stabilizing MALAT1 expression in colorectal cancer cells. Cell Death Dis. 2021;12:24.
Acknowledgements
This work was supported by the National Science Foundation of China (82074478 to RQL), the Project of six “1” Project of Jiangsu Province (LGY2016012 to XYW), Social Development Project of Jiangsu Province (BE2019768 to WXY), Maternal and Child Health Research Project of Jiangsu Province (F201763 to XWW) and Jiangsu Provincial Hospital of Traditional Chinese Medicine Research Fund Project (Y19053 to XWW). The funding institutions did not have any roles in the study design, data collection, or analysis.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
WX and JJ have been involved in drafting the manuscript. XW, QR, and MF have made substantial contributions to the revising of the manuscript and the design of the Figures. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, WW., Jin, J., Wu, Xy. et al. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int 22, 126 (2022). https://doi.org/10.1186/s12935-022-02540-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-022-02540-y