Abstract
Background
Cyclic dipeptides are an important class of natural products owing to their structural diversity and biological activities. In fungi, the cyclo-ring system is formed through the condensation of two α-amino acids via non-ribosomal peptide synthetase (NRPS). However, there are few investigations on the functional identification of this enzyme. Additionally, information on how to increase the production of cyclic dipeptide molecules is relatively scarce.
Results
We isolated the Eurotium cristatum NWAFU-1 fungus from Jing-Wei Fu brick tea, whose fermentation metabolites contain echinulin-related cyclic dipeptide molecules. We cloned the cirC gene, encoding an NRPS, from E. Cristatum NWAFU-1 and transferred it into the heterologous host Aspergillus oryzae. This transformant produced a novel metabolite possessing an l-tryptophan-l-alanine cyclic dipeptide backbone (Cyclo-TA). Based on the results of heterologous expression and microsomal catalysis, CriC is the first NRPS characterized in fungi that catalyzes the formation of a cyclic dipeptide from l-tryptophan and l-alanine. After substrate feeding, the final yield reached 34 mg/L. In this study, we have characterized a novel NRPS and developed a new method for cyclic dipeptide production.
Conclusions
In this study we successfully expressed the E. Cristatum NWAFU-1 criC gene in A. oryzae to efficiently produce cyclic dipeptide compounds. Our findings indicate that the A. oryzae heterologous expression system constitutes an efficient method for the biosynthesis of fungal Cyclic dipeptides.
Similar content being viewed by others
Background
Cyclic dipeptides ring systems are achieved by the fusion of two α-amino acids, and the fungi are well-known primary producers of a diversity of cyclic dipeptides [4,5,4]. They have a wide range of biological activities, including antimicrobial, antiviral, anticancer, and proangiogenic activity [4,5,6]. This type of natural product contains numerous therapeutically promising compounds [7], such as plinabulin, the derivative of phenylahistin which was isolated from the fungus Aspergillus ustus [8], plinabulin has been advanced to Phase III clinical trials as an antitumor drug candidate [9]. Even though medicinal chemists have widely developed and synthesized cyclic dipeptides, the understanding and manipulating their biosynthetic pathways results in the formation of new chemical structures, that may lead to the production of new active compounds [10].
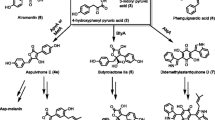
The l-tryptophan-l-alanine cyclic dipeptide (Cyclo-TA (1) series) is a structural moiety containing an indole diketopiperazine (DKP) scaffold. A reverse C2 prenylation (preechinulin (2), echinulin series) and unsaturated derivatives between C10 and C11 (Δ10) (neoechinulin A (3) series) and C14 and C17 (Δ14) (neoechinulin B (4) series) constitute the main structural modifications to the Cyclo-TA backbone [11]. Prenylation of the backbone by prenyltransferase is the most frequent modification occurring in tryptophan-containing cyclic dipeptides, and the varying degrees (one to four prenyl moieties) and positions (C-2/4/5/6/7 or N-1) of phenyl modifications that appear on the backbone expand the structural diversity [12, 13]. Reverse and regular prenylated compounds, such as rubrumline O (5) and 3-methyl-6-[[1-(3-methyl-2-butenyl)-1H-indol-3-yl]methyl]-2,5-piperazinedione (6) with an N-1 modification of the Cyclo-TA backbone are inhibitors of influenza viruses [14]. Cyclo-TA that has not undergone prenylation can be closed with a C-N bond between C-2 and N-12 to form a 6-5-5-6 ring system and further form a heterodimer, such as cristatumin C (7) [15]. Talathermophilin D (8) represents a class of pyranoindole modifications at the C-6 and C-7 positions, which are uncommon in natural DKP products [16]. Variecolorin L (9) [17] and echinulin (10) both contain two dimethylallyl (DMA) moieties at the C-4/5 and C-5/7 positions, respectively, as well as an isopentenyl moiety at C-2. Hydroxylation and oxy-methylation (e.g., rubrumazines A (11)) [18] or acetylation (e.g., rubrumline C (12) [14]) of DMA moieties and the presence of one (dehydroechinulin (13)) [19] or two (cryptoechinuline G (14) [20], rubrumline E (15), and neoechinulin C (16)) [11, 14] backbone double bonds enrich the diversity of compounds. Furthermore, post-modifications occurring on the prenyl moieties increase the structural diversity of Cyclo-TA containing compounds (Fig. 1). These compounds have been mainly isolated from different Penicillium [21, 22], Aspergillus [20, 23], and Eurotium [14, 18, 19] species.
