Abstract
Background
Sugars and triglycerides are common carbon sources for microorganisms. Nonetheless, a systematic comparative interpretation of metabolic changes upon vegetable oil or glucose as sole carbon source is still lacking. Selected fungi that can grow in acidic mineral salt media (MSM) with vegetable oil had been identified recently. Hence, this study aimed to investigate the overall metabolite changes of an omnipotent fungus and to reveal changes at central carbon metabolism corresponding to both carbon sources.
Results
Targeted and non-targeted metabolomics for both polar and semi-polar metabolites of Phialemonium curvatum AWO2 (DSM 23903) cultivated in MSM with palm oil (MSM-P) or glucose (MSM-G) as carbon sources were obtained. Targeted metabolomics on central carbon metabolism of tricarboxylic acid (TCA) cycle and glyoxylate cycle were analysed using LC–MS/MS-TripleQ and GC–MS, while untargeted metabolite profiling was performed using LC–MS/MS-QTOF followed by multivariate analysis. Targeted metabolomics analysis showed that glyoxylate pathway and TCA cycle were recruited at central carbon metabolism for triglyceride and glucose catabolism, respectively. Significant differences in organic acids concentration of about 4- to 8-fold were observed for citric acid, succinic acid, malic acid, and oxaloacetic acid. Correlation of organic acids concentration and key enzymes involved in the central carbon metabolism was further determined by enzymatic assays. On the other hand, the untargeted profiling revealed seven metabolites undergoing significant changes between MSM-P and MSM-G cultures.
Conclusions
Overall, this study has provided insights on the understanding on the effect of triglycerides and sugar as carbon source in fungi global metabolic pathway, which might become important for future optimization of carbon flux engineering in fungi to improve organic acids production when vegetable oil is applied as the sole carbon source.
Similar content being viewed by others
Background
Glucose is a common carbon source in fermentation. Recently, several studies have been done on plant triglycerides or vegetable oil to replace glucose in fermentation [1,2,3,4,5]. For instance, palm oil was found to not only replace lactose but led to a better cell growth and penicillin production in Penicillium chrysogenum culture [3]. Soybean oil was explored and successfully used to culture Ashbya gossypii for the better production of riboflavin [1]. Recently, Phialemonium curvatum AW02 was shown to grow well in acidic (pH 2 – 3) minimal media with rapeseed oil as the carbon and energy source [6]. These studies showed that plant triglycerides could serve as an alternative carbon source for microbial growth. Knowing that only certain lipase-secreting microorganisms are able to grow on this carbon source, triglycerides is selected as the carbon source in selective minimal media. It can suppress the growth of unwanted bacterial contaminant since they are unable to hydrolyse the plant triglyceride [6].
P. curvatum AW02 has been shown to generate an extracellular lipase, even active at acidic pH of 3 [6]. Lipases which are active at acidic pH are found in mammals but are rare in microorganisms [7]. Recently, P. curvatum AW02 was found to produce several secondary metabolites including 4-hydroxybenzoic acid, a commercial metabolite used as drugs preservative [8]. Furthermore, the study also identified 3-indole acetic acid which is a well-known plant hormone and solaniol, a naphthoquinone derivative [8]. These growing number of studies on P. curvatum highlighted its significance in biotechnology.
Vegetable oil including palm oil is known to contain triglyceride, free fatty acid, and vitamin. Triglycerides are the major component in vegetable oils which consist about 95 – 98% [9]. Genera of filamentous fungi comprise of Aspergillus, Rhizopus, Penicillium, Mucor, Geotrichum, and Fusarium were shown able to degrade triglycerides into free fatty acids through hydrolysis process with extracellular lipases [10]. The liberated free fatty acids can be taken by the microorganism and undergo β-oxidation to produce acetyl-CoA [1]. The acetyl-CoA will then take part in glyoxylate cycle which functions to synthesize malate and via PEP to produce all anabolites which will be needed [11] when glucose is depleted [12]. On the other hand, fatty acid is highly reduced and therefore known to provide six times more energy when compared to polysaccharide if the bound water is taken into consideration [13]. In the central carbon metabolism understanding, many studies have provide insights on the organic acids concentration in TCA cycle for several fungal species grown on glucose [14, 15]. On the contrary, the details on concentration for each organic acid in the glyoxylate pathway is still lacking, although extensive studies have showed that glyoxylate pathway is activated in many fungal species that grown on non-sugar carbon sources [16,17,18,19].
Hereby, taking the advantage of P. curvatum AW02 that able to grow on sugar or plant triglycerides in minimal media, this study aimed to provide the details on the changes in central carbon metabolism for the species. In addition, systematic comparative interpretation of metabolic changes is also possible with current metabolomics platform. Hence, the analysis of metabolic changes for both non-targeted metabolites and targeted organic acids which involved in tricarboxylic acid (TCA) cycle or glyoxylate pathway of P. curvatum AW02 cultivated in minimal media with palm oil replacing glucose as sole carbon source was conducted. For the first time, the effect of triglycerides replacing sugar as carbon source on fungal metabolism was investigated using a systematic comparative metabolomics approach. The findings have provided insights that are important for the development of metabolic engineering, in particular on organic acids production of omnipotent fungi that grow on vegetable oil.
Results
Growth comparison of P. curvatum AW02 in MSM-P and MSM-G media
P. curvatum AW02 showed similar growth rate on MSM-P and MSM-G agar, except the growth for MSM-G culture can be measured starting at day 2, as compared to MSM-P culture at day 3 (Fig. 1a). Nonetheless, similar growth rate of P. curvatum AW02 on MSM-P and MSM-G agar indicates that palm oil is an efficient carbon source comparable to glucose (Table 1).
a Colony diameter profile of P. curvatum AW02 on MSM-P agar and MSM-G agar at 33 °C. Colony diameters were determined every 24 h. Values are given as mean of three independent experiments. Standard error was below 20%. b Biomass of P. curvatum AW02 after 5 days culture in MSM-P and MSM-G broth. c Biomass profile of P. curvatum AW02 cultivated in MSM-P and MSM-G. d Lipase activity at day 5 from media of MSM-P or MSM-G cultured with P. curvatum AW02. Lipid droplets inside the cells of P. curvatum AW02 after 5 days cultured in media: e Cells harvested from MSM-P culture; f Cells harvested from MSM-G culture. Cells were stained with Nile red and observed using Axiostar fluorescence microscope equipped with filter combination of excitation filter BP 550/25 and 470/40 nm. Final magnification was ×400. The arrows indicate the lipid bodies. The box (a) showed the abundance of lipid bodies with elongated shape in hyphae from MSM-P culture while box (b) showed the scattered and rounded lipid bodies in hyphae from MSM-G culture
Biomass production of P. curvatum AW02 in both MSM-P and MSM-G were found similar (4.49 ± 1.6 g/L and 4.35 ± 0.2 g/L, respectively after 5 days of cultivation (Fig. 1b). Nonetheless, prolonged cultivation of P. curvatum AW02 in MSM-P up to stationary phase (12 days) produced 10 g/L of biomass, which is 2.5-fold higher as compared to MSM-G culture, which started its stationary phase at day 6 (Fig. 1c). Palm oil as carbon source in defined media has been used for various microorganism cultivation and was found to produce high biomass. For example, Penicillium chrysogenum Hl107 was grown up to 10 g/L at 28 °C on basal media [3].
