Abstract
Background
L-phenylalanine is an essential amino acid with various promising applications. The microbial pathway for L-phenylalanine synthesis from glucose in wild strains involves lengthy steps and stringent feedback regulation that limits the production yield. It is attractive to find other candidates, which could be used to establish a succinct and cost-effective pathway for L-phenylalanine production. Here, we developed an artificial bioconversion process to synthesize L-phenylalanine from inexpensive aromatic precursors (benzaldehyde or benzyl alcohol). In particular, this work opens the possibility of L-phenylalanine production from benzyl alcohol in a cofactor self-sufficient system without any addition of reductant.
Results
The engineered L-phenylalanine biosynthesis pathway comprises two modules: in the first module, aromatic precursors and glycine were converted into phenylpyruvate, the key precursor for L-phenylalanine. The highly active enzyme combination was natural threonine aldolase LtaEP.p and threonine dehydratase A8HB.t, which could produce phenylpyruvate in a titer of 4.3 g/L. Overexpression of gene ridA could further increase phenylpyruvate production by 16.3%, reaching up to 5 g/L. The second module catalyzed phenylpyruvate to L-phenylalanine, and the conversion rate of phenylpyruvate was up to 93% by co-expressing PheDH and FDHV120S. Then, the engineered E. coli containing these two modules could produce L-phenylalanine from benzaldehyde with a conversion rate of 69%. Finally, we expanded the aromatic precursors to produce L-phenylalanine from benzyl alcohol, and firstly constructed the cofactor self-sufficient biosynthetic pathway to synthesize L-phenylalanine without any additional reductant such as formate.
Conclusion
Systematical bioconversion processes have been designed and constructed, which could provide a potential bio-based strategy for the production of high-value L-phenylalanine from low-cost starting materials aromatic precursors.
Similar content being viewed by others
Background
L-phenylalanine is a valuable amino acid with multiple industrial applications [1]. It is extensively used in dietary supplements, feed, cosmetics, and chemical industries [2,3,4]. In addition, L-phenylalanine is widely used in the synthesis of pharmaceutically active compounds, such as cephalosporin antibiotics, anticancer metallodrugs, and HIV protease inhibitors [5,6,7], resulting in a worldwide steadily increasing demand for L-phenylalanine.
Given the widespread applications and growing demands of L-phenylalanine, various strategies have been proposed for the production of L-phenylalanine. L-phenylalanine is primarily produced through chemical, microbial, or enzymatic processes [8]. Traditional chemical synthesis methods rely on expensive transition metals as catalysts and result in the accumulation of toxic by-products [9]. The microbial processes generate less environmental pollution than chemical synthesis [10,11,12]. Production of L-phenylalanine from glucose with relatively high titers in E. coli has been achieved in previous reports [7, 13,14,15]. However, the typical biosynthesis pathway in E. coli (Fig. 1 left) involves lengthy reaction steps (more than 15 steps) and tightly complex feedback regulation, which limits the practical yield of L-phenylalanine production [16,17,18,19]. Therefore, there is a need to find alternative substrates and establish a succinct, cost-effective pathway for L-phenylalanine production.
The aromatic chemicals (benzaldehyde and benzyl alcohol) are attractive candidates for L-phenylalanine production. These two chemicals are inexpensive with a price of ~$2/kg (benzyl alcohol) and ~$2.5/kg (benzaldehyde), respectively [20, 21]. The aromatic ring could provide the bulky carbon backbone for L-phenylalanine [22, 23]. In addition, the theoretical yield of aromatic precursor to L-phenylalanine is 100 mol%. Some researchers have reported that using threonine aldolases (TAs) and threonine deaminases (TDs) combination could catalyze the conversion of aldehydes and small amino acids into keto acids [24,25,26]. The conversion of phenylpyruvate to L-phenylalanine by the L-amino acid dehydrogenase or aminotransferase system has been investigated by previous works [27, 28]. One report demonstrated that benzaldehyde could be used to synthesize L-phenylalanine by multi-enzyme-coupled reactions [24]. However, the production of L-phenylalanine with benzyl alcohol as the precursor has not been published to date.
Compared with benzaldehyde, benzyl alcohol is less toxic to cells and has less effect on enzyme activity and cell growth, making the process more feasible [29]. In addition, Benzyl alcohol is more stable than benzaldehyde, which is readily oxidized to benzoic acid on exposure to air at room temperature [30]. Moreover, benzyl alcohol has better solubility than benzaldehyde, which can avoid the use of cosolvent such as DMSO, and reduce production costs. Benzyl alcohol dehydrogenase from Pseudomonas putida (XylBP.p) has been reported to be highly active in converting benzyl alcohol to benzaldehyde, while also providing cofactor NADH for L-phenylalanine production by reducing NAD+ [31].
Here, the work aimed to develop the biosynthetic processes of L-phenylalanine from inexpensive and readily available aromatic precursors (as shown in Fig. 1 right). We first enable the key intermediate phenylpyruvate production by screening and co-expressing the enzymes threonine aldolase LtaEP.p and L-phenylalanine dehydratase A8HB.t. Introducing the enamine deaminase RidA could further improve the titer of phenylpyruvate by 16.3%, reaching up to 5 g/L. The conversion rate of phenylpyruvate to L-phenylalanine was 93% by a recombinant redox cycle including phenylalanine dehydrogenase (PheDH) and formate dehydrogenase (FDHV120S). Synthesis of L-phenylalanine from benzaldehyde was performed, resulting in an L-phenylalanine titer of 1.7 g/L and a benzaldehyde conversion rate of 69%. Moreover, we constructed a cofactor self-sufficient pathway for L-phenylalanine production from benzyl alcohol, a process that NADH/NAD+ in different redox states are interconverted via the enzymes pair XylBP.p and PheDH without another regenerating enzyme, and does not require the reductant formate, which results in a cleaner bioconversion system. In summary, this work describes a succinct and feasible biosynthesis of L-phenylalanine from inexpensive aromatic precursors (benzyl alcohol or benzaldehyde).
