Abstract
Background
Without targets, triple negative breast cancer (TNBC) has the worst prognosis in all subtypes of breast cancer (BC). Recently, eukaryotic translation initiation factor 3 m (eIF3m) has been declared to be involved in the malignant progression of various neoplasms. The aim of this study is to explore biological functions of eIF3m in TNBC.
Methods
Multiple databases, including Oncomine, KM-plotter and so on, were performed to analyze prognosis and function of eIF3m in TNBC. After transfection of eIF3m-shRNA lentivirus, CCK-8, colony formation assay, cell cycle analysis, wound healing assay, transwell assays, mitochondrial membrane potential assay and cell apoptosis analysis were performed to explore the roles of eIF3m in TNBC cell bio-behaviors. In addition, western blotting was conducted to analyze the potential molecular mechanisms of eIF3m.
Results
In multiple databases, up-regulated eIF3m had lower overall survival, relapse-free survival and post progression survival in BC. EIF3m expression in TNBC was obviously higher than in non-TNBC or normal breast tissues. Its expression in TNBC was positively related to differentiation, lymph node invasion and distant metastasis. After knockdown of eIF3m, cell proliferation, migration, invasion and levels of mitochondrial membrane potential of MDA-MB-231 and MDA-MB-436 were all significantly suppressed, while apoptosis rates of them were obviously increased. In addition, eIF3m could regulate cell-cycle, epithelial–mesenchymal transition and apoptosis-related proteins. Combined with public databases and RT-qPCR, 14 genes were identified to be modulated by eIF3m in the development of TNBC.
Conclusions
eIF3m is an unfavorable indicator of TNBC, and plays a vital role in the process of TNBC tumorigenesis.
Similar content being viewed by others
Background
Breast cancer (BC), one of the most common diseases in women, has severely threatened human health and social development [1]. Pathologically, invasive breast carcinoma is characterized by at least four molecular subtypes: Luminal A, Luminal B, HER2-positive and triple negative breast cancer (TNBC) [2]. TNBC, accounting for 15–25% of all breast cancer cases, is characterized by the lack of expression of estrogen receptor (ER), progesterone receptor (PR) and c-erbB-2 (HER2) [2]. Despite well-established benefits of chemotherapy after surgery on tumor growth, TNBC still has the worst prognosis in all subtypes of BC in the result of no targeted therapy [2, 3]. Due to obscure pathogenesis of TNBC, rapid progress, recurrence and drug resistance also become obstacles in the process of anti-cancer treatment [4]. Therefore, it is urgent to understand genomics and explore mechanisms of carcinogenesis in order to improve the diagnosis and treatment of TNBC.
Protein synthesis has been considered as a critical step from gene expression to cell biological behavior [5]. Abnormal translations of oncogenes, tumor suppressors and eukaryotic translation initiation factors (EIFs) are essential for tumorigenesis [5]. Among eIFs, eukaryotic translation initiation factor 3 (eIF3) is the most complex eukaryotic translation initiation factor [6]. This complex composed of 13 subunits (eIF3a to m) is necessary for the initiation of protein synthesis [7]. Among these subunits, 5 core subunits (eIF3a, b, c, g and i) are conserved in all eukaryotes, while the rest only exist in mammal [8]. Recent studies have reported that several subunits are essential for the development of various tumors, including BC [9,10,11]. Notably, as the last defined subunit, eIF3m plays an indispensable role on maintaining the integrity of the eIF3 complex by stabilizing the core subunits [12]. Therefore, the ablation of eIF3m severely suppresses the translation and cancer cell proliferation [13]. In addition, eIF3m is a critical factor for cell cycle progression via regulating cell division cycle 25A (CDC25A) [13]. However, its function in BC or TNBC is not yet characterized.
In this research, eIF3 subunits were detected in breast tumors and corresponding adjacent normal breast tissues as well as public databases. After selection of suitable breast cancer cell lines with higher eIF3m expression, stable transfected cells with eIF3m knockdown were constructed for investigating the significance of eIF3m in vitro and exploring the potential pathogenesis of TNBC tumorigenesis. This article revealed possible roles of eIF3m in TNBC and provided a novel and reliable bio-marker for clinic.
Methods
Data mining of datasets
First, five cohorts selected from Oncomine (https://www.oncomine.org/) were used to analyze differences of eIF3m expression between breast tumors and normal breast tissues: “Curtis breast 2012” (1989 tumors, 3 benign breast neoplasms and 144 normal breast tissues), “Sorlie breast 2003” (92 tumors and 4 normal breast tissues), “Ma breast 2009” (9 tumors and 14 normal breast tissues), “Richardson breast 2006” (40 tumors and 7 normal breast tissues) and “TCGA breast 2012” (639 tumors and 111 normal breast tissues). To assess the prognostic role of eIF3m in BC, KM-plotter (http://kmplot.com/analysis/index.php?p=service&cancer=breast) and BCIP database (http://www.omicsnet.org/bcancer/database) were performed. The survival parameters included overall survival (OS), relapse-free survival (RFS) and post progression survival (PPS). BCIP database also showed co-expressed genes of eIF3m in the development of breast normal tissues and TNBC. In addition, a GEO database (GEO accession: GSE45827) consisting of four BC subtypes (29 cases of Luminal A, 30 cases of Luminal B, 30 cases of HER2-positive and 41 cases of TNBC) and breast normal-like tissues (11 cases) was conducted to investigate the expression of eIF3 subunits. Another two bioinformation databsaes, STRING (https://string-db.org/) and PDBsum entry (https://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html), were used for identifying the structure of eIF3 and the location of eIF3m. Furthermore, KEGG pathway (https://www.kegg.jp/) was used for exploring potential carcinogenic mechanisms of eIF3m.