Lipase activity of P. curvatum AW02 in MSM-P culture
Lipase activity was detected in MSM-P culture. However, when glucose was used as carbon source, no lipase activity was detectable (Fig. 1d). Since the detection limit was < 3 U per 100 mL, an induction factor of the respective lipase gene of at least 100 can be concluded.
Lipid bodies visualization
P. curvatum AW02 culture in MSM-P was found to pose abundance of lipid bodies with elongated shape compared to MSM-G culture (Fig. 1E and F). This observation suggested that free fatty acid from palm oil that had been liberated by the fungal lipase was likely uptaken and accumulated in P. curvatum AW02 as triglyceride containing lipid bodies in intracellular compartment. The abundance of lipid bodies in intracellular compartment of MSM-P culture may contribute to the high biomass production. This was supported by total cellular lipid quantification, in which the amount of cellular lipids in MSM-P culture was found 13-fold higher in compared to MSM-G culture (Additional file 1: Fig. S1). It has been known that cells can accumulate the fatty acids and form lipid bodies under high carbon concentration and thus increase the cell biomass [20]. Free fatty acid is known to undergo β-oxidation to produce acetyl-CoA for carbon metabolism in TCA cycle [21]. It is possible that the liberated free fatty acid from palm oil might be accumulated and used to synthesise lipid in intracellular compartment as energy storage.
Untargeted metabolite profiling
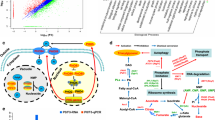
Untargeted profiling of intracellular metabolites of both MSM-P and MSM-G culture using LC–MS-TOF had detected 144 metabolites based on FMF algorithm (Additional file 1: Table S1), with 30 metabolites were successfully identified by cross-checking with QC (Table 2, Additional file 1: Fig. S2). These metabolites that belongs to the classes of sugars, lipids, nucleotides, organic acids, and coenzymes were subjected for multivariate analysis to compare the metabolome differences of MSM-P and MSM-G culture. Principal component analysis (PCA) score plot with a good fit of R2X (0.83) clustered MSM-P and MSM-G into two separate groups (Fig. 2a), indicating that P. curvatum AW02 undergoes metabolic changes when palm oil replaces glucose as carbon source. This observation was validated using QC with the intensity’s deviation was < 2SD as shown in Additional file 1: Fig. S3.
Unlike intracellular, untargeted profiling of extracellular metabolites of both MSM-G and MSM-P cultures have detected trehalose as the only molecule with above the limit of detection. This result indicates that P. curvatum is not active in secreting polar molecules outside the cells. Trehalose is a non-reducing sugar and an abundance compatible solute in fungal cells is known for its functions as stress protectant [22]. The finding suggests that trehalose is likely an important stress protectant molecule for P. curvatum, similar to Xanthophyllomyces dendrorhous strains that accumulate trehalose as stress protectant against reactive oxygen species [23].
To distinguish specific intracellular metabolites that significantly differ between MSM-P and MSM-G cultures, partial least squared-discriminant analysis (PLS-DA) was carried out. The PLS-DA plot shows the difference of MSP-P and MSM-G metabolome as explained by PLS 1 with 0.658 (Fig. 2b). Further analysis based on the VIP > 0.95, p > 0.01, and FDR cut-off 0.01 have identified seven 7 intracellular metabolites that have significant difference between MSM-P and MSM-G (Table 2). These metabolites include trehalose, glycerophosphocholine, sn-glycero-3-phospho-1-inositol, citric acid, and two unknown metabolites with less abundance in MSM-P culture. Only one unknown metabolite was found highly abundant in MSM-P culture.
Changes in targeted organic acids concentration upon glucose substitution of palm oil in minimal media cultures
To understand the central carbon metabolism of an omnipotent fungus upon catabolising carbohydrates and triglycerides as sole carbon source, all organic acids involved in TCA and glyoxylate cycle were targeted and quantified from intracellular extracts of MSM-P and MSM-G cultures (Fig. 3) (Additional file 1: Table S2).
The organic acids and enzyme activities in TCA and glyoxylate cycle from intracellular extracts of P. curvatum AW02. Organic acid concentration was indicated with mg/g biomass and enzymatic activity was measured in U/g biomass. Asterisk indicated significant difference enzymatic activity with p < 0.05. LOD: limit of detection, Trace: limit of quantitation (LOQ). Details on the LOD and LOQ for each targeted organic acid was described in Additional file 1: Table S4
In MSM-G culture, the organic acids from TCA cycle were all detected and quantified. The results show that P. curvatum AW02 grown on glucose produced high malic acid (159 mg/g) compared to other TCA intermediates such as citric acid (84 mg/g) and succinic acid (21 mg/g). Ratio of organic acids composition in the TCA cycle of P. curvatum AW02 was found similar with Y. lipolytica that grown on glucose, in which high concentration of malic acid was observed, followed by fumaric acid and citric acid [24]. Nonetheless, this ratio of organic acids composition profile was shown to be different than S. cerevisiae. It was proposed that intracellular metabolite concentrations in TCA cycle is species-specific [14].
In MSM-P culture, only trace concentration of alpha-ketoglutaric acid was observed while the fumaric acid was under limit of detection. These showed that the TCA cycle for MSM-P that we observed was incomplete, suggesting that MSM-P culture recruit glyoxylate cycle as the key metabolic pathway at central carbon metabolism. This was supported by higher abundance of glyoxylic acid in MSM-P compared to the extract of MSM-G culture (Additional file 1: Fig. S4B). Furthermore, the citric acid, succinic acid, malic acid, and oxaloacetic acid were found at ~ 4- to 8-fold lower compared to the MSM-G culture. This observation is in agreement with the previous Y. lipolytica study that showed high concentration of malic acid, fumaric acid, succinic acid, and pyruvic acid in glucose-grown when compared to oleic acid-grown [24].