Recently, Nies et al. successfully identified a biosynthetic gene cluster (BGC) for echinulin in A. ruber [11]. This cluster comprises four genes that are responsible for the backbone's components, including the coding region for a non-ribosomal peptide synthetase (NRPS, echPS), a cytochrome P450 enzyme (echP450), and two prenyltransferases (echPT1 and echPT2). In an in vitro enzyme activity experiment, EchPT1 was responsible for indole ring C2 reverse-prenylation, EchP450 for forming a double bond, and EchPT2 for the multi-prenylation at the C-5, 6, or 7 positions [11, 12]. Although NRPS is speculated to be the responsible enzyme for cyclic dipeptide production, its function has not been independently analyzed and verified.
In this study, we isolated a fungus from Jing-Wei Fu brick tea, which was identified by internal transcribed spacer (ITS) sequence analysis and named Eurotium cristatum NWAFU-1. The ability to generate cyclic dipeptide-related molecules was discovered upon analysis of the metabolites from fungal fermentation. Through analysis of the genomic data, we identified a gene cluster containing NRPS, which was predicted to be involved in the synthesis of Cyclo-TA. Furthermore, we describe the production of a cyclic dipeptide from transformants containing the core NRPS gene (criC) in the heterologous expression host A. oryzae.
Results and discussion
Identification of the Cri gene cluster for echinulin biosynthesis in E. cristatum
Jing-Wei Fu brick tea is a unique post-fermented tea product that is naturally co-fermented by microorganisms and has gained global popularity due to its potential health benefits. It naturally produces golden particles, commonly referred to as “golden flowers”, and contains a symbiotic fungus named E. cristatum [24]. We isolated the fungus E. cristatum NWAFU-1 from a brick tea sample collected from Xianyang City, Shaanxi Province, China (Additional file 1: Fig. S1). ITS sequence analysis of the ribosomal DNA was highly similar (99.43%) to the GenBank sequences from the fungus E. cristatum YKY807.
First, we examined the metabolites of E. cristatum NWAFU-1 by ultraperformance liquid chromatography–electrospray ionization–high resolution mass spectrometry (UPLC–ESI–HRMS) via classical or feature-based molecular networking workflows with the Global Natural Products Social Molecular Networking (GNPS, http://gnps.ucsd.edu) web platform. The molecular masses of 326.19, 394.25, 462.31, and 490.34 were detected, suggesting that these compounds could be echinulin and its derivatives (Additional file 1: Fig. S2).
To determine the BGC responsible for the production of cyclic dipeptide related compounds, we employed an independent study for genome mining on E. cristatum YKY807, using gene cluster search methods (a local BLAST search and the 2ndFind program [25]). We identified a putative BGC (cri), localized on a continuous DNA region of 27.4 kb that encoded seven putative enzymes: one NRPS (CriC), two annotated prenyltransferases (CriA and CriF), one cytochrome P450 (CriE), one FMN oxidoreductase (CriG), one transporter (CriB), and one functionally unknown protein (Additional file 1: Fig. S3).
Sequence similarity network analysis showed that CriC is grouped with NRPSs, suggesting a common role in the biosynthesis of cyclic dipeptides. Orthologous CriC proteins are involved in the biosynthesis of the structurally related cyclic dipeptide (AtaP/GliP/SirP/AclP/VerP) [26,27,28,29,30]. These observations indicate that sequence similarity network analysis can be used to predict the function of CriC family proteins with close similarity (Additional file 1: Fig. S4).