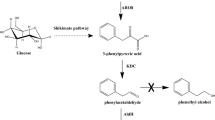
Design the artificial biosynthetic pathways for L-phenylalanine production from aromatic precursors (benzaldehyde or benzyl alcohol) and glycine. Gray arrows indicated the natural biosynthesis pathway of L-phenylalanine in E. coli. The green and red arrows demonstrated the novel pathway. ADH alcohol dehydrogenase, LTA threonine aldolase, LTD threonine dehydratase, PheDH phenylalanine dehydrogenase, FDH formate dehydrogenase. The dotted gray arrows  indicate the repression and inhibition of the relevant genes in native pathway. Metabolites abbreviations: G6P glucose 6-phosphate, DAHP 3-deoxy-d-arabino- heptulosonate-7-phosphate, L-Glu L-glutamate, 2-KG 2-ketoglutarate
indicate the repression and inhibition of the relevant genes in native pathway. Metabolites abbreviations: G6P glucose 6-phosphate, DAHP 3-deoxy-d-arabino- heptulosonate-7-phosphate, L-Glu L-glutamate, 2-KG 2-ketoglutarate
Results and discussion
Identification of enzymes to produce the key intermediate phenylpyruvate
The validation of this L-phenylalanine pathway started with building a well-behaved chassis for phenylpyruvate production, which is the key precursor for L-phenylalanine production. We used benzaldehyde and glycine as substrates to screen natural high-active enzymes threonine aldolase (LTA) and threonine dehydratase (LTD) (Fig. 2a). To convert benzaldehyde and glycine to phenylserine, we screened three LTAs from Pseudomonas putida (LtaEP.p), E. coli (LtaEE.c), and Caulobacter crescentus CB15 (LtaEC.c), and cloned them into plasmids pPLA-1, pPLA-2, and pPLA-3, respectively (Table 1). These plasmids also carried a threonine/serine dehydratase from Burkholderia thailandensis (A8HB.t), for screening based on phenylpyruvate production, and were transformed into wild-type BW25113 individually, to obtain strains M1, M2, and M3 (Table 1). We used 4.2 g/L (40mM) benzaldehyde and 20 g/L glycine as cosubstrate, as shown in Fig. 2b, the strains expressing LtaEP.p, LtaEE.c, and LtaEC.c produced phenylpyruvate at a titer of 4.3 g/L (26 mM), 3.5 g/L (21 mM), and 2.7 g/L (16.4 mM) within 24 h, respectively (column #1, #2, #3 in Fig. 2b). Notably, all of these enzymes are promiscuous enough to catalyze the bioconversion of benzaldehyde and glycine into phenylpyruvate. These results indicated that LtaEP.p, among the three investigated enzymes, was the best natural LTA for benzaldehyde conversion.
In addition to LTAs, we also investigated other LTDs for α,β-elimination to see which combination would produce the maximal titer of phenylpyruvate. We cloned genes coding L-threonine dehydratase from Burkholderia ambifaria (ilvAB.a) and ammonia-lyase from Burkholderia thailandensis (TAAB.t) individually after the gene encoding LtaEP.p to build an expression cassette on a high-copy plasmid, named pPLA-4, and pPLA-5, respectively. The strains transformed with pPLA-4 and pPLA-5 (Table 1, strains M4 and M5) could produce 3.4 g/L (21 mM) and 0.3 g/L (1.8 mM) phenylpyruvate within 24 h, respectively (column #2, #3 in Fig. 2c). Overall, the highly active natural enzyme combination was LtaEP.p and A8HB.t for the phenylpyruvate production from benzaldehyde and glycine, resulting in a benzaldehyde conversion rate of 65%. In a previous report, the conversion of benzaldehyde and glycine to phenylpyruvate was improved from 23% (wide-type threonine deaminase) to 88% (mutated by rational protein engineering) [24]. In this respect, further directed evolution in A8HB.t could significantly enhance the production of phenylpyruvate in our future work.
Phenylpyruvate production using different combinations of threonine aldolase (LTA) and threonine dehydratase (LTD). a Phenylpyruvate production pathway from benzaldehyde and glycine catalyzed by LTA and LTD. b The effect of different threonine aldolases (LtaEP.p, LtaEE.c, LtaEC.c) on phenylpyruvate production. c The effect of different L-phenylalanine dehydratase (A8HB.t, IlvAB.a, TAAB.t) on phenylpyruvate production. Error bars are the standard deviation for three independent experiments
Effect of temperature and pH on phenylpyruvate production
The reaction conditions of the enzyme cascade catalytic system are important for production performance. To achieve a good conversion of reactants, the temperature and pH of the reaction system need to be optimized [32]. Preliminary analysis of the plasmid pPLA-1 showed the highest activity toward phenylpyruvate production, and we used strain M1 to analyze the effects of different reaction conditions on phenylpyruvate production. As shown in Fig. 3a, the activity of this enzyme cascade was optimum at 30 ℃. When the assay temperature was set at 20 ℃, 25 ℃, 35 ℃ and 40 ℃, the phenylpyruvate titer was reduced to 2.84 g/L (17 mM), 4 g/L (24.1 mM), 2.5 g/L (15.2 mM) and 2.1 g/L (12.8 mM), respectively, about 66%, 93%, 58% and 48% of that value at 30 ℃. Under different pHs in the reaction mixture (Fig. 3b), enzyme activity of LtaEP.p and A8HB.t combination was optimum at pH 8.0, with 4.3 g/L (26 mM) phenylpyruvate detected. The enzyme activities were almost not affected when the pH value was up to 8.5, 4.0 g/L (24.3 mM) phenylpyruvate was produced. The titer of phenylpyruvate decreased to 3.2 g/L (19.4 mM) at pH 7.5, and 2.3 g/L (14 mM) at pH 6.5, respectively. These results indicated that the enzyme activities of LtaEP.p and A8HB.t were highly dependent on temperature and pH. Further experiments were conducted at the optimum conditions of 30 ℃ and pH 8.0.