Enzymatic assay of ICL, IDH, and SDH
To further understand the correlation of organic acids concentration and key enzymes involved in the activation of glyoxylate cycle upon growing on different carbon source, the activities of selected enzymes in TCA and glyoxylate cycle were determined by enzymatic assays. Isocitrate lyase (ICL) and isocitrate dehydrogenase (IDH) involved in regulating TCA and glyoxylate cycle were shown significantly up-regulated (3-fold) and down-regulated (−3-fold), respectively when palm oil was used as carbon source (Fig. 3). These results are in agreement with the low concentration of α-ketoglutaric acid observed in MSM-P culture. Up-regulation of ICL and down-regulation of IDH is known for the activation of glyoxylate cycle. ICL cleaves isocitric acid to glyoxylic acid and succinic acid, which limits the production of α-ketoglutaric. The low activity of IDH may be due to the phosphorylation of IDH, which could happen when the fungus grows under glucose starvation condition [25].
Metabolic pathway analysis
The analysis was carried out to identify the changes in metabolic pathway of P. curvatum AWO2 grown on two different carbon sources. Saccharomyces cerevisiae metabolome (Kyoto Encyclopaedia of Genes and Genomes) was used as the model organism for metabolic pathway topology and enrichment analysis. Combinations of metabolites obtained from identification Level 1 and 2 of untargeted metabolite profiling (Table 2) and targeted organic acid (Additional file 1: Table S2) were used in the analysis.
The results show that glyoxylate and dicarboxylate metabolism, tricarboxylic acid cycle (TCA cycle), and pyruvate metabolism were regulated differently when palm oil replaced glucose as carbon source (-log(p) > 3, pathway impact > 0.1, Fig. 4, Additional file 1: Table S3). This revealed that consumption of palm oil as carbon source results in metabolic changes at central carbon metabolism, which involves glyoxylate and dicarboxylate metabolism, TCA cycle, and pyruvate metabolism. Nonetheless, other metabolism like amino acids metabolism (alanine, aspartate, and glutamate metabolism), glycerophospholipid metabolism, glycolysis or gluconeogenesis, starch and sucrose metabolism, and purine metabolism were also significantly affected but with lower impact value.
Summary of metabolic pathway analysis using MetaboAnalyst 3.0. The analysis is a combination of pathway enrichment analysis and pathway topology analysis. Pathway enrichment analysis provided the significant value of -log(p) (axis-y) for each pathway based on value of each metabolite. Pathway topology analysis gave pathway impact value (axis-x) based on the centrality of a metabolite within a pathway. Red colour represents the most significant regulated pathway while the size of spot represents pathway impact value
Discussion
Proposed metabolic pathway network of P. curvatum AW02 grown on palm oil replacing glucose as carbon source
To obtain better insights on the possible metabolic pathway network of P. curvatum AW02 that utilised palm oil, data from enzymatic assay (lipase, IDH, ICL, and SDH), untargeted metabolite profiling, and targeted organic acid analysis were integrated using Vanted (Visualization and Analysis of Networks containing Experimental Data) software [26] (Fig. 5).
In MSM-P medium that contains palm oil as the sole carbon source, high activity of extracellular lipase was detected (Fig. 1D & 5). The recorded lipase activity of 300 U/100 mL in MSM-P was found, which is 2.8-fold higher as compared to previous study (34 – 170 U/100 mL), in which P. curvatum AW02 was cultivated on rapeseed oil as carbon source [6]. When glucose was the sole source of carbon and energy, lipase activity was found to be 10−2-fold lower. The mechanism of regulation occurs probably on the level of transcription as shown in other ascomycetes including Beauveria bassiana that recently reported [27] where deletion of the gene Bbctf1β, which encoding a zinc finger transcription factor, led to a significant reduction of the expression of nine lipase genes.
The lipase activity hydrolyses triglyceride from palm oil into glycerol and free fatty acids. Fatty acids are known to be transported into the intracellular compartment and are further degraded during β-oxidation process [21]. To grow on the obtained acetyl-CoA, isocitrate lyase will need to provide malate synthase with glyoxylate. The detected increase in isocitrate lyase activity is expected because ICL is known to be repressed by transcription factor cre A in Aspergillus species [28], which ortholog also identified as mig 1 in Saccharomyces cerevisiae [29, 30]. This, regulation is well known as Carbon Catabolite Repression [31].
The concentration of organic acids in TCA/glyoxylate cycle was found to be 4- to 8-folds lower in palm oil-grown as compared to glucose-grown (Fig. 3 & 5). The low concentration of organic acids from fungi that grow on hydrophobic substrate such as vegetable oil might due to the accumulation of lipid [24, 32]. In agreement to this, this study observed abundance of lipid bodies in the intracellular compartment of palm oil-grown when compared to the glucose-grown.
This study also revealed that palm oil substitution affects the metabolisms that are highly dependent on carbon metabolism. For instance, trehalose concentration in palm oil-grown mycelium was found to be 4-fold lower compared to glucose-grown culture (Additional file 1: Table 2, Additional file 1: Fig. S5). Two explanations are possible: (i) Due to the limited supply of glucose, biosynthesis of trehalose have to run via gluconeogenesis after catabolism of triglyceride in palm oil-grown whereas glucose-grown can simply take up the monomer and convert it into trehalose, (ii) Since osmostress is higher in glucose-grown hyphae, more trehalose is needed and synthesised by glucose-grown in comparison with a medium containing the weakly soluble lipid. The function of trehalose as the compatible solute in fungi is well-known [33]. Trehalose protects proteins and membranes from the osmotic dehydration [34]. A previous study showed that an increase of trehalose concentration in both, intra- and extracellular room, was due to the increased osmotic pressure on Corynebacterium glutamicum [35]. To reveal the possible role of trehalose that was 4-fold higher in MSM-G grown media, the osmolality of MSM-G and MSM-P was measured. The results showed that MSM-G medium indeed had higher osmolality (91 ± 6 mOsm/kg) than MSM-P medium (71.8 ± 7 mOsm/kg). The osmolality of MSM-G was then significantly reduced after 5-days cultivation most probably caused by an up-take of glucose by P. curvatum AW02 (Additional file 1: Table S5 & Fig. S6) and followed by the increase of trehalose concentration. Note that, the MSM-G and MSM-P media after 5-days cultivation reached a similar osmolality level. Since osmoregulation works fast the remains of trehalose appear as an anabolic memory.
On the other hand, the metabolism that is less influenced by carbon metabolism were found not to be affected by palm oil substitute glucose as carbon source. For instance, glutathione concentration was found similar in both palm-oil grown and glucose-grown biomass extracts (Table 2, Additional file 1: Fig. S5). Previous study showed that glutathione was highly affected by the source of nitrogen [36]. Thus, the replacement of glucose with palm oil as carbon source did not affect glutathione biosynthesis since both cultures were supplemented with potassium nitrate as nitrogen source.
