Abstract
Background
Long non-coding RNA (lncRNA) is a class of endogenous RNA with a length of more than 200 nucleotides, which is emerging as a pivotal player in cancer development and progression. However, the functional roles of many members in this class remain largely uncharacterized. In the present study, we explored the biological relevance of linc02042 in esophageal squamous cell carcinoma (ESCC).
Methods
qRT-PCR was used to detect the levels of linc02042 and c-Myc. Western blot was used to assess protein expression level. CCK-8 and Transwell assays were employed to test ESCC cell proliferation and invasion, respectively. The mice study including xenograft tumor and lung metastasis models was used to determine the role of linc02042 in vivo. RNA pull-down, ChIP and luciferase reporter assays were employed to test the relationship between linc02042, YBX1 and c-Myc.
Results
Linc02042 was found to be markedly upregulated in ESCC cell lines, tissues and plasma, and was closely correlated with malignant clinical features. Knockdown of linc02042 significantly inhibited ESCC cell viability and invasion in vitro as well as tumor growth and lung metastasis in vivo, whereas overexpression of linc02042 resulted in the opposite results. Mechanistically, linc02042 acted as a scaffold for YBX-1 binding to the 3′-UTR of c-Myc mRNA, leading to enhanced c-Myc mRNA stability, thereby facilitating ESCC growth and metastasis. Moreover, in turn, c-Myc was able to transcriptionally elevate linc02042 by directly binding to the E-box motif proximal to the transcription start site (TSS) of linc02042 promoter. Clinically, linc02042 was identified as an effective diagnostic and prognostic biomarker for ESCC patients, and its expression was strongly positively correlated with c-Myc expression in ESCC tissues.
Conclusion
Our data suggest that linc02042 plays an important tumor-promoting role in ESCC, which lays a foundation for considering it as a potential target for ESCC patients.
Similar content being viewed by others
Background
Esophageal cancer is a common aggressive malignancy with the seventh highest morbidity and mortality [1]. Every year, a large number of patients die from the disease worldwide, especially in China, with an average annual of about 150,000 deaths [2]. Esophageal squamous cell carcinoma (ESCC) is the major histological type of esophageal cancer, its 5-year survival rate is poor due to recurrence, metastasis or chemotherapy resistance [3]. Therefore, it is urgent to elucidate the pathogenesis of ESCC to provide new diagnostic and therapeutic targets for ESCC patients.
Long non-coding RNA (lncRNA) refers to a kind of endogenous RNA that is over 200 nucleotides in length and lacks protein coding potential [4]. LncRNA was initially considered as transcriptional noise, but subsequent studies have shown that many lncRNAs are only expressed in specific parts of the body in specific physiological states, or only in certain biological processes [5]. Knocking down of specific lncRNA could lead to phenotypic changes, thus proving that it has important biological functions. In fact, the current research on lncRNA covers almost all physiological and pathological processes, including the occurrence and development of cancer [6, 7].
It has been well documented that the mechanism of lncRNA functioning is mainly as a molecular sponge of microRNAs (miRNAs), in which lncRNA can directly bind to miRNAs and reduce the repressive effects of miRNAs on their different target genes [8]. However, in addition to this, a growing number of studies have shown that lncRNA can also directly bind to proteins to regulate gene expression, thus participating in the progression of cancer [9]. For instance, lncRNA GIAT4RA was shown to interact with LSH and block UCHL3-mediated deubiquitination of LSH, thereby inhibiting non-small cell lung cancer progression [10]. Zhang et al. reported that MEG3 directly bound to p-STAT3, resulting in p-STAT3 ubiquitination and degradation and inhibition of proliferation of cervical cancer cells [11]. However, this functional model of lncRNA/protein interaction in ESCC remains poorly understood.
A recent study performed genome-wide screening in ESCC and normal tissues and found that linc02042 was significantly upregulated in ESCC tissues [12], however, its molecular function and biological role are still not clear. Herein, we confirmed the overexpression of linc02042 in ESCC, and further elucidated its carcinogenic role as a protein binding partner.