Effect of reaction conditions on the phenylpyruvate production by the enzyme cascade of LtaEP.p and A8HB.t combination. a The effects of reaction temperatures on the phenylpyruvate production. b The effects of reaction pH on the phenylpyruvate production. The following buffer systems were used: 100 mM Tris-HCl for pH 6.5, 7.5, 8.0, and 8.5. Error bars are the standard deviation for three independent experiments
Overexpression of RidA could further increase phenylpyruvate production
Threonine dehydratase, as a Pyridoxal 5’-phosphate (PLP) dependent enzyme, will generate active imine/enamine intermediates that are converted into keto acid by a protein of the members of the RidA family, which were recently shown to be enamine deaminases [33,34,35]. Lacking RidA would decrease the activity of the PLP-dependent transaminase enzyme IlvE in S. enterica strains [36]. It has been reported that the presence of RidA could increase the rate of 2-ketobutyrate formation from threonine by the enzyme IlvA [37]. However, there was no previous report about the effect of gene ridA on phenylpyruvate production catalyzed by threonine dehydratase.
To address this issue, we cloned gene ridA after the gene encoding LtaEP.p and A8HB.t to build the plasmid pPLA-6 (Fig. 4a), and transformed it into BW25113, yielding strain M6. The fermentation results showed that after overexpression of gene ridA, the titer of phenylpyruvate was increased to 5.0 g/L (30.1 mM) within 24 h (Fig. 4b), and the practical conversion rate of benzaldehyde was up to 77%. These results confirmed that overexpression of gene ridA could provide more enamine deaminase to enhance phenylpyruvate formation. However, even with the optimized cascade system, it still cannot lead to a complete conversion of benzaldehyde, which is possibly due to the relatively low activity of dehydratase A8HB.t for the second reaction. For enzyme cascade processes, the second step with high activity can pull the substrates flux into the desired synthesis pathway, improving the overall conversion of the reaction system [38]. We can further improve the dehydratase activity through directed evolution in the future [24, 39]. Besides, we can remove the target genes in the host to minimize the effect of competing endogenous pathways and improve the introduced synthetic pathway performance [40, 41].
Production of L-phenylalanine from phenylpyruvate
Phenylalanine dehydrogenase from Bacillus badius (PheDH, encoded by gene pdh) was selected to promote the conversion of phenylpyruvate into L-phenylalanine [42]. Enzymatic dehydrogenation is usually performed in the presence of stoichiometric amounts of coenzyme (NADH or NADPH), implying the coenzyme cofactor must be recycled during the conversion. Candida boidinii formate dehydrogenase (FDH) has been employed as a workhorse for efficient NADH regeneration for decades, which can be used in a broad pH range of 6 ~ 9 [43,44,45]. In a previous report, the mutant V120S has been shown to greatly improve the activity and stability of enzyme FDH in the catalytic reaction [46]. To ensure sufficient cofactor NADH supply, a recombinant plasmid pPLA-7 expressing enzymes PheDH and FDHV120S was constructed and transformed into BW25113, yielding strain M7. The conversion performance of E. coli with pPLA-7 (strain M7) was investigated with 0.07 M phenylpyruvate, 0.35 M formate, and 0.14 M NH4Cl at 30 ℃ (Fig. 5a). 11 g/L (0.066 M) L-phenylalanine was produced within 10 h, amounting to a conversion rate of 93% of phenylpyruvate. The results showed that the enzyme cascade of PheDH and FDHV120S display high efficacy in the conversion of phenylpyruvate to L-phenylalanine.
Then, the tolerance of the key enzymes PheDH and FDHV120S to benzaldehyde was tested. We performed the same conversion of phenylpyruvate to L-phenylalanine with the addition of varying concentrations of benzaldehyde, as shown in Fig. 5b. As the benzaldehyde concentration increased to 5 mM and 15 mM, the relative activity of enzyme cascade PheDH and FDH toward phenylpyruvate could keep at 93% and 87%, respectively. When benzaldehyde concentration increased up to 25 mM, the L-phenylalanine titer was remarkably decreased by 67% compared with the control experiments (column #1 in Fig. 5b). These results indicated that the efficiency of enzyme cascade PheDH-FDHV120S would be negatively affected by high concentrations of benzaldehyde. Therefore, concentration-limited feeding of benzaldehyde could be adopted to maintain high-yield conversion. In addition, detoxification can be achieved through evolution to improve the tolerance of enzymes to the substrate benzaldehyde [47].