This study identified sn-glycero-3-phospho-1-inositol and sn-glycero-3-phosphoethanolamine, a candidate of glycerophospholipids that may play a role in the lipid bodies formation of P. curvatum AW02 (Fig. 5). It is known that oleaginous fungi commonly form a monolayer of phospholipids coating intracellular lipid bodies [37]. Hence, this study hypothesised that the identified glycerophospholipids may have contributed to the formation of lipid bodies, which was visualized in palm oil-grown hyphae (Fig. 1E & F). Furthermore, this molecule had been reported to correlate with membrane integrity during adaptation of Candida glabrata towards osmotic stress [38]. The level of membrane integrity was also proposed to be correlated with the co-existence of fatty acid such as palmitoleic acid, as has been shown in S. cerevisiae [39, 40]. Note that, palmitoleic acid is one of the fatty acids found in palm oil [41]. Taken all together, this study hypothesised that the glycerophospholipids may have taken part in: (i) Lipid bodies formation and (ii) Membrane cells integration, which cause the glycerophospholipids to be less abundant in the palm oil-grown intracellular sample.
Conclusions
In summary, this study investigated the metabolite and central carbon metabolism changes of P. curvatum AW02 based on the mineral salts medium with different carbon sources; palm oil versus glucose. The replacement of glucose with palm oil was shown to induce physiology changes in P. curvatum AW02 including longer lag phase, lipase activity, and contain abundance of elongated lipid bodies in the hyphae. Comparative metabolomics reveals significant changes at the central carbon metabolism involving glyoxylate and TCA cycles as well as pathways that are related to the carbon metabolism. As expected, an activation of glyoxylate pathway was shown in lipid-degrading hyphae through the detection of higher abundance of glyoxylic acid and up-regulation of isocitrate lyase activity. Concerning trehalose, lower contents in the extracts of palm oil-grown hyphae were expected and found. Trehalose, a stress response metabolite indicated less stress in palm oil-grown; i.e. due to its reduced osmotic activity. The strong staining response of intracellular lipid suggested that it is the reserve. This study suggested that palm oil is a potential alternative carbon source for a minimal sterilised fermentation process. More importantly, the results showed that the concentration among each organic acid in the TCA and glyocylic pathway varied significantly. As malic and oxaloacetic acids concentration were found higher in glucose-grown cultures, citric acid was found the most abundance at palm oil-grown conditions. This provides evidence that central carbon metabolism of the omnipotent filamentous fungus P. curvatum undergone significant changes in response towards the triglyceride and sugar-based carbon source. It also suggests that a carbon flow optimisation may have taken place in the cells during palm oil as carbon source. This observation is important for the development of strategies in fermentation processes that can be implemented on organic acids production or secondary metabolite such as 4-hydroxybenzoic acid.
Methods
Fungus
P. curvatum AW02 (DSM 23903) was pre-cultured on rich media HA agar prior to culture in mineral salt media (MSM) with different carbon sources. Cultivations were carried out in 500 mL Duran baffled flasks (Schott, Germany) containing 100 mL medium at 33 °C with agitation of 120 rpm [6].
Medium composition
Rich media (HA) contained 10 g/L yeast extract (YE), 10 g/L glucose, and 20 g/L agar. Modified mineral salt media (MSM) contained 1.5 g/L KNO3, 0.5 g/L MgSO4·7H2O, 0.5 mg/L FeSO4·7H2O, 0.5 mg/L ZnSO4·7H20, 0.02 mg/L CuSO4·5H2O, 0.02/L mg MnCl2·4H2O, and 1.5 g/L KH2PO3. MSM was added with 18 mL of crude palm oil/L (CPO) as MSM-P or added with 10 g/L glucose as MSM-G. No growth factors including vitamins were included. To prepare MSM-P or MSM-G agar plate, 20 g of bacteriological agar was applied per litre media. All media was adjusted to pH 3 using HCl [6].
Colony growth rate determination for P. curvatum AW02
The growth rate of P. curvatum AW02 in HA medium (control), MSM-P, and MSM-G was determined according to [6]. In short, 10 µL of glycerol stock was inoculated on HA, MSM-P, and MSM-G agar. The experiment was prepared in triplicate. Duplicated experiment was performed. Visible colony diameter was measured on daily basis. The growth rate, Kr was determined by calculating slope of exponential phase, where Kr = (R1-R0)/(t1-t0) [42, 43] which R0 and R1 represent the colony diameter at time t0 and t1, respectively.
Biomass production
About 10 µL of mycelia glycerol stock of P. curvatum AW02 previously cultured in HA broth for 5 days was inoculated on HA agar and incubated at 30 °C for minimum 3 days as described previously [6]. Young mycelia (the outer colony) were collected and weighted before transferred into 50 mL falcon tube that contained 30 glass beads (5 mm diameter) and 10 mL of 0.9% (w/v) NaCl. The sample was vortexed until fine dispersed mycelia suspension was obtained. The final inoculums at concentration of 2 mg/mL was prepared.
Two mL of P. curvatum AW02 inoculums (2% v/v) was inoculated into baffle flask (500 mL) that contained 100 mL of MSM-P or MSM-G broth. The cultures were incubated for 12 days. Sample was harvested for every 2 days. The experiment was prepared in triplicate. To harvest the biomass, two steps filtration was applied. Firstly, the excess palm oil that may still stick on mycelium with organic solvent ethyl acetate was removed. Secondly, the biomass was obtained (data not published). In short, the culture was filtered through a 0.2 µm nylon filter membrane (Sartorius Stedim, Germany). The retained mycelium was then mixed with 1:1 of ethyl acetate by vigorous shaking. The mixture was rested ~ 10 min until clear separation between organic and aqueous phase was obtained. Later, the organic phase that contained crude palm oil was removed and the aqueous phase that contains mycelium was filtered through a 0.2 µm nylon filter paper (Sartorius Stedim, Germany). The retained mycelium was dried overnight in incubator oven at 70 °C before weighted as dry biomass.