Materials and methods
ESCC samples and cell lines
ESCC tissues and plasma in our study were all collected from Henan Provincial Chest Hospital, and patients who had received preoperative chemoradiotherapy were excluded. The detailed clinical data of patients are presented in Table 1. Routine follow-up was conducted every 3 months. All patients signed the informed consent. This study was approved by the Ethics Committee of Henan Provincial Chest Hospital. Five ESCC cell lines and one normal esophageal squamous epithelium HET-1A cells were commercially obtained from Shanghai Institute of Biological Sciences, Chinese Academy of Sciences. They all cultured in DMEM medium supplemented with 10% fetal bovine serum in the incubator.
Identification of the subcellular localization of linc02042
The subcellular localization of linc02042 was determined by Nuclear-Cytoplasmic isolation and fluorescence in situ hybridization (FISH) assays, which were respectively performed by using the Cytoplasmic & Nuclear RNA Purification (Norgen Biotek Corp, ON, CAN) and RiboTM Fluorescent In Situ Hybridization (RiboBio, Guangzhou, China) kits in accordance with the instructions from manufacturers.
Reverse transcription quantitative polymerase chain reaction (qRT-PCR)
Total RNA from ESCC tissues and cultured cells was extracted by Trizol reagent (Invitrogen, CA, USA) according to the standard protocol. Then, cDNA was synthesized using Superscript First-Strand Synthesis System (Invitrogen), followed by PCR amplification and quantification using SYBR® Green qPCR SuperMix (Invitrogen) with specific primers. The expression level of genes relative to GAPDH were calculated by 2−ΔΔCt method. The assay was repeated three times independently.
CCK-8 and Transwell assays
Cell viability was detected by CCK-8 assay using CCK-8 solution (Dojindo, Kumamoto, Japan) in accordance with the manufacturer’s instruction. For cell invasion assay, the indicated cells were seeded onto 24-well culture plate mounted with Transwell chamber. After incubation for 2 days, the cells on the upper surface of the chamber were removed, and the cells on the lower surface were stained with crystal violet. The analysis was performed based on five random field under the microscope.
In vivo tumorigenicity and lung metastasis
The animal experiment was approved by the Committee on Animal Care of Henan Provincial Chest Hospital. For the xenograft tumor model, a total of 10 nude mice were randomly divided into two groups (n = 5 per group), followed by subcutaneous injection of 1 × 107 linc02042-depleted or control KYSE30 cells into nude mice. Tumors were measured every week. In the fifth week, all mice were sacrificed and tumor tissues were collected and weighed. For the lung metastasis model, 1 × 106 linc02042-depleted or control KYSE30 cells were tail vein injected into nude mice (n = 5 per group), and the lung metastatic nodules were counted 6 weeks after injection.
Western blot
Total protein from ESCC tissues and cultured cells was extracted by lysis buffer on the ice and separated on 10% SDS-PAGE gel. Then, the protein was transferred onto PVDF member and blocked by 5% non-fat milk powder for 30 min. The member was incubated with anti-c-Myc (#9402, CST, 1:1000 dilution) and anti-YBX1 (#9744, CST, 1:2000 dilution) primary antibodies at 4 ℃ overnight. The next day, the member was incubated with anti-rabbit IgG secondary antibody for 1 h at room temperature. Lastly, the member was exposed with ECL solution in the darkroom.
Luciferase reporter assay
The promoters of c-Myc and linc02042 were respectively cloned into pGL3-basic vector (Promega, WI, USA) and co-transfected with 5ng pRL-TK-Renilla into KYSE-30 and KYSE-150 cells using Lipofectamine 2000 (Invitrogen) as per manufacturer’s protocol. After 48 h of transfection, the luciferase activity was detected by Dual-Luciferase Reporter Assay System (Promega) as per manufacturer’s protocol.
RNA pull-down and RNA immunocoprecipitation (RIP) assays
The linc02042 and anti-sense biotin-labeled probes were in vitro synthesized and labeled by using T7 High Yield RNA Synthesis Kit (Ambion, TX, USA) and RNA 3′ End Biotinylation Kit (Themo, Waltham, MA), respectively. After that, the probes were incubated with whole protein lysate extracted from KYSE-30 and KYSE-150 cells at 4 ℃ overnight. Subsequently, the protein-probe complex was incubated with BeaverBeads™ Streptavidin magnetic beads (Beaver, Suzhou, China) for 2 h at room temperature. Then, the bead-probe-protein complex was washed six times and subjected for Western blot analysis. RIP assay was performed using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA) according to the manufacturer’s instructions using anti-YBX1 (#9744, CST) antibody.