Patients and tissue specimens
Specimens from a total of 184 female invasive breast cancer patients who underwent modified radical mastectomy, breast reservation radical correction or mastectomy (only for distant metastasis) in Kunshan First People’s Hospital (63 cases), Kunshan Hospital of Traditional Chinese Medicine (37 cases), Wuxi Xishan People’s Hospital (49 cases) and Wuxi Fifth People’s Hospital (35 cases) from January, 2016 to January, 2019 were collected, with a mean age of 53.47 ± 13.02 years. According to the pathological diagnosis, specimens were divided into four subtypes: 42 Luminal A, 62 Luminal B, 43 HER2-positive and 37 TNBC. Every case consisted of breast tumor and corresponding adjacent normal tissue. No one received any radiotherapy or chemotherapy before surgery. All cases had paraffin-embedded consecutive sections that were used for Immunohistochemistry (IHC). Among them, 158 samples including 32 TNBC and 126 Non-TNBC with corresponding adjacent normal tissues were available for reverse transcription-quantitative PCR (RT-qPCR), while we failed to extract total RNA from the rest. This research had received the approval of Kunshan First People’s Hospital Ethics Committee, Kunshan Hospital of Traditional Chinese Medicine Ethics Committee, Wuxi Xishan People’s Hospital Ethics Committee and Wuxi Fifth People’s Hospital Ethics Committee. Every patient signed the informed consent form.
Cell lines and cell culture
Five human breast cancer cell lines (SK-BR3, MCF-7, BT-474, MDA-MB-231 and MDA-MB-436) were obtained from FuDan IBS Cell Center, China. And the human mammary epithelial cell line (MCF-10A) was obtained from Shanghai Bioleaf Biotech Co.,Ltd, China. Cell lines were cultured in appropriate media supplemented with 10% fetal bovine serum (FBS, GIBCO Life Technologies, USA) and 1% antibiotic/antimycotic solution (Sigma-Aldrich, USA) at 37 °C in a humidified atmosphere containing 5% CO2: Dulbecco’s modified Eagle’s medium (DMEM; GIBCO Life Technologies, USA) for SK-BR3, MCF-7 and MCF-10A; RPMI-1640 (GIBCO Life Technologies, USA) for BT-474 and MDA-MB-231; and Leibovitz’s L-15 (GIBCO Life Technologies, USA) for MDA-MB-436.
IHC
Paraffin-embedded consecutive sections were subjected to IHC staining for the expression of eIF3m by the rabbit polyclonal anti-eIF3m antibody (bs-9033R, Beijing BIOSS, China) diluted at 1:100 in phosphate-buffered saline (PBS, GIBCO Life Technologies, USA), with a SP Rabbit & Mouse HRP Kit (CWBIO, China). PBS without primary antibodies was used as negative control. Two pathologists independently evaluated the scores of the staining intensity of eIF3m expression (x = 0, no staining of cells; 1, mild staining; 2, moderate staining; and 3, marked staining). Any disagreement was resolved by discussion. Scores of the percentage of cells were measured by ImageJ 1.52r (y = 0%–100%). The total score (immunoreactivity score, IRS = x×y × 100) ranged from 0 to 300.
Cell transfection
To knock down eIF3m expression, pLKO.1-eIF3m-shRNA-puro was used, and three shRNAs were designed as listed in Table 1. The titer of lentivirus was 1x108 TU/ml. MDA-MB-231 and MDA-MB-436 were cultured for 24 h in a 24-well plate, and then cells were transfected with shRNAs-Puro lentivirus (100 μl/ml) in a medium of 8 mg/ml polybrene (Sigma-Aldrich, USA) when a confluence of 30–50%. After 24 h, transfected cells were cultured in a new medium with 1.0 μg/ml puromycin (TargetMol, China) for about 12 days. To select the most efficient shRNA, the transfection efficiency of each shRNA was analyzed through western blotting and RT-qPCR. Cells transfected with this shRNA were named as “eIF3m-shRNA” groups, and cells transfected with empty vector were named as “Vector” groups.