Lipase assay
P. curvatum AW02 that cultured in MSM-P and MSM-G were harvested and centrifuged at 6000 × g for 6 min at room temperature. To detect the extracellular lipase of P. curvatum AW02, 100 μL supernatant of MSM-P or MSM-G culture were used to incubate with 900 μL substrate containing pNPP (p-nitrophenylpalmitate) for 4 min at room temperature to measure lipase activity. The substrate was freshly prepared by mixing solution A (30 mg of pNPP dissolved in 10 mL isopropanol) and solution B (0.8 mg of Triton X-100 and 0.1 mg gum arabicum that dissolved in 100 mL 0.1 M Tris–HCl, pH 8) in a ratio of 1: 9. The lipase activity was measured according to the release of p-nitrophenol from pNPP upon hydrolysis process by extracellular lipase through spectrophotometer absorbance at 405 nm. One unit (U) was defined as the amount of lipase needed to liberate 1 µmol p-nitrophenol per minute under the previous described conditions [6].
Lipid bodies visualization
Lipid bodies visualization was carried out to observe the possible changes in lipid bodies presence in P. curvatum AWO2 cells grew on palm oil as compared to glucose as carbon source. About 100 µL of P. curvatum AW02 that cultured in MSM-P and MSM-G broth were mixed with 10 mL Nile red solution (1 mg/mL acetone). The samples were mounted on a glass slide and viewed using Zeiss fluorescence microscope (ZEISS, Germany) with the filter combination BP 375-425/FT 425/LP. Lipid bodies were visualised as intense yellow fluorescence [20, 44].
Total cellular lipid
Lipid was extracted out from intracellular compartment by using chloroform–methanol method [45]. Briefly, samples from MSM-P and MSM-G cultures were harvested to obtain biomass. The biomass was freeze dried and suspended in mixture of chloroform/methanol solvent (2:1), before vortexed with glass beads for 20 min. Then, the organic phase was washed with 0.4 mL of 0.9% NaCl (w/v) before being dried at 60 °C for overnight. The total cellular lipid content was expressed as gram of lipid per gram of biomass (%).
Enzymatic assay of isocitrate lyase, isocitrate dehydrogenase, and succinate dehydrogenase assay
The enzymatic activity of isocitrate lyase was assayed using ICL assay kit (MyBioSource, USA) as manufacturer’s instructions. One unit of ICL activity is defined when the enzyme decomposes of 1 µmol of the NADH per minute. Meanwhile, the level of isocitrate dehydrogenase was determined using IDH activity assay kit (Sigma-Aldrich, Malaysia) as per manufacturer’s instructions. One unit of IDH is the amount of enzyme that will generate one μmol of NADH or NADP per minute at pH of 8.0 and at 37 °C. The activity of succinate dehydrogenase was determined by a colorimetric method using succinate dehydrogenase activity colorimetric assay kit (BioVision, USA). One unit of SDH is the amount of enzyme that generates 1 µmol of dichlorophenolindophenol (DCIP) per minute at pH of 7.2 and at 25 °C.
All reactions were carried out in microtiter plates and scanned at each respective wavelength using microtiter plate reader (Tecan, Switzerland).
Determination of osmolality
The osmolality of the medium MSM-G and MSM-P were analysed by using a Micro-Osmometer Model 3320 (Advanced Instruments Inc., USA) [46, 47]. Briefly, 20 μL samples of medium before cultivation and after 5 days cultivation were analysed in triplicates using the osmometer with distilled water as a reference. The results were given as mOsm/kg.
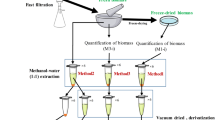
Metabolomics analysis
To perform metabolomics analysis of P. curvatum AW02 samples, instruments including LC–MS-TOF/LC–MS/MS-QTOF, LC–MS/MS-TripleQ, and GC–MS were employed, similar to previous study by [48] and the workflow is summarised in Additional file 1: Fig. S7.
Metabolites extraction
The following steps were carried out as described previously [49, 50]. One volume of sample culture was mixed with 5 volumes of quenching solution (60% methanol, 10 mM HEPES, pH 7.5, −40 °C). Later, the mixture was kept at −40 °C for 3 – 5 min. The mixtures were centrifuged (5000 g, 6 min at 0 °C) to separate pellet/mycelia (intracellular) and supernatant (extracellular). The extracellular sample was used directly for metabolite analysis. The intracellular metabolites sample from mycelia was extracted using 5 mL extraction solution (75% ethanol, 10 mM HEPES, pH 7.5) at 80 °C and incubated for 5 min. Then, the lysate was chilled in ice for 5 min and subsequently centrifuged for 5000 g at 4 °C for 10 min. The supernatants of cell lysate or extract of polar and semi-polar metabolites were used for the analysis. Finally, camphorsulfonic acid and gallic acid were spiked into the cell lysate as internal standard with the final concentrations of 10 ppm.
LC–MS-TOF condition and data acquisition
Sample extracts from previous section were analysed using UPLC-ESI–MS Micro Time of Flight (Bruker, Germany). Samples were separated with reverse phase chromatography using C18 column at 45 °C. The mobile phase was water with 0.1% formic acid (A) and acetonitrile (B). The gradient flow of mobile phase composition for liquid chromatography (LC) was programmed as follows: 5 to 40% B (v/v) in 3 min, then to 95% B (v/v) until 5 min and hold for 10 s, sharply decreased to 5% B (v/v) and maintained until 15 min. The flow rate was 0.3 mL/minute and injection volume was 3 μL [51].
In mass spectrometry (MS), mass spectra were generated by electrospray ionization (ESI) in negative mode with a range of m/z 50 to 1000 for scanning. The acquisition parameters were as follows: 4 kV of capillary voltage, 8 L/minute of dry gas, 200 °C of dry gas heater, nebulizer at 1.2 bar. The calibration of MS was done in every sample injection using 180 μL/hour of a sodium formate cluster mix which contained minimum 7 calibration points with mass range 112.9856 – 996.8221 m/z to check mass precision in each run.
Data processing, multivariate, and statistical analyses
The mass spectra data were processed using Compass software of DataAnalysis and ProfileAnalysis (Bruker, Germany), which served to align the recorded m/z and retention from each sample in a bucket form. Find molecular features (FMF) algorithm was used by performing the advanced bucketing, where each bucket will represent metabolites and contains intensity value [52]. The data matrix that contain the list of all buckets or metabolites along with their intensity values were generated and exported to metaboanalyst online programme (http://www.metaboanalyst.ca) for normalisation of intensities and statistical analysis. Normalisation was done based on spikes with an internal standard in each sample.
For statistical analysis, Student’s t test was performed using both MSM-P and MSM-G cultures, treated as paired sample. The significant changes in metabolites between these culture set were identified at p < 0.01 and false discovery rate (FDR) cut-off at 0.01.