Functional analysis of CriC
To examine the function of NRPS, the criC gene was amplified from E. cristatum NWAFU-1 genomic DNA, the purified PCR fragment was cloned into a pUSA2 plasmid [31], and transformed into A. oryzae NSAR1 to form AO-criC transformants. HPLC analysis of the partially purified fraction of AO-criC mycelial extracts showed a new peak, which was not found in the control culture of the wild-type strain (Fig. 2A). To determine the structure of this novel compound, the mycelial ethyl acetate extract of AO-criC was obtained by large-scale incubation in MPY medium. The crude extract was purified with silica gel column chromatography and partitioned with hexane–ethyl acetate. HPLC analysis resulted in the pure product Cyclo-TA (1) at a concentration of 7.5 mg/L. The obtained UV/Vis spectrum (maxima at λ = 195, 209, and 275 nm) was in agreement with the absorption characteristics of the indole ring (Fig. 2B). The molecular formula was determined to be C14H16N3O2 (calcd: 258.1238 [M + H]+, 280.1057 [M + Na]+ and found: 258.1241, 280.1062) by high resolution–electrospray ionization–mass spectrometry (HR-ESI–MS) analysis. A molecular weight of [2M + H]+ and [2M + Na]+ was also detected. MS/MS data showed a specific fragment of 130.0657 (an indole core ion) that was identified as a fragment of Cyclo-TA (Fig. 2C).
Heterologous expression of criC and characterization of metabolites. A HPLC trace of criC heterologous expression in A. oryzae NSAR1. (I) AO-WT (A. oryzae wild type), (II) metabolites produced by transformant AO-criC, (III) AO-criC feed by substrates (l-Trp and l-Ala). B UV absorption spectroscopy of criC product. C MS and MS/MS spectrum of CriC product. D Summary of HMBC, COSY, and NOESY experiments of the AO-criC product.
13C nuclear magnetic resonance (NMR) indicated two ketones (δc 169.5 & δc 170.6) carbonyl groups, suggesting that the structure of the compound is closely related to that of cyclic dipeptides. Extensive NMR data analysis, including HSQC, HMBC, COSY, and NOESY, confirmed the structure of 1, as shown in Fig. 2D (Additional file 1: Figs. S5–10, Table S1).
Biochemical characterization of CriC and improving Cyclo-TA production
The substrate specificity of CriC was also investigated. We performed an in vitro analysis using microsomes of AO-criC strains. Along with ATP, l-Trp and l-Ala were used as substrates in the reaction, which confirmed CriC's substrate specificity for these two amino acids (Fig. 3A). Using a time-dependent in vitro assay with both l-Trp and l-Ala as substrates, the final product increased over time (Additional file 1: Fig. S11). Using l-Trp or l-Ala as a fixed substrate to react with 19 other native amino acids from microsomes yielded no product (Additional file 1: Fig. S12).
Biochemical characterization and improved production capacity of CriC. A Biochemical characterization of CriC microsome, (I) Standard cyclo-TA, (II) reaction mix containing CriC microsome and substrates (l-Trp and l-Ala), (III) reaction mix containing denatured CriC microsome and substrates, (iv) reaction mix containing substrates and microsome prepared from AO host. B Substrate feeding increases the yield of AO-criC. The substrate l-Trp and l-Ala was added in equal amounts for each gradient
Next, we investigated the potential of A. oryzae expressing recombinant criC for the industrial production of Cyclo-TA. As AO-criC transformants can efficiently produce the target product, we investigated the effects of increased yields of feed substrates (l-Trp and l-Ala) during culture. When different concentrations of the two substrates were added to the fermentation medium, the amount of the final product was significantly increased; when 20 μM of the two substrates were added separately, the yield reached 34 mg/L (Fig. 3B, Additional file 1: Fig. S13).
Phylogenetic analysis of cyclic dipeptide synthetase
Based on the PKS/NRPS analysis website, CriC encodes an NRPS protein of 2127 amino acids that consist of two sets of domains for adenylation (A), thiolation (T), and condensation (C) (Additional file 1: Fig. S14A, B). The A domain chooses and activates a carboxylic acid substrate through adenylation following consumption of ATP, and then forms a thioester linkage by transferring the acyl group to the phosphopantetheinyl arm linked to the T domain. The C domain is involved in the formation of peptide bonds between two adjacent modules [32]. Correlation analysis for the adenylation domain of CriC and TaqA, an enzyme that binds anthranilate and two amino acids (l-Trp and l-Ala), showed that they act as substrates in the synthesis of fumiquinazoline F [33]. A comparison of the two proteins revealed that the domains of binding amino acids were predicted to have similar functions. The proposed biosynthetic reaction mechanism is shown in Additional file 1: Figure S14C.