RT-qPCR
Cells and tissues were collected to isolate total RNA through Trizol regent (Thermo Fisher Scientific, USA). A total of 2 µg RNA of each sample was reverse transcribed using the SuperScript II RNase-Reverse Transcriptase system (Thermo Fisher Scientific, USA). cDNA was subjected to quantitative PCR using primers specific for eIF3 subunits, co-expressed genes and GAPDH. PCR primers were designed as listed in Table 2 and Additional file 1: Table S1. The PCR cycling conditions were as follows: (1) 94 °C for 4 min; (2) 40 cycles of 95 °C for 1 min; (3) 60 °C for 1 min; and (4) 72 °C for 1 min. Amplifed DNA was measured by the SYBR Premix Ex Taq™ kit (Takara Bio, Japan), and qPCR was performed using an iQ5 real-time PCR detection system (Bio-Rad, USA). Then, we used 2−ΔΔCt value to calculate the relative expression. The formula of ΔΔCt could refer to the previous method [14].
Cell viability assay
To assess cell proliferation, 2000 stable transfected cells per well were seeded in 96-well plates for 1–5 days, and then were treated with CCK-8 (cell counting kit-8, Immunoway, USA) for 2 h at the incubator. The absorbance of each group was read at 450 nm of the microplate reader (OD value). Cell viability was calculated by the formula: Cell viability (%) = OD value of “eIF3m shRNA”/OD value of “Vector”.
Colony formation assay
Cells were seeded in 6-well plates at a density of 200 cells. They were cultured for about 10 days, until the clones were visible (≥ 50 cells). The colonies were fixed for 15 min with methanol and then stained with crystal violet for 15 min after airing. Finally, numbers of colonies were counted.
Cell cycle analysis
Centrifugated cells were washed 2 times with pre-cooling PBS. And then, they were fixed in 80% ethanol for about 12 h at 4 °C. Cells were washed 2 times again with pre-cooling PBS prior to incubation for 30 min at 37 °C in dark with propidium iodide and RNAse A (KeyGen Biotech, China). Fluorescence-activated cell sorting analysis was performed in the flow cytometer (Becton, Dickinson and Company, USA).
Wound healing assay
Transfected cells were planted into 6-well plates to complete confluence in order to assess the role of eIF3m on the migration. Cells were wounded by scratching with a sterile plastic 200 µL micropipette tip and the loose cells were removed by PBS. Subsequently, Opti-MEM medium (GIBCO Life Technologies, USA) was added to the wells for incubation at 37 °C with 5% CO2 for 24 h. Finally, images were captured by the inverted microscope (IX73, Olympus, Shanghai, China). The relative proportion of wound = the width of the wound/the initial width.
Transwell assays
8 mm transwell inserts (JETbiofil, China) were placed into 24-well plates, separating upper and lower chambers. The upper side of the membrane was precoated with Matrigel (Corning, USA) for gel formation. New medium was added to the lower chamber and 105 cells/well in Opti-MEM were added to the upper chamber. After 72 h at 37 °C, the number of invading cells was counted through a counting chamber under the inverted microscope.
Mitochondrial membrane potential assay
This assay could be divided into four parts:
-
1.
Microscopic observation: Transfected cells were centrifugated and add with JC-1 working solution (Beyotime Biotechnology, China) for cultivation at 37 °C for 20 min. After washing and centrifugation by JC-1 diluent (1 ×) for 2 times, the red and green fluorescence were detected by fluorescence microscope (Olympus Corporation, China).
-
2.
Flow cytometric analysis: Then, centrifugated cells that were treated by JC-1 were analyzed with the flow cytometer.
-
3.
Purified mitochondria: Mitochondrial protein and cytosol were separated by Minute™ Mitochondria Isolation Kit (Invent Biotechnologies, Inc., USA). JC-1 kit (Beyotime Biotechnology, China) was used to analyze the level of red fluorescence and green fluorescence in mitochondrial proteins through the microplate reader. The ratio of red to green fluorescence was used to measure the level of depolarization of mitochondria.
-
4.
Cyto-c releasing: Cyto-c (1:1000, ABclonal Technology, China) in mitochondria or cytosol was investigated by western blotting. In addition, COX-IV and Tubulin (1:1000, ABclonal Technology, China) were used as the reference protein of mitochondria and cytosol, respectively.
Cell apoptosis analysis
Centrifugated cells were washed 2 times with PBS and stained with PE and 7-AAD for 15 min at room temperature in dark (KeyGen Biotech, China). After diluted in PBS, stained cells were analyzed with the flow cytometer.
Western blot analysis
Proteins of tissues or cells were extracted by RIPA extraction buffer with PMSF (Beyotime Biotechnology, China). 20 µL per sample were loaded in pre-cast gels (Thermo Fisher Scientific, USA) for electrophoresis. And then, proteins were transferred into PVDF membranes (Beyotime Biotechnology, China) for exposure. eIF3m, CyclinD1, CyclinE, CDK2, CDK4, CDK6, Bax, Bcl-2, Bad, E-cadherin, N-cadherin, Vimentin, Snail, Twist (1:1000, Beijing BIOSS, China), P21, P27 (1:500, ImmunoWay Biotechnology, USA), Caspase3 (1:1000, Abcam, USA), Cleaved-Caspase3 (1:500, Abcam, USA), Caspase9 (1:1000, Abcam, USA) and Cleaved-Caspase9 (1:300, Abcam, USA) were primary antibodies, while GAPDH (1:1000, Cell Signaling Technology, USA) was used as the reference protein. The gray value of each protein was investigated by ImageJ 1.52r. The relative level of each protein was deduced from the ratio of the mean value of each band to that of GAPDH.