Chromatin immunoprecipitation (ChIP) assay
KYSE-30 and KYSE-150 cells were fixed with 1% formaldehyde for 10 min for the cross-link between protein and DNA, followed by quenching with 0.125 M glycine for 5 min. Then, DNA was sonicated into 200–1000 bp in length. The sample was incubated with anti-c-Myc (#9402, CST, 1:50) at 4 ℃ overnight, followed by incubation with ChIP-Grade Protein G Agarose Beads (#9007, CST) at 4 ℃ for 1 h, and the complex was washed and eluted by Elution buffer (1% SDS, 0.1 M NaHCO3). Lastly, the enriched fragments were subjected for qPCR analysis.
Statistical analysis
All statistical results were analyzed by SPSS 22.0 software (IBM, NY, USA) and figures were generated by Graphpad Prism v5 software (Graphpad, CA, USA). Differences were determined by Student’s t test for two groups and by one-way ANOVA for three or more groups. p ≤ 0.05 was considered significant. *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Linc02042 expression is significantly upregulated in ESCC
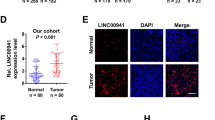
We collected 98 pairs of ESCC and adjacent normal tissues to detect linc02042 expression. As shown in Fig. 1a, linc02042 was notably increased in ESCC tissues as compared to normal tissues. And this upregulation was also observed in five ESCC cell lines (Fig. 1b). We then evaluated the relationship between linc02042 expression and clinicopathological features of ESCC patients, the results showed that high linc02042 was positively correlated with malignant features, including larger tumor size, poor differentiation, advanced TNM stage and lymph node metastasis (Table 1). Further, we found that linc02042 was also overexpressed in ESCC plasma in comparison to healthy control plasma (Fig. 1c), and the receiver operating characteristic (ROC) curve showed that the area under the curve (AUC) value was 0.9294 (95%CI 0.8638 to 0.9949) (Fig. 1d), implying that plasma linc02042 has an excellent diagnostic efficacy for ESCC. Kaplan–Meier plotter displayed that patients with high linc02042 had shorter overall survival time than those with low linc02042 (Fig. 1e). Besides, we assessed the subcellular localization of linc02042 by using qRT-PCR and FISH assays, and the results showed that linc02042 was predominantly located in the cytoplasm (Fig. 1f). These above data suggest that linc02042 is frequently overexpressed in ESCC and may play an essential role in the pathogenesis of ESCC.
Linc02042 is upregulated in ESCC. a, b qRT-PCR analysis of linc02042 expression in ESCC tissues (n = 98) and cell lines. c qRT-PCR analysis of linc02042 expression in plasma samples from ESCC patients and healthy controls (n = 30). d The ROC curve showing the diagnostic value of plasma linc02042 level. e The survival curve showing the prognostic value of linc02042 in ESCC. f The location of linc02042 detected by qRT-PCR analysis and FISH assay in HET-1A cells. *p < 0.05, **p < 0.01, ***p < 0.001
Linc02042 promotes ESCC cell proliferation and invasion both in vitro and in vivo
To explore the biological function of linc02042, we generated the stable linc02042-overexpressed and -silenced ESCC cell lines by using pCDH-CMV-MCS-EF1-Puro lentiviral vector (Fig. 2a, b). The CCK-8 results showed that overexpression of linc02042 markedly strengthened cell viability (Fig. 2c), while linc02042 knockdown dramatically weakened cell viability (Fig. 2d). Similarly, exogenous linc02042 expression enhanced cell invasiveness (Fig. 2e), whereas depletion of linc02042 resulted in an opposite effect (Fig. 2f, g). Further, we established in vivo tumorigenesis and metastasis models by subcutaneous and caudal vein injection of linc02042-depleted KYSE30 cells into nude mice, respectively. The results showed that the tumor volume/weight and lung metastasis nodules in sh-linc02042 group were less than those in control group (Fig. 2h, i). These findings indicate that linc02042 enhances ESCC cell aggressive phenotype.