We constructed a phylogenetic tree using cyclic dipeptide proteins from BLAST analysis. The cyclic peptides ring system is generated by the condensation of two amino acids via two different pathways: via NRPS, which uses and activates free amino acids through adenylation [34, 35], or via cyclic dipeptide synthases (CDPs). CDPs kidnap aminoacyl-transfer RNAs (aa-tRNAs) from their primary use in the translation process [36]. According to phylogenetic tree analysis, enzymes that form cyclic dipeptides using tryptophan and/or alanine as substrates can be divided into two groups: bacterial-derived aa-tRNA-dependent cyclic dipeptide synthases and fungal-derived NRPSs. CriC differs from other reported NRPSs using tryptophan or alanine as substrates in that it forms a new branch with 14 other sequences from the National Center for Biotechnology Information (NCBI) whose function has not been reported (Fig. 4, Additional file 1: Table S2). Based on genome mining of a BGC for echinulin biosynthesis, we identified 15 fungal strains, depending on the presence of BGCs for non-ribosomal peptide biosynthesis, and a possible cluster for cyclic dipeptide biosynthesis. Additional file 1: Figure S15 shows 15 other gene clusters from Eurotium, Aspergillus, and Penicillium species.
Phylogenetic analysis of CriC. Phylogenetic analysis of CriC and Cyclic dipeptide synthase with tryptophan or alanine as a substrate. cWW, cWL, cWA, cWP, cWV refer to cyclic dipeptides formed by tryptophan with tryptophan, leucine, alanine, proline, and valine, respectively, while cAA and cAG refer to cyclic dipeptides formed by alanine with alanine and glycine, respectively. Phylogenetic trees were constructed using MEGA X the maximum likelihood method
Conclusions
In summary, we showed that the criC gene, located in the echinulin BGC in E. cristatum, encodes an NRPS that catalyzes the condensation of l-Trp and l-Ala to produce Cyclo-TA (1) [37]. Phylogenetic trees show that it belongs to a distinct family, and similar gene clusters are found in Eurotium, Penicillium, and Aspergillus sp. Additionally, we developed a new method for the directional production of cyclic dipeptide backbones. The criC gene was successfully expressed in A. oryzae, which produced 34 mg/L Cyclo-TA when substrates were provided in the culture medium. Microsomal experiments further demonstrated the catalytic effects of CriC in vitro. Overall, we not only elucidated the function of large fragments gene NRPS, but also demonstrated that A. oryzae is a powerful tool for the production of complex natural products.
Material and methods
General experimental procedures
All reagents commercially supplied were used as received. HPLC analysis was performed using an Agilent 1260 Series with a DAD detector (California, CA, USA). 1H- and 13C-NMR spectra were recorded on Bruker AVAN CEIII HD 500. Chemical shifts were reported as δ scale in ppm as an internal reference (CD3OD; 1H NMR = 3.31 ppm, 13C NMR = 49.0 ppm). Mass spectra were obtained with an AB SCEIX Triple TOF 6600. Column chromatography was carried out on C18 silica gel (Agilent Technologies. USA). Oligonucleotides for polymerase chain reaction (PCR) were purchased from Tsingke Biotechnology Co., Ltd.
Escherichia coli DH5α was used for cloning and following standard recombinant DNA techniques. This study used a fungal host strain A. oryzae NSAR1, a quadruple auxotrophic mutant (niaD-, sC-, ∆argB, adeA-) for fungal expression [38]. The transformant was grown on DPY (dextrin-polypeptone-yeast extract: 2 % dextrin, 1 % polypeptone, 0.5 % yeast extract, 100 mL) medium supplemented with appropriate nutrients [39].