Biotransformation of phenylpyruvate into L-phenylalanine. a L-phenylalanine production from phenylpyruvate by coexpressing Bacillus badius phenylalanine dehydrogenase (PheDH) and Candida boidinii formate dehydrogenase (FDHV120S). FDH was used for cofactor NADH regeneration. b The tolerance of the key enzymes PheDH and FDHV120S to benzaldehyde. Error bars are the standard deviation for three independent experiments
Biosynthesis of L-phenylalanine from aromatic precursor benzaldehyde
Synthesis of L-phenylalanine from benzaldehyde and glycine was performed by using these two modules LtaEP.p-A8HB.t-RidA and PheDH- FDHV120S in one host cell (as shown in Fig. 6a). Plasmids pPLA-6 and pPLA-7 were co-transformed into E. coli BW25113 to obtain strain M8. The L-phenylalanine production performance of strain M8 (Table 1) was investigated with 15 mM benzaldehyde, 150 mM glycine, and 150 mM formate at 30 ℃. As shown in Fig. 6b, without cofactor NAD+ addition, 0.84 g/L (5 mM) L-phenylalanine was detected after 24 h (column #1in Fig. 6b). The conversion rate of benzaldehyde was relatively low, only 34%. Employing the multi-enzyme cascade, the formation of the NAD+/NADH cofactor equilibrium was not as efficient as in the two-enzyme system [48]. To prove this hypothesis, we added 0.5 mM NAD+ under the same conversion condition, as expected, the titer of L-phenylalanine was significantly increased to 1.7 g/L (10 mM) after 24 h with a conversion of 69% (more than 2-fold increase, column #2 in Fig. 6b), implying that the addition of feasible amounts of cofactor could increase the yield and productivity for L-phenylalanine.
In a previous report [24], the titer of L-phenylalanine production from benzaldehyde via an enzyme cascade was up to 1.5 g/L by semisaturated mutation of CgTD for α,β-elimination and reaction conditions optimization. Therefore, we also tested the engineered threonine deaminase CgTDF114A,R229T for L-phenylalanine production under our experimental conditions (Additional file 1: Table S1). It can be seen that the activity of wild-type A8HB.t can be comparable to that of the CgTDF114A,R229T for L-phenylalanine production. Therefore, L-phenylalanine production can be further improved in the future by the wild-type protein A8HB.t adaptive evolution.
Biosynthesis of L-phenylalanine from aromatic precursor benzaldehyde. a Scheme showing the one-pot L-phenylalanine production from benzaldehyde with co-substrate formate for cofactor NADH regeneration. Production enzymes indicated the enzymes LtaEP.p, A8HB.t, and RidA that converted benzaldehyde to phenylpyruvate. b Biotransformation of the L-phenylalanine production in strain M8. Strain M8: BW25113 transformed with plasmids pPLA-6 and pPLA-7 to overexpress genes ltaEP.p, A8HB.t, ridA, pdh, and fdh. Error bars are the standard deviation for three independent experiments
Cofactor self-sufficient system for L-phenylalanine production from precursor benzyl alcohol
As can be seen from Fig. 6a, the enzyme FDHV120S was exploited for NADH balance by enzymatically oxidizing the sacrificial substrate formate into CO2, which not only complicates the L-phenylalanine production system but also increases the production cost. Therefore, based on these benzaldehyde conversion processes, we expanded the aromatic precursors to produce L-phenylalanine from another inexpensive aromatic compound benzyl alcohol. Benzyl alcohol can be oxidized into benzaldehyde by aryl-alcohol dehydrogenase, while obtaining NADH equivalents for reducing phenylpyruvate into L-phenylalanine. In addition, Benzyl alcohol is less toxic to enzyme activity and cell growth than benzaldehyde, making the process more feasible. Therefore, we tried to develop a bioconversion process for synthesizing L-phenylalanine from benzyl alcohol by rational design, which does not require additional cosubstrate as a reductant or another cofactor regenerating enzyme, achieving an important self-sufficient cofactor regeneration system (as shown in Fig. 7a).
Benzyl alcohol dehydrogenase from Pseudomonas putida (XylBP.p) has been reported to have a high activity toward benzyl alcohol [31]. We cloned the genes xylBP.p and pdh into the plasmid pPLA-8. BW 25113 transformed with plasmids pPLA-6 and pPLA-8 (strain M9) was used for the conversion of benzyl alcohol and glycine into L-phenylalanine. With 15 mM benzyl alcohol and 150 mM glycine as co-substrates, this new enzyme cascade can accumulate 0.82 g/L (4.8 mM) and 1.1 g/L (6.7 mM) L-phenylalanine without or with 0.5 mM NAD+ addition, respectively (column #1, #2 in Fig. 7b). These results for the first time demonstrated a cofactor self-sufficient system to produce L-phenylalanine from aromatic precursor without any addition of reductant like formate. In the future, we can further optimize the bioconversion pathway by screening and evolving target enzymes with higher activity to further increase the efficiency of L-phenylalanine production from benzyl alcohol. In addition, metabolic engineering of the chassis cell could also be adopted to further improve the performance of L-phenylalanine production, such as increasing the uptake of substrate, and reducing substrate and product degradation [50].