The effect of linc02042 on ESCC cell phenotype. a, b qRT-PCR analysis confirming the overexpression or knockdown efficiency in ESCC cells. c, d CCK-8 assays in linc02042-overexpressed or silenced cells, the absorbance at 450 nm was detected. e–g Transwell invasion assays in linc02042-overexpressed or silenced cells. h The tumor volume and weight in control and linc02042-depleted groups. i The representative image of H&E staining of lung metastasis nodule in control and linc02042-depelted groups. **p < 0.01, ***p < 0.001
Linc02042 increases the stability of c-Myc mRNA
Through analyzing the TCGA ESCC database using cBioPortal software, we found that linc02042 was positively linked with c-Myc, a well-known proto-oncogene. Consistently, c-Myc mRNA as well as protein levels were significantly decreased in linc02042 knockdown ESCC cells in comparison with control cells (Fig. 3a, b). Moreover, the IHC staining results showed that less c-Myc positive cells were observed in linc02042-depleted transplanted tissues compared to control tissues (Fig. 3b). To test whether linc02042 affected c-Myc expression at the transcriptional level, we performed the luciferase reporter assay by inserting the promoter of c-Myc into pGL3-basic vector. As shown in Fig. 3c, the promoter activity of c-Myc was not affected after depletion of linc02042, implying that c-Myc was controlled by linc02042 at the post-transcriptional level. We then tested the mRNA stability via treating cells with Actinomycin D, a transcription inhibitor. Surprisingly, knockdown of linc02042 shortened the half-life of c-Myc mRNA from more than 60 min to about 35 min (Fig. 3d). Besides, we found that c-Myc expression was substantially overexpressed in ESCC tissues (Fig. 3e) and strongly positively correlated with linc02042 expression (r = 0.755) (Fig. 3f). Importantly, overexpression of linc02042 could not enhance cell viability and invasiveness in the absence of c-Myc (Fig. 3g). These data suggest that c-Myc is indispensable for the tumor-promoting function of linc02042 in ESCC.
Linc02042 increases c-Myc stability. a qRT-PCR analysis of c-Myc expression in linc02042-silenced ESCC cells. b Western blotting and IHC staining analysis of c-Myc protein expression in linc02042-silenced ESCC cells and transplanted tumor tissues, respectively. c Luciferase reporter assay detecting the promoter activity of c-Myc in linc02042-silenced ESCC cells. d qRT-PCR analysis of remaining c-Myc expression in linc02042-silenced ESCC cells treated with 1 μM Actinomycin D for the indicated time. e qRT-PCR analysis of c-Myc expression in ESCC adjacent normal tissues. f The correlation between linc02042 and c-Myc expression in ESCC tissues. g The assessment of cell viability and invasion respectively detected by CCK-8 and Transwell assays in linc02042-overexpressing cells transfected with c-Myc siRNA. **p < 0.01
YBX1 mediates the regulation of linc02042 on c-Myc
Given that YBX1 acts as an important regulator of c-Myc mRNA stability via binding to its 3′-UTR [13] and that numerous lncRNAs have been found to be bound by YBX1 [14], we thus inferred that linc02042 might increase c-Myc mRNA stability via YBX1. To confirm this inference, we first test the possibility of binding between linc02042 and YBX1 by using RPISeq database, the results showed that linc02042 has a strong possibility of direct interaction with YBX1 (RF classifier = 0.88, SVM classifier = 0.9) (Fig. 4a). Next, we performed the RNA pull-down assay, as shown in Fig. 4b, endogenous YBX1 was abundantly enriched by linc02042 probe. And RIP assay coupled qRT-PCR analysis further confirmed this interaction, in which more linc02042 was immunoprecipitated by YBX1 antibody as compared with IgG antibody (Fig. 4c). However, linc02042 knockdown did not affect YBX1 protein expression (Fig. 4d). Importantly, we found that YBX1 was abundantly enriched on the 3′-UTR of c-Myc mRNA, but this phenomenon was eliminated in the absence of linc02042 (Fig. 4e), suggesting that linc02042 is required for the binding of YBX1 to c-Myc mRNA. Further, overexpression of linc02042 could not increase c-Myc expression after depletion of YBX1 (Fig. 4f), and the enhanced aggressive phenotype caused by linc02042 overexpression was substantially blocked by YBX1 knockdown (Fig. 4g). These results indicate that linc02042 and YBX1 form a dimer complex that regulates c-Myc expression.