Fungal material, preparation of expression plasmids, and transformation of A. oryzae
Eurotium cristatum NWAFU-1 was identified by morphological observation and by analysis of the ITS regions of its rDNA (GenBank accession No: OM276864). Genomic DNA of E. cristatum NWAFU-1 was prepared according to the literature procedure. The criC was amplified with a primer set as shown in Additional file 1: Table S3. PCR reactions were performed with the KOD-Plus-Neo (TOYOBO). PCR product was inserted into the appropriate restriction site KpnI using ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech Laboratories) to construct expression plasmids, pUSA2-CriC. The expression vector was sequenced by Sangon BioTech using KBseq technology to obtain the DNA sequence of criC, which was submitted to NCBI under the GenBank accession No. OM307404. Transformation of A. oryzae NSAR1 was performed by the protoplast-polyethylene glycol method reported previously [39]. pUSA2-criC was used for the transformation to construct AO-criC.
Biotransformation and substrate addition
Mycelia of A. oryzae transformants were inoculated into 10 mL of MPY (maltose-peptone-yeast extract: 3 % maltose, 1 % polypeptone, 0.5 % yeast extract) medium containing appropriate nutrients in 50 mL Erlenmeyer flasks. After an additional 3 days of incubation at 30 °C, the mycelia were collected by filtration and soaked in acetone (20 mL). The organic layer was then concentrated in vacuo. The crude extracts were analyzed by Agilent 1260 Series HPLC equipped with an Agilent EC-C18 (PorosheII 120, 150 mm × 4.6 mm) at the following conditions: flow rate; 0.6 mL/min, Detection; 280 nm, Solvent system; methanol in H2O, 0–16 min, from 5% to 75% linear; 17 min, 100%; 18–22 min 5%.
Substrate addition experimental procedures and subsequent product extraction procedures were the same as described above. Equimolar concentrations of alanine and tryptophan were added to the transformants before induction culture. Quantification of conversion products was achieved by the standard curve method with HPLC. The HPLC analytical method is as described above.
Microsomal activity assay for AO-criC transformant
A. oryzae NSAR1 transformant AO-criC was grown in 100 mL of MPY medium without methionine at 200 rpm and 30 °C for two days. The mycelium was then collected by centrifugation at 5000g for 10 min and ground to a powder in liquid nitrogen. The powder was resuspended in buffer B (0.6 M sorbitol, 0.1 M KCl, 1.0 mM EDTA, 2.0 mM DTT, 1.0 mM PMSF, 50 mM Tris-HCl, pH 7.5) and lysed using an ultrasonic cell disruptor (0 °C for 30 min). The suspension was centrifuged at 8000g and 4 °C for 10 min and the supernatant was further separated by ultracentrifugation at 100,000g and 4 °C for 1 h. The microsomal precipitate that had settled at the bottom of the centrifuge tube was then resuspended in 1 mL of buffer C (20 % glycerol, 50 mM Tris-HCl, 1.0 mM MgCl2, 1.0 mM EDTA, 1.0 mM DTT, pH 7.5) and stored at −80 °C. The mycelium of A. oryzae NSAR1 was used to isolate the microsomal fragments and stored at −80 °C.
The catalytic activity test of microsomes containing CriC protein was performed in a total volume of 200 μL reaction mixture, containing 180 μL microsomal fraction of the transformant harboring CriC, 1 mM ATP, and 0.5 mM substrates were incubated at 30 °C for 24 h. As a negative control, the reaction catalyzed by microsomal fraction of A. oryzae NSAR1 was also performed with the same reaction mixture and reaction conditions. Subsequently, the reaction mixture was quenched with equal methanol. The reaction mixture is subjected to HPLC detection after 15,000g centrifugation for 10 min and filtration (0.22 μM). The reactions were quenched at 4 h, 8 h, 12 h, 16 h, 20 h, and 24 h in time-course experiments.
Large scale fermentation AO-criC transformant
Mycelia of transformant AO-criC was inoculated into 100 mL of MPY medium containing appropriate l-Trp and l-Ala in 500 mL Erlenmeyer flasks, and a total of six Erlenmeyer flasks. After incubation at 30 °C for 3 days, the mycelia were collected by filtration and extracted with acetone (400 mL). After filtration, the filtrates were concentrated in vacuo. The residues were resolved in ethyl acetate, and the organic layer was washed with brine and concentrated in vacuo. The crude extracts were purified using silica gel column chromatography (hexane:ethyl acetate, 4:1 to 2:1) to isolate the products.