SIMCA 14.1 (Umetrics, Sweden) was used for multivariate analysis. The normalised data matrix was scaled using Pareto before analysed using principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA). Pareto scaling was used to overcome the problem of strong variations of intensity between different metabolites. To identify the metabolites that contribute to metabolites variation, they were arranged according to their importance in projecting the variations. Metabolites with variable importance in the projection (VIP) values more than 0.95 were identified as the metabolites that significantly changes between MSM-P and MSM-G cultures [53].
Method validation, quality control, and data treatment
Internal standard of gallic acid was used to demonstrate precision, stability, and repeatability of the method according to the variation of peak intensities and retention times. The average recorded retention time and m/z were 5.5 min: 169.013 m/z in negative ion mode. The relative standard deviations (RSDs) of peak intensities and retention times (RT) were estimated to be 6 and 0%, respectively.
Pooled sample of MSM-P and MSM-G culture were prepared for each set of intracellular and extracellular and treated as quality control (QC). Later, the generated data matrix from ProfileAnalysis (Bruker, Germany) software were filtered by cross-checking each sample of MSM-P or MSM-G culture with QC sample to ensure a good repeatable analysis [54].
LC–MS/MS-QTOF and metabolite identification
Automated fragmentation had been applied for all acquit precursor ion mass in negative mode using UPLC-ESI–MS/MS Micro Quadrupole-Time of Flight (Bruker, Germany). The metabolites were tentatively identified at fragmentation level. The mass of precursor ion during MS were corroborated with their product ion during MS/MS fragmentation. Later, the information was used to identify the metabolite based on fragments hit and neutral loss using online database, METLIN (https://metlin.scripps.edu) and MassBank (http://www.massbank.jp). The neutral loss calculation was performed according to loss of functional group such as carboxyl during fragmentation or ion adduct that react to precursor ion during ionization process [55]. In-silico fragmentation using MetFrag (https://msbi.ipb-halle.de/MetFragBeta/) was applied when both fragments hit and neutral loss provided no information from online database.
The level of identification for metabolites identified in this study was ranged based on criteria described previously [56] with some modifications. Identification Level 1 was assigned for metabolites that validated with authentic standard, whereas Level 2 was for tentatively identified metabolites that could not be validated by authentic standard. In the case of daughter ions that were not unavailable, parent ions were used for the identification and assigned as Level 3. The identification at this level was stated as tentatively characterised class. Identification Level 4 applied for unknown metabolites.
UPLC-ESI–MS/MS TripleQ condition and data acquisition for targeting organic acids
Targeted organic acids in intracellular of MSM-P and MSM-G culture were quantified using UPLC-ESI–MS/MS Triple quadropole (Waters, USA). The intracellular metabolites samples from metabolite extraction section were used and separated using Acquity UPLC HSS T3 1.8 µm, C18 column at temperature of 45 °C. The mobile phase, gradient flow, and mass spectra acquisition parameter were same as LC–MS-TOF as described previously.
Organic acids quantification
Multiple reactions monitoring (MRM) was used to target organic acid in intracellular samples of MSM-P and MSM-G culture along with their daughter ions during fragmentation. Collision energy and optimal depolarization potentials of targeted organic acids was first determined by injecting organic acids standards into the mass spectrometer (Table 3). The chromatogram of each organic acid standard with their respective RT is shown in Additional file 1: Fig. S8.
The concentration of targeted organic acids was determined according to the standard curve of organic acids standards with concentration of 0.2, 0.4, 0.6, 0.8, 1.0, 2, 4, 6, 8, and 10 ppm using TargetLynx™ software (Waters, USA).
Detection and quantification of glyoxylic acid via GC–MS
The intracellular metabolites samples from metabolite extraction section were incubated with 50 µL methoxyamine solution (25 mg/mL methoxyamine hydrochloride in pyridine) for 30 min at 60 °C. Subsequently, 50 µL of TMS reagent (BSTFA/TMCS, 99:1) was added into the mixture and incubated for 60 min at 60 °C. In this derivatization step, the keto group of glyoxylic acid will be methoxylated into methoxyamino groups, whereas hydroxyl group will be added with trimethysilyl during trimethysilation [48].
Glyoxylic acid detection and quantification was performed using GC–MS (Agilent, USA) coupled with BP-20 polar column (30 m × 250 μm, 0.25 μm film thickness). Helium gas was used as carrier at 1.3 mL/min. One μL sample was injected in split-less mode via autosampler. The inlet temperature was set at 280 °C. The temperature program of oven was started with 40 °C for 3 min and gradually increased with the rate of 5 °C/min for 36 min until the temperature achieved 220 °C, and maintained for 3 min. Mass spectra were collected with the range of 45 to 600 m/z with 7 min solvent delay.
Single ion monitoring (SIM) mode was used to detect glyoxylic acid from sample. Initially, glyoxylic acid standard was analysed through GC–MS using scan mode to obtain mass spectra. The gathered mass spectra (Additional file 1: Fig. S4A) was cross checked with previous study [48] and used as reference in SIM mode to detect glyoxylic acid from sample.
Standard curve was built according to glyoxylic acids standards measured at various concentration (62.5, 100, 125, 500, and 1000 ppm). Later, the quantification of glyoxylic acid in sample was based on the developed standard curve.
Statistical analysis
Student’s t-test was performed in lipase and enzymatic assay to verify the differences between MSM-P and MSM-G cultures from the obtained data. Each experiment was repeated at least 2 times with each experiment run in triplicate. The probability was set at p < 0.05. Statistical analyses were performed using the software Statistical Package for the Social Sciences (SPSS) 18.0 (IBM, USA).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files.
Abbreviations
- MSM-P:
-
Minimal medium with palm oil as carbon source
- MSM-G:
-
Minimal medium with glucose as carbon source
- PCA:
-
Principal component analysis
- PLS-DA:
-
Partial least square discriminate analysis
- VIP:
-
Variable importance in projection
- ICL:
-
Isocitrate lyase
- IDH:
-
Isocitrate dehydrogenase
- SDH:
-
Succinate dehydrogenase
- QC:
-
Quality control
References
Lim S, Choi J, Park E. Microbial production of riboflavin using riboflavin overproducers, Ashbya gossypii, Bacillus subtilis, and Candida famate: an overview. Biotechnol Bioproc E. 2001;6(2):75–88.
Park EY, Ming H. Oxidation of rapeseed oil in waste activated bleaching earth and its effect on riboflavin production in culture of Ashbya gossypii. J Biosci Bioeng. 2004;97(1):59–64.
Tan IK-P, Ho C-C. Growth and the production of penicillins in Penicillium chrysogenum with palm oil and its various fractions as carbon sources. Appl Microbiol Biotechnol. 1991;36(2):163–6.