Biosynthesis of L-phenylalanine from aromatic precursor benzyl alcohol. a Scheme showing the one-pot L-phenylalanine production from benzyl alcohol with an engineering cofactor NADH self-regeneration system. Production enzymes indicated the LtaEP.p, A8HB.t, and RidA that converted benzaldehyde to phenylpyruvate. b Biotransformation of the L-phenylalanine production in strain M9. Strain M9: BW25113 transformed with plasmids pPLA-6 and pPLA-8 to overexpress genes ltaEP.p, A8HB.t, ridA, pdh, and xylBP.p. Error bars are the standard deviation for three independent experiments
Conclusions
In this work, we developed and constructed an artificial biosynthesis pathway for L-phenylalanine production from aromatic precursors. To establish the pathway to phenylpyruvate, we first screened and expressed a high-activity natural enzyme combination including LtaEP.p and A8HB.t, 4.3 g/L of phenylpyruvate was produced. Then, we confirmed that further overexpression of enzyme RidA could increase phenylpyruvate production by 16.3%, up to 5 g/L. The conversion of phenylpyruvate into L-phenylalanine was up to 93% by co-expressing PheDH and FDHV120S. Furthermore, the engineered E. coli overexpressing LtaEP.p, A8HB.t, RidA, PheDH, and FDHV120S could produce 1.7 g/L L-phenylalanine from benzaldehyde, and the conversion rate of benzaldehyde was 69%. Finally, we constructed a new pathway for L-phenylalanine production from benzyl alcohol in a cofactor self-sufficient system. Based on overexpression LtaEP.p, A8HB.t, RidA, PheDH, and XylBP.p, 1.1 g/L L-phenylalanine was produced without the addition of reductant formate. The yield and productivity of the L-phenylalanine bioconversion system developed in this study still need to be improved in the future. Further work will focus on systematic protein screening or directed evolution to increase the activities of these key enzymes. Overall, we demonstrated a promising approach for the biosynthesis of L-phenylalanine from low-cost aromatic precursors.
Materials and methods
Strains and media
The details for all strains used in this study are shown in Table 1. The E. coli DH5α was used as the cloning host for plasmid construction. Unless otherwise specified, all strains were cultivated at 37 ℃ in LB media (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L sodium chloride) medium with appropriate antibiotics.
Biotransformation procedures
Three colonies of recombinant E. coli strain were cultivated for 12 h at 37 ℃ in LB medium with appropriate antibiotics. The culture was then inoculated (1% vol/vol) into a 250 mL flask with 50 ml fresh LB culture containing appropriate antibiotics. When the OD600 of the culture broth reached 0.6 ~ 0.8, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce gene expression. The cells were inducted at 16 °C for 20 h and collected at 4 ℃ by centrifugation (7500×g, 5 min). Then, the strains were resuspended in the appropriate buffer to the desired density as resting cells for biotransformation.
Biotransformation of benzaldehyde and glycine to phenylpyruvate was conducted with resting cells of E. coli (M1, M2, M3, M4, M5, M6, the OD600nm of the condensed cell was 50) in 2 ml Tris buffer (100 mM, pH 8.0), 50 µM PLP, at 220 rpm and 30 °C for 24 h. Biotransformation of phenylpyruvate to L-phenylalanine was conducted with resting cells of E. coli M7 (the OD600nm of the condensed cell was 50) in 2 ml Tris buffer (100 mM, pH 8.0), 0.07 M phenylpyruvate, 0.35 M formate, and 0.14 M NH4Cl at 220 rpm and 30 °C for 24 h. Biotransformation of benzaldehyde and glycine to L-phenylalanine was conducted with resting cells of E. coli M8 (the OD600nm of the condensed cell was 50) in 2 ml Tris buffer (100 mM, pH 8.0), 15 mM benzaldehyde and 150 mM glycine, 150 mM M formate, and 140 mM NH4Cl, with or without 0.5 mM NAD+ at 220 rpm and 30 °C for 24 h. Biotransformation of benzyl alcohol and glycine to L-phenylalanine was conducted with resting cells of E. coli M8 (the OD600nm of the condensed cell was 50) in 2 ml Tris buffer (100 mM, pH 8.0, 15 mM benzyl alcohol, and 150 mM glycine, and 140 mM NH4Cl, with or without 0.5 mM NAD+) at 220 rpm and 30 °C for 24 h.
Measurement of metabolites analysis
Metabolites were analyzed using an Agilent 1260 Infinity HPLC system. The target samples were collected within 24 h. The concentrations of phenylpyruvate, benzyl alcohol, and benzaldehyde, were analyzed using an Aminex HPX 87 H column (Bio-Rad, USA) and a refractive-index detector. The mobile phase is 5 mM H2SO4 with a flow rate of 0.6 mL/min. The column temperature and detection temperature are 35 ℃ and 50 ℃, respectively. The concentration of L-phenylalanine was analyzed using an Agilent C18 column (4.6 × 100 mm, 3.5 mm) and a DAD detector. The mobile phase gradient program and automated liquid sampler program were performed as the manufacturer’s instructions (http://www.chem.agilent.com/Library/applications/ 5990-4547EN.pdf).
Plasmids construction
Primers (Table 2) were ordered from Tsingke. PCR reactions were carried out with Phanta DNA polymerase according to the manufacturer’s instructions. The sequences of all the plasmids produced were verified by DNA sequencing. The details for all plasmids are shown in Table 1. A gene fragment encoding lac repressor LacI [51] was inserted into the EcoRI site of plasmid pZE12 and pZA24 [52] to yield plasmid pZElac with ampicillin resistance, and pZAlac with kanamycin resistance, respectively.
pZE-P LlacO1 -ltaE P.p -A8H B.t
Genes ltaEP.p, and A8HB.t were amplified based on Pseudomonas putida and Burkholderia thailandensis genomic DNA. Primers ltaEP.p-F and ltaEP.p-R were used to amplify gene ltaEP.p. Primers A8HBt-F and A8HBt-R were used to amplify gene A8HBt. The vector fragment of pZE was amplified from plasmid pIVC3 with primers VecpZE-F and VecpZE-R. Then these two fragments and the vector fragment of pZElac were digested with Acc65I-SpeI, SpeI-XbaI, and Acc65I -XbaI, respectively. These digested genes were ligated with T4 DNA ligase respectively to form the plasmid pZE- PLlacO1- ltaEP.p-A8HB.t.