Statistical analysis
All experiments were done in triplicates. Continuous variables were expressed as the‾x ± SD and experiments were performed in triplicate. All results were analyzed by t test through SPSS 20.0 software with a p < 0.05 considered statistically significant. The differences of eIF3 subuntis expression between breast tumors and matched normal tissues were analyzed by a paired-t test, while the differences of biological behaviors between “Vector” group and “eIF3m-shRNA” group were analyzed by an unpaired-t test. An ANOVA followed by Tukey’s multiple comparisons test was used to compare eIF3m expression in cell lines. Kaplan–Meier analysis and the log rank test were also conducted to analyze survival data of public databases. All graphs were generated by GraphPad Prism 6.0.
Results
Abnormal up-regulation of eIF3m had worse prognosis in BC in multiple databases
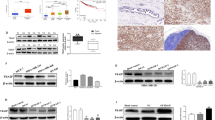
Oncomine analysis of tumor versus. normal samples revealed that eIF3m was significantly elevated in invasive breast carcinoma, such as invasive ductal breast carcinoma, mixed lobular and ductal breast carcinoma, invasive lobular breast carcinoma, and so on (Table 3). In addition, there were no substantial differences of eIF3m between normal breast tissues and benign breast neoplasm (p > 0.05, Table 3). KM-plotter and BCIP database revealed that high eIF3m was correlated with a poorer prognosis, including OS, RFS and PPS in BC (Fig. 1).
eIF3m became a unique and valuable indicator for TNBC
To analyze differences of eIF3m expression among different types of breast cancer, one GEO database (GEO accession: GSE45827) showing all subunits expression in TNBC, non-TNBC and normal-like tissues was performed (Fig. 2a). In Table 4, expression levels of eIF3b, d, f, g, i, j, k, l and m in breast tumors were significantly higher than in normal-like tissues (p < 0.05). Among these subunits, there were no differences of eIF3b and k expression between TNBC and normal-like tissues, or between TNBC and non-TNBC (p > 0.05). Only eIF3m expression in TNBC was obviously higher than in non-TNBC tissues (p = 0.000253). In addition, similar results were found in 158 BC cases via RT-qPCR (Fig. 2b–d). These results reflected that eIF3 was a vital protein molecule in BC, and eIF3m was involved in the occurrence and development of TNBC.
Expression of eIF3 subunits in different subtypes of breast cancer and breast normal tissues. a GEO database (GEO accession: GSE45827); b Comparison of 13 subunits expression between breast tumors and corresponding adjacent normal tissues by RT-qPCR (n = 158); c Comparison of subunits expression between TNBC and corresponding adjacent normal tissues by RT-qPCR (n = 32); d Comparison of subunits expression between TNBC (n = 36) and non-TNBC (n = 126) by RT-qPCR. TNBC: triple negative breast cancer. *p < 0.05, **p < 0.01, ***p < 0.001
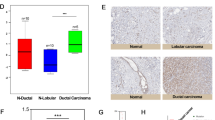
Expression of eIF3m was detected in the cytoplasm through IHC (Fig. 3). There were no differences of eIF3m expression between tumors and corresponding adjacent normal breast tissues in non-TNBC. But, its expression levels in TNBC were significantly higher compared with adjacent normal tissues (p < 0.001). In addition, expression of eIF3m in TNBC was also higher than non-TNBC (p < 0.001).
eIF3m expression in breast tumors and normal tissues via immunohistochemistry. a, c, e. eIF3m expression in poor (a) and well-moderate (c) differentiation of TNBC and normal tissues (e) were investigated at × 200 magnification; b, d, f ImageJ analysis of eIF3m expression (× 200); g Analysis of eIF3m expression in different subtypes of breast tumor and normal tissues, as well as in differentiation, lymph node invasion and distant metastasis. *p < 0.05, **p < 0.01, ***p < 0.001
Poor differentiation group of TNBC had a higher level of eIF3m in comparison of well-moderate differentiation group (p < 0.01, Fig. 3g). Cases with lymph node metastasis positive or distant metastasis positive group also had a higher level of eIF3m compared with metastasis negative group (p < 0.05, p < 0.01, Fig. 3g).
eIF3m expression in breast cell and breast cancer cell lines
Among five BC cell lines and MCF-10A, MDA-MB-231 and MDA-MB-436, two TNBC cell lines, showed highest expression of eIF3m (Fig. 4a, b). Therefore, these two cell lines were chosen to be transfected with lentivirus. The transfection efficiency was accessed by western blotting (Fig. 4c) and RT-qPCR (Fig. 4d). Among shRNAs, shRNA2# had the lowest eIF3m expression. Thus, we used shRNA2# for the next assays.