Elimer E. Citric Acid Production from Rape Seed Oil by Aspergillus niger. Food Technol Biotechnol. 1998;36 189–92.
Darvishi F, Nahvi I, Zarkesh-Esfahani H, Momenbeik F. Effect of plant oils upon lipase and citric acid production in Yarrowia lipolytica yeast. J Biomed Biotechnol. 2009;2009.
Barig S, Alisch R, Nieland S, Wuttke A, Gräser Y, Huddar M, et al. Monoseptic growth of fungal lipase producers under minimized sterile conditions: cultivation of Phialemonium curvatum in 350 L scale. Eng Life Sci. 2011;11(4):387–94.
Mhetras NC, Bastawde KB, Gokhale DV. Purification and characterization of acidic lipase from Aspergillus niger NCIM 1207. Bioresour Technol. 2009;100(3):1486–90.
Rathnayake GRN, Kumar NS, Jayasinghe L, Araya H, Fujimoto Y. Chemical investigation of metabolites produced by an endophytic fungi Phialemonium curvatum from the leaves of Passiflora edulis. Nat Prod Res. 2018;32(20):2483–6.
Cert A, Moreda W, Pérez-Camino MC. Chromatographic analysis of minor constituents in vegetable oils. J Chromatogr A. 2000;881(1):131–48.
Colla LM, Primaz AL, Benedetti S, Loss RA, de Lima M, Reinehr CO, et al. Surface response methodology for the optimization of lipase production under submerged fermentation by filamentous fungi. Braz J Microbiol. 2016;47(2):461–7.
Piekarska K, Hardy G, Mol E, van den Burg J, Strijbis K, van Roermund C, et al. The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional β-oxidation pathway: evidence for reduced metabolite transport across the peroxisomal membrane. Microbiology. 2008;154(10):3061–72.
Kornberg HL, Krebs HA. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature. 1957;179(4568):988–91.
Berg JM, Tymoczko JL, Stryer L. Triacylglycerols are highly concentrated energy stores. In: Freeman WH, ed. Biochemistry, 5th edn. New York: Springere; 2002.
Christen S, Sauer U. Intracellular characterization of aerobic glucose metabolism in seven yeast species by 13C flux analysis and metabolomics. FEMS Yeast Res. 2011;11(3):263–72.
Imura M, Nitta K, Iwakiri R, Matsuda F, Shimizu H, Fukusaki E. Comparison of metabolic profiles of yeasts based on the difference of the Crabtree positive and negative. J Biosci Bioeng. 2020;129(1):52–8.
Dutta S, Das TK. Acetate as inducer for enzymes of glyoxylate cycle in Aspergillus oryzae. Indian J Microbiol. 2004;43:257–60.
Kamzolova SV, Lunina JN, Morgunov IG. Biochemistry of citric acid production from rapeseed oil by Yarrowia lipolytica Yeast. J Am Oil Chem Soc. 2011;88(12):1965–76.
Kim H-J, Kim T-H, Kim Y, Lee H-S. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J Bacteriol. 2004;186(11):3453–60.
Kunze M, Kragler F, Binder M, Hartig A, Gurvitz A. Targeting of malate synthase 1 to the peroxisomes of Saccharomyces cerevisiae cells depends on growth on oleic acid medium. Eur J Biochem. 2002;269(3):915–22.
Stahmann K-P, Kupp C, Feldmann SD, Sahm H. Formation and degradation of lipid bodies found in the riboflavin-producing fungus Ashbya gossypii. Appl Microbiol Biotechnol. 1994;42(1):121–7.
Clark D, Cronan J. Two-carbon compounds and fatty acids as carbon sources. EcoSal Plus. 2005.
Gibson BR, Lawrence SJ, Leclaire JPR, Powell CD, Smart KA. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev. 2007;31(5):535–69.
Martinez-Moya P, Niehaus K, Alcaino J, Baeza M, Cifuentes V. Proteomic and metabolomic analysis of the carotenogenic yeast Xanthophyllomyces dendrorhous using different carbon sources. BMC Genomics. 2015;16:289.
Zhao C, Gu D, Nambou K, Wei L, Chen J, Imanaka T, et al. Metabolome analysis and pathway abundance profiling of Yarrowia lipolytica cultivated on different carbon sources. J Biotechnol. 2015;206:42–51.
Cronan J, Laporte JP. Tricarboxylic acid cycle and glyoxylate bypass. EcoSal Plus: Springer; 2006.
Rohn H, Junker A, Hartmann A, Grafahrend-Belau E, Treutler H, Klapperstück M, et al. VANTED v2: a framework for systems biology applications. BMC Syst Biol. 2012;6(1):139.
Wang Z-L, Pan H-B, Huang J, Yu X-P. The zinc finger transcription factors Bbctf1α and Bbctf1β regulate the expression of genes involved in lipid degradation and contribute to stress tolerance and virulence in a fungal insect pathogen. Pest Manage Sci. 2020;76(8):2589–600.
Bowyer P, De Lucas JR, Turner G. Regulation of the expression of the isocitrate lyase gene (acuD) of Aspergillus nidulans. MGG. 1994;242(4):484–9.
Caza M, Hu G, Price M, Perfect JR, Kronstad JW. The zinc finger protein Mig1 regulates mitochondrial function and Azole drug susceptibility in the pathogenic fungus Cryptococcus neoformans. mSphere. 2016;1(1):e00080–15.
Adnan M, Zheng W, Islam W, Arif M, Abubakar YS, Wang Z, et al. Carbon catabolite repression in filamentous fungi. Int J Mol Sci. 2018;19(1):48.
Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62(2):334.
Papanikolaou S, Chevalot I, Komaitis M, Marc I, Aggelis G. Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures. Appl Microbiol Biotechnol. 2002;58(3):308–12.
Rubio-Texeira M, Van Zeebroeck G, Thevelein JM. 10 trehalose metabolism: enzymatic pathways and physiological functions. In: Hoffmeister D, editor. Biochemistry and molecular biology. Cham: Springer International Publishing; 2016. p. 191–277.
Welsh DT. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol Rev. 2000;24(3):263–90.
Varela CA, Baez ME, Agosin E. Osmotic stress response: quantification of cell maintenance and metabolic fluxes in a lysine-overproducing strain of Corynebacterium glutamicum. Appl Environ Microbiol. 2004;70(7):4222–9.
Wang D, Wei G, Nie M, Chen J. Effects of nitrogen source and carbon/nitrogen ratio on batch fermentation of glutathione by Candida utilis. Korean J Chem Eng. 2010;27(2):551–9.