pZE-P LlacO1 -ltaE E.c -A8H B.t
Genes ltaEE.c, and A8HB.t were amplified based on E. coli and Burkholderia thailandensis genomic DNA. Primers ltaEE.c-F and ltaEE.c-R were used to amplify gene ltaEE.c. Primers A8HB.t-F and A8HB.t-R were used to amplify gene A8HB.t. Then these two fragments were digested with Acc65I-SpeI, SpeI-XbaI. Then these three fragments and the vector fragment of pZElac were ligated with T4 DNA ligase to form plasmid pZE- PLlacO1- ltaEE.c-A8HB.t.
pZE-P LlacO1 -ltaE C.c -A8H B.t
Genes ltaEC.c, and A8HB.t were amplified based on Caulobacter crescentus CB15 and Burkholderia thailandensis genomic DNA. Primers ltaEC.c-F and ltaEC.c-R were used to amplify gene ltaEC.c. Primers A8HB.t-F and A8HB.t-R were used to amplify gene A8HB.t. Then these two fragments were digested with Acc65I-SpeI, SpeI-XbaI. Then these three fragments and the vector fragment of pZElac were ligated with T4 DNA ligase to form plasmid pZE- PLlacO1- ltaEC.c-A8HB.t.
pZE-P LlacO1 -ltaE P.p -ilvA B.a
Genes ltaEP.p, and IlvAB.a were amplified based on Pseudomonas putida and Burkholderia ambifaria genomic DNA. Primers ltaEP.p-F and ltaEP.p-R were used to amplify gene ltaEP.p. Primers ilvAB.a-F and ilvAB.a-R were used to amplify gene IlvAB.a. Then these two fragments were digested with Acc65I-SpeI, SpeI-XbaI. Then these three fragments and the vector fragment of pZElac were ligated with T4 DNA ligase to form plasmid pZE-PLlacO1- ltaEP.p-IlvAB.a.
pZE-P LlacO1 -ltaE P.p -TAA B.t
Genes ltaEP.p, and TAAB.t were amplified based on Pseudomonas putida and Burkholderia thailandensis genomic DNA. Primers ltaEP.p-F and ltaEP.p-R were used to amplify gene ltaEP.p. Primers TAAB.t-F and TAAB.t-R were used to amplify gene TAAB.t. Then these two fragments were digested with Acc65I-SpeI, SpeI-XbaI. Then these three fragments and the vector fragment of pZElac were ligated with T4 DNA ligase to form plasmid pZE-PLlacO1- ltaEP.p-TAAB.t.
pZE-P LlacO1 -ltaE P.p -A8H B.t-ridA
Genes ridA, ltaEP.p, and A8HB.t were amplified based on E. coli, Pseudomonas putida and Burkholderia thailandensis genomic DNA, respectively. Primers ridA-F and ridA-R were used to amplify gene ltaEP.p. Primers ltaEP.p-F-1 and ltaEP.p-R-1 were used to amplify gene ltaEP.p. Primers A8HB.t-F and A8HB.t-R were used to amplify gene A8HB.t. Then these two fragments were digested with Acc65I-SpeI, SpeI-NheI, NheI-XbaI. Then these three fragments and the vector fragment of pZElac were ligated with T4 DNA ligase to form plasmid pZE-PLlacO1- ltaEP.p-A8HB.t-ridA.
pZA-P LlacO1 -fdh-pdh
Genes phedh, fdh were amplified based on Bacillus badius, and Candida boidinii genomic DNA, respectively. Primers phedh-F and phedh-R were used to amplify gene phedh. Primers fdh-F and fdh-R were used to amplify gene fdh. The vector fragment of pZA was amplified from plasmid pIVC3 with primers VecpZA-F and VecpZA-R. Then these two fragments and the vector fragment of pZAlac were digested with Acc65I-SpeI, SpeI-NheI, and Acc65I-NheI, respectively. Then these three fragments and the vector fragment of pZAlac were ligated with T4 DNA ligase to form plasmid pZA-PLlacO1-fdh-phedh.
pZA-P LlacO1 -xylB P.p -pdh
Genes xylBP.p, fdh were amplified based on Pseudomonas putida, and Candida boidinii genomic DNA, respectively. Primers xylBP.p-F and xylBP.p-R were used to amplify gene xylBP.p. Primers fdh-F and fdh-R were used to amplify gene fdh. Then these two fragments were digested with Acc65I-SpeI, and SpeI-NheI, respectively. Then these two fragments and the vector fragment of pZAlac were homologous recombined with Exnase to form plasmid pZA-PLlacO1- xylBP.p-fdh.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Change history
10 February 2024
A Correction to this paper has been published: https://doi.org/10.1186/s12934-024-02327-y
References
Qian Y, Lynch JH, Guo L, Rhodes D, Morgan JA, Dudareva N. Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants. Nat Commun. 2019;10:15.
Yuan P, Cao W, Wang Z, Chen K, Li Y, Ouyang P. Enhancement of L-phenylalanine production by engineered Escherichia coli using phased exponential L-tyrosine feeding combined with nitrogen source optimization. J Biosci Bioeng. 2015;120:36–40.
Ding D, Liu Y, Xu Y, Zheng P, Li H, Zhang D, Sun J. Improving the production of L-phenylalanine by identifying key enzymes through multi-enzyme reaction system in vitro. Sci Rep. 2016;6: 32208.