Cyclo-AT: HR-ESI-MS analysis; calcd. for C14H16N3O2 [M+H]+ calcd: 258.1238, found: 258.1241. [α]D25 = +17.2 (c 0.05, EtOH). The NMR data and spectrums are shown in Additional file 1: Table S2 and Figs. S5–10.
Availability of data and materials
All data for this study are included in this published article and its additional file.
Abbreviations
- aa-tRNA:
-
Aminoacyl-transfer RNA
- BGC:
-
Biosynthesis gene cluster
- CDP:
-
Cyclodipeptide synthase
- DMA:
-
Dimethylallyl
- DKP:
-
Diketopiperazine
- FMN:
-
Flavin mononucleotide
- GNPS:
-
Global natural products social molecular networking
- HPLC:
-
High performance liquid chromatography
- HR–ESI–MS:
-
High resolution–electrospray ionization–mass spectrometry
- ITS:
-
Internal transcribed spacer
- NCBI:
-
National Center for Biotechnology Information
- NMR:
-
Nuclear magnetic resonance
- NRPS:
-
Nonribosomal peptide synthetase
- UPLC–ESI–HRMS:
-
Ultra-high performance liquid chromatography–electrospray ionization–high resolution mass spectrometer
References
Ye Y, Du L, Zhang X, Newmister SA, McCauley M, Alegre-Requena JV, Zhang W, Mu S, Minami A, Fraley AE, et al. Fungal-derived brevianamide assembly by a stereoselective semipinacolase. Nat Catal. 2020;3:497–506.
Klas KR, Kato H, Frisvad JC, Yu F, Newmister SA, Fraley AE, Sherman DH, Tsukamoto S, Williams RM. Structural and stereochemical diversity in prenylated indole alkaloids containing the bicyclo[2.2.2]diazaoctane ring system from marine and terrestrial fungi. Nat Prod Rep. 2018;35:532–58.
Wang X, Lin M, Xu D, Lai D, Zhou L. Structural diversity and biological activities of fungal cyclic peptides excluding cyclodipeptides. Molecules. 2017;22:2069.
Ma Y-M, Liang X-A, Kong Y, Jia B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J Agric Food Chem. 2016;64:6659–71.
Borthwick AD. 2,5-Diketopiperazines: synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem Rev. 2012;112:3641–716.
Yan LH, Li PH, Li XM, Yang SQ, Liu KC, Wang BG, Li X. Chevalinulins A and B, proangiogenic alkaloids with a Spiro[bicyclo[222]octane-diketopiperazine] skeleton from deep-sea cold-seep-derived fungus Aspergillus chevalieri CS-122. Org Lett. 2022;24:2684–8.
Ortiz A, Sansinenea E. Cyclic dipeptides: secondary metabolites isolated from different microorganisms with diverse biological activities. Curr Med Chem. 2017;24:2773–80.
Kanoh K, Kohno S, Asari T, Harada T, Katada J, Muramatsu M, Kawashima H, Sekiya H, Uno I. (-)-Phenylahistin: A new mammalian cell cycle inhibitor produced by Aspergillus ustus. Bioorg Med Chem Lett. 1997;7:2847–52.
Jimenez PC, Wilke DV, Branco PC, Bauermeister A, Rezende-Teixeira P, Gaudencio SP, Costa-Lotufo LV. Enriching cancer pharmacology with drugs of marine origin. Br J Pharmacol. 2020;177:3–27.
Canu N, Moutiez M, Belin P, Gondry M. Cyclodipeptide synthases: a promising biotechnological tool for the synthesis of diverse 2,5-diketopiperazines. Nat Prod Rep. 2020;37:312–21.
Nies J, Li SM. Prenylation and dehydrogenation of a C2-reversely prenylated diketopiperazine as a branching point in the biosynthesis of echinulin family alkaloids in Aspergillus ruber. ACS Chem Biol. 2021;16:185–92.