Capus A, Monnerat M, Ribeiro LC, de Souza W, Martins JL, Sant’Anna C. Application of high-content image analysis for quantitatively estimating lipid accumulation in oleaginous yeasts with potential for use in biodiesel production. Bioresour Technol. 2016;203(Supplement C):309–17.
Wu C, Zhang J, Zhu G, Yao R, Chen X, Liu L. CgHog1-Mediated CgRds2 phosphorylation alters glycerophospholipid composition to coordinate osmotic stress in Candida glabrata. Appl Environ Microbiol. 2019;85(6):e02822–918.
Ding M-Z, Tian H-C, Cheng J-S, Yuan Y-J. Inoculum size-dependent interactive regulation of metabolism and stress response of Saccharomyces cerevisiae revealed by comparative metabolomics. J Biotechnol. 2009;144(4):279–86.
Mannazzu I, Angelozzi D, Belviso S, Budroni M, Farris GA, Goffrini P, et al. Behaviour of Saccharomyces cerevisiae wine strains during adaptation to unfavourable conditions of fermentation on synthetic medium: cell lipid composition, membrane integrity, viability and fermentative activity. Int J Food Microbiol. 2008;121(1):84–91.
Back A, Rossignol T, Krier F, Nicaud J-M, Dhulster P. High-throughput fermentation screening for the yeast Yarrowia lipolytica with real-time monitoring of biomass and lipid production. Microb Cell Fact. 2016;15(1):147.
Trinci APJ. Influence of the width of the peripheral growth zone on the radial growth rate of fungal colonies on solid media. J Gen Microbiol. 1971;67(3):325–44.
Reeslev M, Kjoller A. Comparison of biomass dry weights and radial growth rates of fungal colonies on media solidified with different gelling compounds. Appl Environ Microbiol. 1995;61(12):4236–9.
Bellou S, Triantaphyllidou IE, Mizerakis P, Aggelis G. High lipid accumulation in Yarrowia lipolytica cultivated under double limitation of nitrogen and magnesium. J Biotechnol. 2016;234(Supplement C):116–26.
Rakicka M, Lazar Z, Dulermo T, Fickers P, Nicaud JM. Lipid production by the oleaginous yeast Yarrowia lipolytica using industrial by-products under different culture conditions. Biotechnol Biofuels. 2015;8(1):104.
Amran MHH, Danik MF, Abdullah MSP, Zulfakar MH, Shamsuddin AF. Superolein based intravenous lipid emulsion 20% W/W physicochemical characterization, stability and its effect on liver status. 2019;48(5):1043–54.
Liu X, Lv J, Xu J, Xia J, He A, Zhang T, et al. Effects of osmotic pressure and pH on citric acid and erythritol production from waste cooking oil by Yarrowia lipolytica. Eng Life Sci. 2018;18(6):344–52.
Yang S, Sadilek M, Synovec RE, Lidstrom ME. Liquid chromatography–tandem quadrupole mass spectrometry and comprehensive two-dimensional gas chromatography–time-of-flight mass spectrometry measurement of targeted metabolites of Methylobacterium extorquens AM1 grown on two different carbon sources. J Chromatogr A. 2009;1216(15):3280–9.
Hajjaj H, Blanc PJ, Goma G, François J. Sampling techniques and comparative extraction procedures for quantitative determination of intra- and extracellular metabolites in filamentous fungi. FEMS Microbiol Lett. 1998;164(1):195–200.
Pinu FR, Villas-Boas SG, Aggio R. Analysis of intracellular metabolites from microorganisms: quenching and extraction protocols. Metabolites. 2017;7:4.
Neoh BK, Teh HF, Ng TLM, Tiong SH, Thang YM, Ersad MA, et al. Profiling of metabolites in oil palm mesocarp at different stages of oil biosynthesis. J Agric Food Chem. 2013;61(8):1920–7.
Krug D, Zurek G, Schneider B, Garcia R, Müller R. Efficient mining of myxobacterial metabolite profiles enabled by liquid chromatography–electrospray ionisation-time-of-flight mass spectrometry and compound-based principal component analysis. Anal Chim Acta. 2008;624(1):97–106.
Azizan KA, Baharum SN, Ressom HW, Noor NM. GC-MS analysis and PLS-DA validation of the trimethyl silyl-derivatization techniques. Am J Appl Sci. 2012;9(7):1124–36.
Sangster T, Major H, Plumb R, Wilson AJ, Wilson ID. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst. 2006;131(10):1075–8 (Epub 2006 Aug 15).
Abu-Reidah IM, Contreras MM, Arráez-Román D, Segura-Carretero A, Fernández-Gutiérrez A. Reversed-phase ultra-high-performance liquid chromatography coupled to electrospray ionization-quadrupole-time-of-flight mass spectrometry as a powerful tool for metabolic profiling of vegetables: Lactuca sativa as an example of its application. J Chromatogr A. 2013;1313:212–27.
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3(3):211–21.
Acknowledgements
The authors would like to acknowledge Universiti Kebangsaan Malaysia for financial support through the Economic Transformation Program Grant, ETP-2013-064 and also German Federal Ministry of Education and Research (BMBF) for the grant, 031A275. Arief Izzairy Zamani is supported by MyPhD scholarship under Ministry of Higher Education Malaysia. Susann Barig was supported by the project “Bioeconomy international-Microbial conversion of palm oil” funded by the German Federal Ministry of Education and Research (BMBF; sponsorship no. 031A275). We thank Kamalrul Azlan Azizan (INBIOSIS, UKM), Norzamzurina Ismail (CRIM, UKM), Emelda Rosseleena Rohani (INBIOSIS, UKM), and Farahayu Khairuddin (Malaysia Genome Institute) for useful discussion and technical support.
Funding
This study was funded by Universiti Kebangsaan Malaysia through the Economic Transformation Program Grant (ETP-2013-064) and German Federal Ministry of Education and Research (BMBF; sponsorship no. 031A275).
Author information
Authors and Affiliations
Contributions
AIZ, SB, HMY, SNB, K-P.S and CLN designed the study. AIZ carried out most of the experiments. SB involved in providing the strains from her previous study. SI carried out the untargeted metabolomics analysis while both JYSL and KSF carried out targeted organic acids profiling. AIZ, SB, JI, SNB, K-P.S and CLN analyzed and interpreted the obtained data. AIZ, SNB, K-P.S and CLN wrote the manuscript. All authors read and approved the final manuscript
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Additional tables and figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zamani, A.I., Barig, S., Ibrahim, S. et al. Comparative metabolomics of Phialemonium curvatum as an omnipotent fungus cultivated on crude palm oil versus glucose. Microb Cell Fact 19, 179 (2020). https://doi.org/10.1186/s12934-020-01434-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-020-01434-w