Linc02042 interacts with YBX1. a RPISeq database predicting the interaction probability between linc02042 and YBX1. Both RF and SVM classifier greater than 0.5 mean positive. b RNA pull-down coupled Western blotting analysis in ESCC cells. c RIP assays in ESCC cells with IgG and YBX1 antibodies, followed by qRT-PCR analysis of linc02042 expression. d Western blotting analysis of YBX1 protein expression in linc02042-depleted ESCC cells. e RIP assays in ESCC cells with IgG and YBX1 antibodies in linc02042-depleted ESCC cells. f qRT-PCR analysis of c-Myc expression in linc02042-overexpressed cells treated with YBX1 siRNA. g The assessment of cell viability and invasion respectively detected by CCK-8 and Transwell assays in linc02042-overexpressing cells transfected with YBX1 siRNA. **p < 0.01, ***p < 0.001
c-Myc trans-activates linc02042 expression
Intriguingly, three E-box motifs (c-Myc binding site) were found on the linc02042 promoter, we thus assumed that linc02042 might be also regulated by c-Myc. To test this hypothesis, we constructed a series of pGL3-basic luciferase vector (Fig. 5a) and performed the luciferase reporter assay. The results showed that silencing of c-Myc significantly reduced the promoter activity of wild-type vector in both KYSE30 and KYSE150 cells, whereas this phenomenon was eliminated after mutation of E-box#3 rather than E-box#1 or E-box#2 (Fig. 5b, c). Further, the ChIP assay results revealed that c-Myc directly bound to E-box#3, but not E-box#1 or E-box#2 (Fig. 5d, e). Consistently, linc02042 expression was evidently decreased in c-Myc-silenced KYSE30 and KYSE150 cells in comparison to control cells (Fig. 5f). These data suggest that linc02042 is transcriptionally controlled by c-Myc.
Linc02042 is activated by c-Myc. a–c Luciferase reporter assay detecting the promoter activity of linc02042 in ESCC cells using the indicated mutant pGL3-basic vector. d, e ChIP assays in ESCC cells using IgG and c-Myc antibodies, followed by qPCR analysis. f qRT-PCR analysis of linc02042 expression in ESCC cells transfected with control or c-Myc siRNA. **p < 0.01, ***p < 0.001
Discussion
ESCC is a common malignant tumor with high heterogeneity, which makes its pathogenesis and progression extremely complicated. In the current study, we found a novel ESCC-related lncRNA, linc02042, which was significantly increased in ESCC tissues, cells and plasma. Gain and loss functional assays showed that linc02042 promoted ESCC cell proliferation and invasion both in vitro and in vivo. Stepwise mechanism studies revealed that linc02042 directly interacted with YBX1 and increased the binding of YBX1 to c-Myc 3′-UTR, resulting in potentiating c-Myc mRNA stability, thereby facilitating ESCC malignant progression. Therefore, our data highlight the importance of lncRNA in ESCC and also advance the understanding of the regulatory mechanism of proto-oncogene c-Myc.
A large number of studies have reported that non-coding RNA is an effective cancer biomarker, especially lncRNA with length over 200 nt. Fox example, lncRNA MALAT1 [15], PIK3CD-AS1 [16], linc-ZNF469-3 [17] and LINC01133 [18] were respectively identified as diagnostic or prognostic biomarkers of colorectal cancer, hepatocellular carcinoma, triple negative breast cancer and gastric cancer. Herein, ESCC patients with high linc02042 expression displayed shorter survival time than those with low linc02042 expression, implicating the prognostic value of linc02042 in ESCC. In addition, we also found that plasma linc02042 was notably overexpressed in ESCC patients in comparison with healthy controls, and the AUC value is 0.9294, indicating that plasma linc02042 level is an excellent non-invasive diagnostic biomarker of patients with ESCC. Further investigation in large-scale samples is needed to confirm the clinical implication of linc02042.