Hou Y, Hossain GS, Li J, Shin HD, Liu L, Du G. Production of phenylpyruvic acid from L-phenylalanine using an L-amino acid deaminase from Proteus mirabilis: comparison of enzymatic and whole-cell biotransformation approaches. Appl Microbiol Biotechnol. 2015;99:8391–402.
Sun Z, Ning Y, Liu L, Liu Y, Sun B, Jiang W, Yang C, Yang S. Metabolic engineering of the L-phenylalanine pathway in Escherichia coli for the production of S- or R-mandelic acid. Microb Cell Fact. 2011;10: 71.
Movassaghi S, Leung E, Hanif M, Lee BYT, Holtkamp HU, Tu JKY, Söhnel T, Jamieson SMF, Hartinger CG. A bioactive l-phenylalanine-derived arene in multitargeted organoruthenium compounds: impact on the antiproliferative activity and mode of action. Inorg Chem. 2018;57:8521–9.
Liu Y, Xu Y, Ding D, Wen J, Zhu B, Zhang D. Genetic engineering of Escherichia coli to improve L-phenylalanine production. BMC Biotechnol. 2018. https://doi.org/10.1186/s12896-018-0418-1.
Liu X, Niu H, Li Q, Gu P. Metabolic engineering for the production of l-phenylalanine in Escherichia coli. 3 Biotech. 2019;9:85.
Maiti TK, Chatterjee SP. L-phenylalanine production by double auxotrophic mutants of Arthrobacter globiformis. Folia Microbiol (Praha). 1991;36:234–9.
Zhou H, Liao X, Wang T, Du G, Chen J. Enhanced l-phenylalanine biosynthesis by co-expression of pheA(fbr) and aroF(wt). Bioresour Technol. 2010;101:4151–6.
Berry A, Ahmad S, Liss A, Jensen RA. Enzymological features of aromatic amino acid biosynthesis reflect the phylogeny of mycoplasmas. J Gen Microbiol. 1987;133:2147–54.
Li H, Lyv Y, Zhou S, Yu S, Zhou J. Microbial cell factories for the production of flavonoids–barriers and opportunities. Bioresour Technol. 2022. https://doi.org/10.1016/j.biortech.2022.127538.
Liu SP, Liu RX, Xiao MR, Zhang L, Ding ZY, Gu ZH, Shi GY. A systems level engineered E. Coli capable of efficiently producing L-phenylalanine. Process Biochem. 2014;49:751–7.
Zhou H, Liao X, Liu L, Wang T, Du G, Chen J. Enhanced l-phenylalanine production by recombinant Escherichia coli BR-42 (pAP-B03) resistant to bacteriophage BP-1 via a two-stage feeding approach. J Ind Microbiol Biotechnol. 2010;38:1219–27.
Sun W, Ding D, Bai D, Lin Y, Zhu Y, Zhang C, Zhang D. Transcriptomics and metabolomics analysis of L-phenylalanine overproduction in Escherichia coli. Microb Cell Fact. 2023. https://doi.org/10.1186/s12934-023-02070-w.
Tohge T, Watanabe M, Hoefgen R, Fernie AR. Shikimate and phenylalanine biosynthesis in the green lineage. Front Plant Sci. 2013;4:62.
Yoo H, Widhalm JR, Qian Y, Maeda H, Cooper BR, Jannasch AS, Gonda I, Lewinsohn E, Rhodes D, Dudareva N. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat Commun. 2013;4:2833.
Liu SP, Zhang L, Mao J, Ding ZY, Shi GY. Metabolic engineering of Escherichia coli for the production of phenylpyruvate derivatives. Metab Eng. 2015;32:55–65.
Prakash P, Pathak N, Hasnain SE. pheA (Rv3838c) of Mycobacterium tuberculosis encodes an allosterically regulated monofunctional prephenate dehydratase that requires both catalytic and regulatory domains for optimum activity. J Biol Chem. 2005;280:20666–71.
Market price of benzyl alcohol. https://sunwisechem.en.made-in-china.com/product/fXEJbNnuYKpC/China-Benzyl-Alcohol-99-CAS-No-100-51-6.html. Accessed 7 Oct 2023.
Market price of benzaldehyde. https://aoks-bio.en.made-in-china.com/product/aNpnJKhobZWP/China-Factory-Supply-Benzaldehyde-CAS-100-52-7-99-Price-in-Stock-100527.html. Accessed 7 Oct 2023.
Song Y, Sanyal U, Pangotra D, Holladay JD, Camaioni DM, Gutiérrez OY, Lercher JA. Hydrogenation of benzaldehyde via electrocatalysis and thermal catalysis on carbon-supported metals. J Catal. 2018;359:68–75.
Dalle-Donne I, Rossi R, Giustarini D, Milzani A, Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin Chim Acta. 2003;329:23–38.
Song W, Wang JH, Wu J, Liu J, Chen XL, Liu LM. Asymmetric assembly of high-value alpha-functionalized organic acids using a biocatalytic chiral-group-resetting process. Nat Commun. 2018;9:3818.
Hernandez K, Bujons J, Joglar J, Charnock SJ, Domínguez de María P, Fessner WD, Clapés P. Combining aldolases and transaminases for the synthesis of 2-Amino-4-hydroxybutanoic acid. ACS Catal. 2017;7:1707–11.
Hernandez K, Zelen I, Petrillo G, Usón I, Wandtke CM, Bujons J, Joglar J, Parella T, Clapés P. EngineeredL-serine hydroxymethyltransferase fromStreptococcus thermophilusfor the synthesis of α,α-dialkyl-α-amino acids. Angew Chem Int Ed. 2015;54:3013–7.