Wohlgemuth V, Kindinger F, Xie X, Wang BG, Li SM. Two prenyltransferases govern a consecutive prenylation cascade in the biosynthesis of echinulin and neoechinulin. Org Lett. 2017;19:5928–31.
Wohlgemuth V, Kindinger F, Li S-M. Convenient synthetic approach for tri- and tetraprenylated cyclodipeptides by consecutive enzymatic prenylations. Appl Microbiol Biotechnol. 2018;102:2671–81.
Chen X, Si L, Liu D, Proksch P, Zhang L, Zhou D, Lin W. Neoechinulin B and its analogues as potential entry inhibitors of influenza viruses, targeting viral hemagglutinin. Eur J Med Chem. 2015;93:182–95.
Lorenzo P, Alvarez R, de Lera AR. Total synthesis and structural revision of (+)-cristatumin C. J Nat Prod. 2014;77:421–3.
Guo J-P, Tan J-L, Wang Y-L, Wu H-Y, Zhang C-P, Niu X-M, Pan W-Z, Huang X-W, Zhang K-Q. Isolation of talathermophilins from the thermophilic fungus Talaromyces thermophilus YM3-4. J Nat Prod. 2011;74:2278–81.
Li D-L, Li X-M, Li T-G, Dang H-Y, Wang B-G. Dioxopiperazine alkaloids produced by the marine mangrove derived endophytic fungus Eurotium rubrum. Helv Chim Acta. 2008;91:1888–93.
Meng L-H, Du F-Y, Li X-M, Pedpradab P, Xu G-M, Wang B-G. Rubrumazines A-C, indolediketopiperazines of the isoechinulin class from Eurotium rubrum MA-150, a fungus obtained from marine mangrove-derived rhizospheric soil. J Nat Prod. 2015;78:909–13.
Zou X, Li Y, Zhang X, Li Q, Liu X, Huang Y, Tang T, Zheng S, Wang W, Tang J. A new prenylated indole diketopiperazine alkaloid from Eurotium cristatum. Molecules. 2014;19:17839–47.
Wang W-L, Lu Z-Y, Tao H-W, Zhu T-J, Fang Y-C, Gu Q-Q, Zhu W-M. Isoechinulin-type alkaloids, variecolorins A-L, from Halotolerant Aspergillus variecolor. J Nat Prod. 2007;70:1558–64.
Du F-Y, Li X, Li X-M, Zhu L-W, Wang B-G. Indolediketopiperazine alkaloids from Eurotium cristatum EN-220, an endophytic fungus isolated from the marine alga Sargassum thunbergii. Mar Drugs. 2017;15:24.
Zhou L-N, Zhu T-J, Cai S-X, Gu Q-Q, Li D-H. Three new indole-containing diketopiperazine alkaloids from a deep ocean sediment-derived fungus Penicillium griseofulvum. Helv Chim Acta. 2010;93:1758–63.
Li Y, Li X, Kim S-K, Kang JS, Choi HD, Rho JR, Son BW. Golmaenone, a new diketopiperazine alkaloid from the marine-derived fungus Aspergillus sp. Chem Pharm Bull. 2004;52:375–6.
Du H, Wang Q, Yang X. Fu brick tea alleviates chronic kidney disease of rats with high fat diet consumption through attenuating insulin resistance in skeletal muscle. J Agric Food Chem. 2019;67:2839–47.
2ndFind : http ://biosyn.nih.go.jp/2ndfind/.
Guo CJ, Yeh HH, Chiang YM, Sanchez JF, Chang SL, Bruno KS, Wang CCC. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J Am Chem Soc. 2013;135:7205–13.
Balibar CJ, Walsh CT. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry. 2006;45:15029–38.
Stack D, Neville C, Doyle S. Nonribosomal peptide synthesis in Aspergillus fumigatus and other fungi. Microbiology-Sgm. 2007;153:1297–306.
Wang Y, Hu PJ, Pan YY, Zhu YX, Liu XZ, Che YS, Liu G. Identification and characterization of the verticillin biosynthetic gene cluster in Clonostachys rogersoniana. Fungal Genet Biol. 2017;103:25–33.