Increasing evidence suggests that lncRNA participates in cancer occurrence development by acting as a protein-binding partner [19]. Herein, we found that linc02042 could directly bind to YBX1 by performing RNA pull-down and RIP assays. YBX1 is a multifunctional oncoprotein involved in cancer cell growth, metastasis and chemotherapy resistance that can modulate mRNA stability by bind to AU-rich elements (ARE) on mRNA 3′-UTR [20]. To date, numerous lncRNAs have been identified to interact with YBX1, such as HOXC-AS3 [21] and LINC00312 [22], and YBX1 was significantly upregulated in human cancers [23]. However, in this study, the interaction between linc02042 and YBX1 did not increase YBX1 protein level, but rather recruited YBX1 to the 3′-UTR of c-Myc mRNA to stabilize c-Myc. This notion was confirmed by the RIP assay in which linc02042 knockdown substantially decreased the binding of YBX1 to c-Myc 3′-UTR. c-Myc is a well-known oncogene that activates a variety of oncogenic signaling pathways [24]. c-Myc is shown to be frequently overexpressed in multiple cancers, and its expression is tightly controlled by different factors at the transcription or post-transcription level [25]. Up to now, only a few studies focused the regulatory role of lncRNA on c-Myc mRNA stability. Here, we found that linc02042 increased c-Myc mRNA stability, and the pro-tumor effect of linc02042 was practically disappeared in the absence of c-Myc, suggesting that c-Myc is required for the function of linc02042 in ESCC. Besides, silencing of YBX1 evidently blocked the increased c-Myc level caused by linc02042 overexpression, indicating that linc02042 regulates c-Myc mRNA stability in an YBX1-dependent mechanism.
Of note, c-Myc is also a key transcription factor that regulates gene expression via directly binding to the E-box motif on the promoter [26]. Many genes have been identified as the downstream targets of c-Myc, including lncRNAs, such as MINCR [27] and IDH1-AS1 [28]. In the present study, we found three E-box motifs on linc02042 promoter, thus speculated that linc02042 might be also regulated by c-Myc. By performing a series of assays, we confirmed above hypothesis, in which c-Myc was able to directly bind to the E-box motif proximal to TSS on linc02042 promoter and increase linc02042 expression. This positive feedback loop between linc02042 and c-Myc magnifies the pro-tumor effect of linc02042 in ESCC.
Conclusion
Taken together, our study for the first time describes the important biological functions of linc02042 in ESCC, and meanwhile provides a novel biomarker and therapeutic target for patients with ESCC. Whether linc02042 also plays the fundamental role in other malignancies is worth further investigation.
Availability of data and materials
Please contact authors for data request.
Change history
14 June 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1186/s12935-024-03397-z
Abbreviations
- LncRNA:
-
Long non-coding RNA
- ESCC:
-
Esophageal squamous cell carcinoma
- miRNAs:
-
microRNAs
- FISH:
-
Fluorescence in situ hybridization
- CCK-8:
-
Cell Counting Kit-8
- RIP:
-
RNA immunocoprecipitation
- ChIP:
-
Chromatin immunoprecipitation
- UTR:
-
Untranslated region
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Peng L, Cheng S, Lin Y, Cui Q, Luo Y, Chu J, Shao M, Fan W, Chen Y, Lin A, Xi Y, Sun Y, Zhang L, Zhang C, Tan W, Gao G, Wu C, Lin D. CCGD-ESCC: a comprehensive database for genetic variants associated with esophageal squamous cell carcinoma in Chinese population. Genomics Proteomics Bioinform. 2018;16:262–8.
Murphy G, McCormack V, Abedi-Ardekani B, Arnold M, Camargo MC, Dar NA, Dawsey SM, Etemadi A, Fitzgerald RC, Fleischer DE, Freedman ND, Goldstein AM, Gopal S, Hashemian M, Hu N, Hyland PL, Kaimila B, Kamangar F, Malekzadeh R, Mathew CG, Menya D, Mulima G, Mwachiro MM, Mwasamwaja A, Pritchett N, Qiao YL, Ribeiro-Pinto LF, Ricciardone M, Schuz J, Sitas F, Taylor PR, Van Loon K, Wang SM, Wei WQ, Wild CP, Wu C, Abnet CC, Chanock SJ, Brennan P. International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann Oncol. 2017;28:2086–93.
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41.
Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–7.
Battaglin F, Naseem M, Puccini A, Lenz HJ. Molecular biomarkers in gastro-esophageal cancer: recent developments, current trends and future directions. Cancer Cell Int. 2018;18:99.
Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL, Kim IM. Crosstalk between long noncoding RNAs and MicroRNAs in health and disease. Int J Mol Sci. 2016;17:356.
Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106–16.
Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C, Liu Y, Wang M, Lai W, Tang H, Gao M, Xiao D, Wang X, Zhou H, Tang CE, Liu W, Yu F, Cao Y, Yan Q, Liu S, Tao Y. GIAT4RA functions as a tumor suppressor in non-small cell lung cancer by counteracting Uchl3-mediated deubiquitination of LSH. Oncogene. 2019;38(1):280.
Zhang J, Gao Y. Long non-coding RNA MEG3 inhibits cervical cancer cell growth by promoting degradation of P-STAT3 protein via ubiquitination. Cancer Cell Int. 2019;19:175.
Wang W, Wei C, Li P, Wang L, Li W, Chen K, Zhang J, Zhang W, Jiang G. Integrative analysis of mRNA and lncRNA profiles identified pathogenetic lncRNAs in esophageal squamous cell carcinoma. Gene. 2018;661:169–75.
Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–15.
Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, Lambowitz AM. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci USA. 2017;114:E8987–95.
Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, Li G, Wang G, Song J, Li Z, Zhang Z, Yuan W, Li X. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627–44.
Song W, Zhang J, Zhang J, Sun M, Xia Q. Overexpression of lncRNA PIK3CD-AS1 promotes expression of LATS1 by competitive binding with microRNA-566 to inhibit the growth, invasion and metastasis of hepatocellular carcinoma cells. Cancer Cell Int. 2019;19:150.
Wang PS, Chou CH, Lin CH, Yao YC, Cheng HC, Li HY, Chuang YC, Yang CN, Ger LP, Chen YC, Lin FC, Shen TL, Hsiao M, Lu PJ. A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer. Oncogene. 2018;37:4662–78.
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126.
Gawronski AR, Uhl M, Zhang Y, Lin YY, Niknafs YS, Ramnarine VR, Malik R, Feng F, Chinnaiyan AM, Collins CC, Sahinalp SC, Backofen R. MechRNA: prediction of lncRNA mechanisms from RNA-RNA and RNA-protein interactions. Bioinformatics. 2018;34:3101–10.
Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5:95–110.
Zhang E, He X, Zhang C, Su J, Lu X, Si X, Chen J, Yin D, Han L, De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154.
Peng Z, Wang J, Shan B, Li B, Peng W, Dong Y, Shi W, Zhao W, He D, Duan M, Cheng Y, Zhang C, Duan C. The long noncoding RNA LINC00312 induces lung adenocarcinoma migration and vasculogenic mimicry through directly binding YBX1. Mol Cancer. 2018;17:167.
Kuwano M, Shibata T, Watari K, Ono M. Oncogenic Y-box binding protein-1 as an effective therapeutic target in drug-resistant cancer. Cancer Sci. 2019;110:1536–43.
Yoshida GJ. Emerging roles of Myc in stem cell biology and novel tumor therapies. J Exp Clin Cancer Res. 2018;37:173.
Wang XN, Su XX, Cheng SQ, Sun ZY, Huang ZS, Ou TM. MYC modulators in cancer: a patent review. Expert Opin Ther Pat. 2019;29:353–67.
Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77.
Chen S, Gu T, Lu Z, Qiu L, Xiao G, Zhu X, Li F, Yu H, Li G, Liu H. Roles of MYC-targeting long non-coding RNA MINCR in cell cycle regulation and apoptosis in non-small cell lung Cancer. Respir Res. 2019;20:202.
Xiang S, Gu H, Jin L, Thorne RF, Zhang XD, Wu M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1alpha via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci USA. 2018;115:E1465–74.
Acknowledgements
None.
Funding
This project was supported by Grants from Henan Province Science and Technology Tackling Project (172102310116).
Author information
Authors and Affiliations
Contributions
WY designed this study and drafted the manuscript; JD, GZ, HQ conducted the experiments and analyzed the results. HY provided technical guidance. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed in accordance with institutional ethical guidelines and was approved by the Ethics Committee of Henan Provincial Chest Hospital. Informed written consent was obtained from each participants.
Consent for publication
All authors approved publication of the manuscript.
Competing interests
The authors report no conflicts of interest in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1186/s12935-024-03397-z
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Du, J., Zhang, G., Qiu, H. et al. RETRACTED ARTICLE: A novel positive feedback loop of linc02042 and c-Myc mediated by YBX1 promotes tumorigenesis and metastasis in esophageal squamous cell carcinoma. Cancer Cell Int 20, 75 (2020). https://doi.org/10.1186/s12935-020-1154-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-1154-x