eIF3m expression in cell lines. a, b Expression of eIF3m protein (a) and mRNA (b) in breast cancer cells and MCF-10A was investigated via western blotting and RT-qPCR, respectively; c, d Regulation of eIF3m protein (c) and mRNA (d) in transfected cells (MDA-MB-231 and MDA-MB-436) was analyzed through western blotting and RT-qPCR, respectively. *p < 0.05, **p < 0.01, ***p < 0.001
Knocking down eIF3m inhibited cell proliferation in TNBC cells
As shown in Fig. 5a, reduced expression of eIF3m obviously suppressed cell viability (p < 0.05). The number of colonies formed on 6-well plates in “eIF3m-shRNA” groups was significantly fewer than in “Vector” groups (p < 0.01, Fig. 5b). In addition, cells transfected with eIF3m-shRNA obtained a higher proportion at G1 phase (Fig. 5c). At the same time, expression of CyclinD1, CyclinE, CDK2, CDK4 and CDK6 decreased obviously in “eIF3m-shRNA” groups in comparison of “Vector” groups; but P21 and P27 in “eIF3m-shRNA” groups were up-regulated (Fig. 5d). These results confirmed that knockdown of eIF3m suppressed proliferation of TNBC cells.
Influence of eIF3m knockdown on cell proliferation in TNBC. a The viability of transfected cells was determined by CCK-8 assay at 450 nm OD value of the microplate reader; b The effects of eIF3m on cell growth were confirmed by colony formation assay; c Cell cycle distributions were detected by flow cytometric analysis in transfected cells; d Cell cycle-related proteins of transfected cells were investigated via western blotting. *p < 0.05, **p < 0.01, ***p < 0.001
Knocking down eIF3m suppressed migration and invasion in TNBC cells
By knocking down eIF3m, the width of scratch area after 24 h was obviously longer compared with “Vector”, (Fig. 6a). Also, the transwell assay showed fewer cells going through chambers when decreasing expression of eIF3m in TNBC (Fig. 6b). Knockdown of eIF3m debased the levels of N-cadherin, Vimentin, Snail and Twist, while it elevated E-cadherin expression (Fig. 6c). Thus, low expression of eIF3m made a sharp decline in the abilities of migration and invasion.
Influence of eIF3m knockdown on migration and invasion in TNBC. a Wound healing assay measured relative proportion rates at 24 h for comparing migration abilities of different cells (× 40); b Transwell assay detected the number of cells passing through chambers in each group by image analysis (× 100); c Epithelial–mesenchymal transition related proteins of transfected cells were investigated via western blotting. *p < 0.05, **p < 0.01, ***p < 0.001
Knocking down eIF3m promoted apoptosis in TNBC cells
The brightness of JC-1 red in MDA-MB-231 and MDA-MB-436 was reduced by using eIF3m-shRNA, while JC-1 green’s brightness was increased obviously (Fig. 7a).The ratios of red/green fluorescence were also significantly decreased (Fig. 7b, c). Through extraction of mitochondrial proteins and cytosol, knockdown of eIF3m down-regulated Cyto-C obviously in mitochondria, but elevated its expression in cytosol (Fig. 7d). In addition, rates of cell apoptosis in “eIF3m-shRNA” groups were increased along with the regulation of Bcl-2 family and the activation of Caspase-3 and Caspase-9 (Fig. 7e, f). These results revealed that knockdown of eIF3m elevated apoptosis rates through releasing Cyto-C from mitochondria into cytosol and activating Caspases.
Influence of eIF3m knockdown on mitochondrial membrane potential and cell apoptosis in TNBC. a JC-1 green and JC-1 red of cells were observed in the fluorescence microscope (× 400); b JC-1 green and JC-1 red distributions were detected by flow cytometric analysis; c ratios of red/green fluorescence in mitochondria were measured through the microplate reader; d Blots and relative protein expression of Cyto-c expression in mitochondria and cytosol of stably transfected cells; e Cell apoptotic rates were detected by flow cytometry; f Apoptosis-related proteins of transfected cells were investigated via western blotting, Cle-Cas3: Cleaved-Caspase-3; Cas3: Caspase-3; Cle-Cas9: Cleaved-Caspase-9; Cas9: Caspase-9. *p < 0.05, **p < 0.01, ***p < 0.001
Functions of eIF3m on the development of breast and TNBC analyzed by databases
PDBsum entry showed the structure of eIF3, and eIF3m was a key subunit maintaining the integrity of the eIF3 complex by stabilizing other subunits (Fig. 8a) [15, 16]. KEGG pathways displayed the vital role of eIF3 on RNA transport in which eIF3 and other eukaryotic translation initiation factors must be combined with the ribosome and mRNAs in order to complete the translation (Fig. 8b). In addition, eIF3m was involved in the development of mammary epithelium and gland (Fig. 8c). During the development of TNBC, eIF3m became an essential factor of the activation of various genes (Fig. 8c). Through RT-qPCR, 14 of top 20 co-expressed genes, most of which were oncogenes, including CSTF3, DPH4, CAPRIN1, PDHX, C11orf46, ELP4, COMMD9, API5, TRIM44, TRAF6, CCDC34, NUP160, TCP11L1 and CCNB3, had lower expression in “eIF3m-shRNA” groups than in “Vector” groups (Fig. 8d and Table 5). Therefore, the carcinogenic mechanism of eIF3m was displayed in Fig. 9.