Hummel W, Weiss N, Kula M-R. Isolation and characterization of a bacterium possessing L-phenylalanine dehydrogenase activity. Arch Microbiol. 1984;137:47–52.
Bulot E, Cooney CL. Selective production of phenylalanine from phenylpyruvate using growing cells ofCorynebacteriumglutamicum. Biotechnol Lett. 1985;7:93–7.
Fiorentino G, Ronca R, Bartolucci S. A novel E. Coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein. Appl Microbiol Biotechnol. 2009;82:67–77.
Sankar M, Nowicka E, Carter E, Murphy DM, Knight DW, Bethell D, Hutchings GJ. The benzaldehyde oxidation paradox explained by the interception of peroxy radical by benzyl alcohol. Nat Commun. 2014;5:3332.
Shaw JP, Harayama S. Purification and characterisation of TOL plasmid-encoded benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase of Pseudomonas putida. Eur J Biochem. 1990;191:705–14.
Arcus VL, Prentice EJ, Hobbs JK, Mulholland AJ, Van der Kamp MW, Pudney CR, Parker EJ, Schipper LA. On the temperature dependence of enzyme-catalyzed rates. Biochemistry. 2016;55:1681–8.
Niehaus TD, Gerdes S, Hodge-Hanson K, Zhukov A, Cooper AJ, ElBadawi-Sidhu M, Fiehn O, Downs DM, Hanson AD. Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (rid) protein family. BMC Genomics. 2015;16:382.
Okuda H, Nagata S, Misono H. A novel phenylserine dehydratase from Pseudomonas pickettii PS22: purification, characterization, and sequence of its phosphopyridoxyl peptide. J Biochem. 1996;119:690–6.
Borchert AJ, Downs DM. Analyses of variants of the Ser/Thr dehydratase IlvA provide insight into 2-aminoacrylate metabolism in Salmonella enterica. J Biol Chem. 2018;293:19240–9.
Lambrecht JA, Schmitz GE, Downs DM. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio. 2013;4:e00033-00013.
Lambrecht JA, Flynn JM, Downs DM. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem. 2012;287:3454–61.
Gu Y, Lv X, Liu Y, Li J, Du G, Chen J, Rodrigo LA, Liu L. Synthetic redesign of central carbon and redox metabolism for high yield production of N-acetylglucosamine in Bacillus subtilis. Metab Eng. 2019;51:59–69.
Tai YS, Xiong M, Jambunathan P, Wang J, Wang J, Stapleton C, Zhang K. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products. Nat Chem Biol. 2016;12:247–53.
Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng. 2013;4:259–88.
Bayer T, Milker S, Wiesinger T, Winkler M, Mihovilovic MD, Rudroff F. In vivo synthesis of polyhydroxylated compounds from a hidden reservoir of toxic aldehyde species. ChemCatChem. 2017;9:2919–23.
Villalonga R, Tachibana S, Perez Y, Asano Y. Increased conformational and thermal stability properties for phenylalanine dehydrogenase by chemical glycosidation with end-group activated dextran. Biotechnol Lett. 2005;27:1311–7.
Mesentsev AV, Lamzin VS, Tishkov VI, Ustinnikova TB, Popov VO. Effect of pH on kinetic parameters of NAD+-dependent formate dehydrogenase. Biochem J. 1997;321(Pt 2):475–80.
Baumchen C, Bringer-Meyer S. Expression of glf Z.m. increases D-mannitol formation in whole cell biotransformation with resting cells of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2007;76:545–52.
Kratzer R, Pukl M, Egger S, Nidetzky B. Whole-cell bioreduction of aromatic alpha-keto esters using Candida tenuis xylose reductase and Candida boidinii formate dehydrogenase co-expressed in Escherichia coli. Microb Cell Fact. 2008;7: 37.
Jiang W, Lin P, Yang R, Fang B. Identification of catalysis, substrate, and coenzyme binding sites and improvement catalytic efficiency of formate dehydrogenase from Candida boidinii. Appl Microbiol Biotechnol. 2016;100:8425–37.
Kunjapur AM, Prather KL. Microbial engineering for aldehyde synthesis. Appl Environ Microbiol. 2015;81:1892–901.
Li Z, Yan J, Sun J, Xu P, Ma C, Gao C. Production of value-added chemicals from glycerol using in vitro enzymatic cascades. Commun Chem. 2018;1:1.
INVALID CITATION [24].
Lin B, Tao Y. Whole-cell biocatalysts by design. Microb Cell Fact. 2017;16:106.
Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci U S A. 2008;105:20653–8.
Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–10.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grant No. 22078267), Westlake University-Muyuan Joint Research Institute, Westlake University Research Center for Industries of The Future, and Westlake Center of Synthetic Biology and Integrated Bioengineering.
Author information
Authors and Affiliations
Contributions
KZ conceived and supervised the project. MN designed and performed the main experiments. ZC and CC participated in plasmid construction. Data was evaluated by all authors. MN and JW wrote the manuscript, and KZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In the Acknowledgements section of this article the grant number relating to National Natural Science Foundation of China was incorrectly given as 10327A012001 and should have been 22078267.
Supplementary Information
Additional file 1:
Table S1. Bioconversion of benzaldehyde and glycine into L-phenylalanine.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nie, M., Wang, J., Chen, Z. et al. Systematic engineering enables efficient biosynthesis of L-phenylalanine in E. coli from inexpensive aromatic precursors. Microb Cell Fact 23, 12 (2024). https://doi.org/10.1186/s12934-023-02282-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-023-02282-0