Chankhamjon P, Boettger-Schmidt D, Scherlach K, Urbansky B, Lackner G, Kalb D, Dahse HM, Hoffmeister D, Hertweck C. Biosynthesis of the halogenated mycotoxin Aspirochlorine in Koji mold involves a cryptic amino acid conversion. Angewandte Chemie-International Edition. 2014;53:13409–13.
Tagami K, Minami A, Fujii R, Liu C, Tanaka M, Gomi K, Dairi T, Oikawa H. Rapid reconstitution of biosynthetic machinery for fungal metabolites in Aspergillus oryzae: total biosynthesis of aflatrem. ChemBioChem. 2014;15:2076–80.
Fischbach MA, Walsh CT. Assembly-Line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–96.
Zhang JR, Liu N, Cacho RA, Gong Z, Liu Z, Qin WM, Tang C, Tang Y, Zhou JH. Structural basis of nonribosomal peptide macrocyclization in fungi. Nat Chem Biol. 2016;12:1001–3.
Walsh CT. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat Prod Rep. 2016;33:127–35.
Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y. Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat Chem Biol. 2012;8:823–30.
Belin P, Moutiez M, Lautru S, Seguin J, Pernodet J-L, Gondry M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat Prod Rep. 2012;29:961–79.
Liu C, Qi J, Liu C: A method for the production of L-tryptophan-L-alanine cyclic dipeptide by Aspergillus oryzae. 2022:No. 202210503663.X.
Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett. 2004;239:79–85.
Tagami K, Liu C, Minami A, Noike M, Isaka T, Fueki S, Shichijo Y, Toshima H, Gomi K, Dairi T, Oikawa H. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J Am Chem Soc. 2013;135:1260–3.
Acknowledgements
We thank Prof. H. Oikawa (Hokkaido University) for providing A. oryzae NSAR1 and the expression vector pUSA2.
Funding
This work was supported by the National Natural Science Foundation of China (Project Nos. 31900064 and 31800031) and the Innovation & Development Joint Fund of Natural Science Foundation from Shandong Province (ZR2021LSW022).
Author information
Authors and Affiliations
Contributions
JQ, JG, and CL designed the study and wrote the manuscript. CX, XX, JG, and CL critically revised the manuscript. JQ, HH, DS, ST, CL, and PW performed the experiments and analyzed the results. XX, JG, and CL designed and supervised the project. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary experimental section. Fig. S1. Mycelial morphology of E. cristatum NWAFU-1. Fig. S2. Molecular network of the metabolic products from E. cristatum NWAFU-1. Fig. S3. Proposed biosynthetic gene clusters of echinulin and function analysis of each gene in the BGC. Fig. S4. Sequence similarity network analysis of CriC. Fig. S5. 1H-NMR spectrum of Cyclo-TA (CD3OD-d4, 500 MHz). Fig. S6. 13C-NMR spectrum of Cyclo-TA (CD3OD-d4, 125 MHz). Fig. S7. HSQC spectrum of Cyclo-TA (CD3OD-d4). Fig. S8. HMBC spectrum of Cyclo-TA (CD3OD-d4). Fig. S9. 1H-1H COSY spectrum of Cyclo-TA (CD3OD-d4). Fig. S10. NOESY spectrum of Cyclo-TA (CD3OD-d4). Fig. S11. HPLC traces of time-course biochemical assays for microsome containing CriC. Fig. S12. Substrate promiscuity analysis of CriC. Fig. S13. HPLC traces of AO-criC product under non-linear increasing concentration gradient substrate feeding. Fig. S14. Domain analysis and speculative reaction mechanism for CriC. Fig. S15. Genome mining-based CriC uncovered serval BGCs responsible for Cyclo-TA containing compounds. Table S1. Primers used for construction of expression plasmids. Table S2. NMR Data of Cyclo-TA in CD3OD-d4 (500 MHz for 1H NMR, 125 MHz for 13C NMR). Table S3. Percent identity matrix of CriC and its homologies in CriC group branch on the evolutionary tree.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Qi, J., Han, H., Sui, D. et al. Efficient production of a cyclic dipeptide (cyclo-TA) using heterologous expression system of filamentous fungus Aspergillus oryzae. Microb Cell Fact 21, 146 (2022). https://doi.org/10.1186/s12934-022-01872-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-022-01872-8