Functions of eIF3m on the development of breast and TNBC in public databases. a PDBsum entry displayed the structure of eIF3; b Schematic model of the role of eIF3 on RNA transport analyzed by KEGG pathways; c BCIP database showed co-expressed genes of eIF3m in the development of breast and TNBC; d RT-qPCR investigated co-expressed genes in different groups. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
It is well known that breast tumor occurs in the process of continuous and cumulative improper regulation of gene expression. This abnormal regulation is caused by various exterior and internal carcinogenic factors. In other words, molecular mechanisms of the BC tumorigenesis are not thoroughly understood.
EIFs are involved in translation and protein synthesis in order to meet the demand of successful embryonic development and maintenance of normal tissue homeostasis [37]. Among them, several proteins or subunits of EIFs are essential for tumorigenesis [5]. Recently, oncologists have focused on the biggest eukaryotic translation initiation factor, eIF3, which is constituted by 13 subunits. This molecule participates in the 43S pre-initiation complex formation and facilitates recruiting GTP-eIF2-tRNA-methionine ternary complex [38]. The complex stimulates mRNA binding with 43S pre-initiation complex, promotes the process of translation and influences cell biological behaviors and functions [39]. Not only each subunit plays an important role in the synthesis and metabolic activities, but also several ones are implicated in carcinogenesis. As the largest subunit, eIF3a has vital biological functions in various carcinomas, including BC [40]. Other core subunits, eIF3c, g and i are also associated with BC tumorigenesis, especially in drug resistance [11, 41, 42]. In addition, several “non-core” subunits (eIF3b, d, e and f) correlated strongly with the tumorigenesis of breast neoplasm [43,44,45,46].
Based on public databases, a unique advantage of eIF3m could be found in TNBC. Among 13 subunits, only eIF3m met the three points: 1. a higher level in breast tumor than in normal tissue; 2. a higher level in TNBC than normal-like tissues; and 3. a higher level in TNBC than in non-TNBC. These results revealed that eIF3m might become a critical molecule in the development and progression of BC, especially in TNBC. One previous study demonstrated that eIF3m was required for cell-cycle and cell proliferation [14]. Silencing eIF3m almost completely turned a caspase substrate, PARP1, into activated form, which led to promoting cell death of HCT-116 [13]. In this research, we verified functions of eIF3m on cell-cycle and cell apoptosis of TNBC. Through regulation of CyclinD1 and other cell-cycle proteins, knockdown of eIF3m arrested cell-cycle at G1 phase and inhibited cell proliferation. Decreased eIF3m also caused a sharp decline of mitochondrial membrane potential, followed by release of Cyto-C from mitochondria to cytosol. As a result, instable mitochondrial membrane activated Caspases and then induced cell apoptosis. In addition, knocking down eIF3m suppressed migration and invasion of TNBC through regulation of epithelial–mesenchymal transition (EMT) related proteins. EMT is the process of epithelial cells transformed into mesenchymal cells. It elevates motility, migration and invasion of cancer cells, leading to metastasis of tumors, especially in TNBC [47]. Thus, we argue that over-expressed eIF3m promotes proliferation and activate EMT in TNBC.
Results of IHC uncovered that high eIF3m was involved in poor differentiation, lymph node metastasis and distant metastasis of TNBC. In addition, high expression of eIF3m had a poorer prognosis of BC patients both in two public databases, KM-plotter and BCIP database. Due to no enough survival data of this cohort, further follow-ups and researches will be performed to analyze the prognostic role of eIF3m in TNBC patients. Thus, we demonstrate that eIF3m is a novel and reliable bio-marker of prognosis and clinicopathology in BC, especially in TNBC.
eIF3m maintained the integrity of the eIF3 complex in order to complete the process of translation which was essential for the development of normal breast tissues and tumors. Compared with other subtypes and normal tissues, TNBC had the highest expression level of eIF3m. This might be caused by complex genetic imbalance involving activation of oncogenes and inhibition of tumor suppressors. This imbalance was influenced by various unfavorable factors from daily life, society and environment [48]. When eIF3m was up-regulated, more oncogenes were expressed and vital signaling pathways related to tumor growth and invasion were activated. As a result, eIF3m expression was elevated again, and then improve oncogenes and pathways one more time. This complex cycle accelerated the carcinogenesis of TNBC. As for specific reasons or factors that led to eIF3m up-regulation, we still need to perform further investigations and explain potential molecular mechanisms in the near future. Due to no animal experiment temporally as a result of the limitation of our funding, the next step will include in vivo assays to identify its molecular mechanisms and roles in regulating expression of oncogenes. In addition, our research discovered a discrepancy that mRNA expression of different oncogenes was not showing the similar kind of response in these two TNBC cells. This difference might be as a result of the specificities of different cell lines or experimental errors. Therefore, further researches should choose the oncogenes with the similar response to investigate their interaction with eIF3m.
Conclusion
eIF3m was an indispensable subunit of eIF3 and played a vital role in the process of TNBC tumorigenesis. High expression of eIF3m predicted poor prognosis in BC patients. Further researches should be conducted to reveal its possible molecular mechanisms and explore eIF3m-related oncogenes, in order to provide clues for confirming potential therapeutic targets in TNBC.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- TNBC:
-
Triple negative breast cancer
- BC:
-
Breast cancer
- eIF3m:
-
Eukaryotic translation initiation factor 3m
- ER:
-
Estrogen receptor
- PR:
-
progesterone receptor
- HER2:
-
c-erbB-2
- EIFs:
-
Eukaryotic translation initiation factors
- eIF3:
-
Eukaryotic translation initiation factor 3
- CDC25A:
-
Cell division cycle 25A
- OS:
-
Overall survival
- RFS:
-
Relapse-free survival
- PPS:
-
Post progression survival
- IHC:
-
Immunohistochemistry
- IRS:
-
Immunoreactivity score
- RT-qPCR:
-
Reverse transcription-quantitative PCR
- EMT:
-
Epithelial–mesenchymal transition
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
He MY, Rancoule C, Rehailia-Blanchard A, et al. Radiotherapy in triple-negative breast cancer: current situation and upcoming strategies. Crit Rev Oncol Hematol. 2018;131:96–101.
Odle TG. Precision medicine in breast cancer. Radiol Technol. 2017;88(4):401M–21M.
Wang WJ, Lei YY, Mei JH, Wang CL. Recent progress in HER2 associated breast cancer. Asian Pac J Cancer Prev. 2015;16(7):2591–600.
Franzmann TM, Alberti S. Protein phase separation as a stress survival strategy. Cold Spring Harb Perspect Biol. 2019;11(6):a034058.
Sadato D, Ono T, Gotoh-Saito S, et al. Eukaryotic translation initiation factor 3 (eIF3) subunit e is essential for embryonic development and cell proliferation. FEBS Open Bio. 2018;8(8):1188–201.
Raabe K, Honys D, Michailidis C. The role of eukaryotic initiation factor 3 in plant translation regulation. Plant Physiol Biochem. 2019;145:75–83.
Cate JH. Human eIF3: from ‘blobology’ to biological insight. Philos Trans R Soc Lond B Biol Sci. 2017;372(1716):176.
Yang C, Zhang Y, Du W, Cheng H, Li C. Eukaryotic translation initiation factor 3 subunit G promotes human colorectal cancer. Am J Transl Res. 2019;11(2):612–23.
Wang X, Wang H, Zhao S, et al. Eukaryotic translation initiation factor EIF3H potentiates gastric carcinoma cell proliferation. Tissue Cell. 2018;53:23–9.
Zhao W, Li X, Wang J, et al. Decreasing eukaryotic initiation factor 3C (EIF3C) suppresses proliferation and stimulates apoptosis in breast cancer cell lines through mammalian target of rapamycin (mTOR) pathway. Med Sci Monit. 2017;23:4182–91.
Zeng L, Wan Y, Li D, et al. The m subunit of murine translation initiation factor eIF3 maintains the integrity of the eIF3 complex and is required for embryonic development, homeostasis, and organ size control. J Biol Chem. 2013;288(42):30087–93.
Goh SH, Hong SH, Hong SH, et al. eIF3m expression influences the regulation of tumorigenesis-related genes in human colon cancer. Oncogene. 2011;30(4):398–409.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Erzberger JP, Stengel F, Pellarin R, et al. Molecular architecture of the 40S eIF1 eIF3 translation initiation complex. Cell. 2014;158(5):1123–35.
des Georges A, Dhote V, Kuhn L, et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525(7570):491–5.
Dufresne J, Bowden P, Thavarajah T, et al. The plasma peptides of ovarian cancer. Clin Proteomics. 2018;15:41.
Xiong Y, Deng Y, Wang K, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: a study based on large-scale sequencing data. EBioMedicine. 2018;36:183–95.
Stahl S, Silva MateusSeidl AR, Ducret A, et al. Loss of diphthamide pre-activates NF-κB and death receptor pathways and renders MCF7 cells hypersensitive to tumor necrosis factor. Proc Natl Acad Sci U S A. 2015;112(34):10732–7.
Hong SH, Lee WJ, Kim YD, et al. APIP, an ERBB3-binding partner, stimulates erbB2-3 heterodimer formation to promote tumorigenesis. Oncotarget. 2016;7(16):21601–17.
Gong B, Hu H, Chen J, et al. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67(7):629–36.
Chen B, Liu Y, Jin X, et al. MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer. 2014;14:443.
Wu J, Zhu H, Wu J, et al. Inhibition of N-acetyltransferase 10 using remodelin attenuates doxorubicin resistance by reversing the epithelial-mesenchymal transition in breast cancer. Am J Transl Res. 2018;10(1):256–64.
Peter CJ, Saito A, Hasegawa Y, et al. In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene. Nat Commun. 2019;10(1):4112.
Voisin S, Almén MS, Zheleznyakova GY, et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103.
Xu Y, Zhou W, Ji Y, et al. Elongator promotes the migration and invasion of hepatocellular carcinoma cell by the phosphorylation of AKT. Int J Biol Sci. 2018;14(5):518–30.
Isogai T, van der Kammen R, Goerdayal SS, et al. Proteomic analyses uncover a new function and mode of action for mouse homolog of Diaphanous 2 (mDia2). Mol Cell Proteomics. 2015;14(4):1064–78.
Zhan W, Wang W, Han T, et al. COMMD9 promotes TFDP1/E2F1 transcriptional activity via interaction with TFDP1 in non-small cell lung cancer. Cell Signal. 2017;30:59–66.
Basset C, Bonnet-Magnaval F, Navarro MG, et al. Api5 a new cofactor of estrogen receptor alpha involved in breast cancer outcome. Oncotarget. 2017;8(32):52511–26.
Zhou Z, Liu Y, Ma M, Chang L. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:98–103.
Yu J, Qin B, Moyer AM, et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest. 2018;128(6):2376–88.
Geng W, Liang W, Fan Y, Ye Z, Zhang L. Overexpression of CCDC34 in colorectal cancer and its involvement in tumor growth, apoptosis and invasion. Mol Med Rep. 2018;17(1):465–73.
Takahara M, Kunii M, Nakamura K, et al. C11ORF74 interacts with the IFT-A complex and participates in ciliary BBSome localization. J Biochem. 2019;165(3):257–67.
Shimozono N, Jinnin M, Masuzawa M, et al. NUP160-SLC43A3 is a novel recurrent fusion oncogene in angiosarcoma. Cancer Res. 2015;75(21):4458–65.
Seabra CM, Quental S, Neto AP, et al. A novel Alu-mediated microdeletion at 11p13 removes WT1 in a patient with cryptorchidism and azoospermia. Reprod Biomed Online. 2014;29(3):388–91.
Astolfi A, Fiore M, Melchionda F, Indio V, Bertuccio SN, Pession A. BCOR involvement in cancer. Epigenomics. 2019;11(7):835–55.
Tahmasebi S, Amiri M, Sonenberg N. Translational control in stem cells. Front Genet. 2019;9:709.
Dong Z, Zhang JT. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit Rev Oncol Hematol. 2006;59(3):169–80.
Yin JY, Dong Z, Liu ZQ, Zhang JT. Translational control gone awry: a new mechanism of tumorigenesis and novel targets of cancer treatments. Biosci Rep. 2011;31(1):1–15.
Devanand P, Sundaramoorthy S, Ryu MS, Jayabalan AK, Ohn T, Lim IK. Translational downregulation of Twist1 expression by antiproliferative gene, B-cell translocation gene 2, in the triple negative breast cancer cells. Cell Death Dis. 2019;10(6):410.
Zheng Q, Liu H, Ye J, et al. Nuclear distribution of eIF3g and its interacting nuclear proteins in breast cancer cells. Mol Med Rep. 2016;13(4):2973–80.
Fang HY, Chen SB, Guo DJ, Pan SY, Yu ZL. Proteomic identification of differentially expressed proteins in curcumin-treated MCF-7 cells. Phytomedicine. 2011;18(8–9):697–703.
Ma F, Li X, Ren J, et al. Downregulation of eukaryotic translation initiation factor 3b inhibited proliferation and metastasis of gastric cancer. Cell Death Dis. 2019;10(9):623.
Fan Y, Guo Y. Knockdown of eIF3D inhibits breast cancer cell proliferation and invasion through suppressing the Wnt/β-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8(9):10420–7.
Grzmil M, Rzymski T, Milani M, et al. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene. 2010;29(28):4080–9.
Cuesta R, Berman AY, Alayev A, et al. Estrogen receptor α promotes protein synthesis by fine-tuning the expression of the eukaryotic translation initiation factor 3 subunit f (eIF3f). J Biol Chem. 2019;294(7):2267–78.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8.
Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017;396:130–7.
Acknowledgements
None.
Funding
This study was supported by the Foundation from Jiangsu University Science and Technology Program of Clinical Medicine (major project, Grant No. JLY20180013), Wuxi Youth Projection of Health and family planning commission (No. Q201756), Wuxi Scientific and Technological Development of Medical and Health Guidance Programs (No. NZ2019014) and Kunshan Major Project of Social Research and Development (No. KS19038).
Author information
Authors and Affiliations
Contributions
HNW, HZD and WH conceived and designed the present study. HNW, WH, CTS, QXS and CJW developed the methodology. WH, CZ, CTS, XJG, MHZ, XJS, FC, YWH, JLY and QHW completed the experiment and analyzed the data. WH, HNW and CZ draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Specimen collection and analysis processes were approved by Kunshan First People’s Hospital Ethics Committee, Kunshan Hospital of Traditional Chinese Medicine Ethics Committee, Wuxi Xishan People’s Hospital Ethics Committee and Wuxi Fifth People’s Hospital Ethics Committee. Every patient signed the informed consent form.
Consent for publication
The authors consent for publication.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1
. The primer design of co-expressed genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Han, W., Zhang, C., Shi, Ct. et al. Roles of eIF3m in the tumorigenesis of triple negative breast cancer. Cancer Cell Int 20, 141 (2020). https://doi.org/10.1186/s12935-020-01220-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-01220-z