Abstract
Background
Limitless self-renewal is one of the hallmarks of cancer and is attained by telomere maintenance, essentially through telomerase (hTERT) activation. Transcriptional regulation of hTERT is believed to play a major role in telomerase activation in human cancers.
Main body
The dominant interest in telomerase results from its role in cancer. The role of telomeres and telomere maintenance mechanisms is well established as a major driving force in generating chromosomal and genomic instability. Cancer cells have acquired the ability to overcome their fate of senescence via telomere length maintenance mechanisms, mainly by telomerase activation.
hTERT expression is up-regulated in tumors via multiple genetic and epigenetic mechanisms including hTERT amplifications, hTERT structural variants, hTERT promoter mutations and epigenetic modifications through hTERT promoter methylation. Genetic (hTERT promoter mutations) and epigenetic (hTERT promoter methylation and miRNAs) events were shown to have clinical implications in cancers that depend on hTERT activation. Knowing that telomeres are crucial for cellular self-renewal, the mechanisms responsible for telomere maintenance have a crucial role in cancer diseases and might be important oncological biomarkers. Thus, rather than quantifying TERT expression and its correlation with telomerase activation, the discovery and the assessment of the mechanisms responsible for TERT upregulation offers important information that may be used for diagnosis, prognosis, and treatment monitoring in oncology. Furthermore, a better understanding of these mechanisms may promote their translation into effective targeted cancer therapies.
Conclusion
Herein, we reviewed the underlying mechanisms of hTERT regulation, their role in oncogenesis, and the potential clinical applications in telomerase-dependent cancers.
Similar content being viewed by others
Background
Replicative capacity is one of the most critical features in cancer cells, which is attained by telomere maintenance [1]. Telomeres protect the ends of chromosomes from degradation and end-to-end fusions, contributing to genomic stability [1, 2]. Telomerase, a specialized DNA polymerase, is responsible for telomere maintenance in the majority of human cancers, but its activity is absent in most normal somatic tissues. This differential role makes telomerase and its regulatory mechanisms attractive cancer biomarkers with relevant implications in clinical practice [3].
Telomeres and telomerase
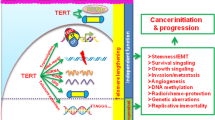
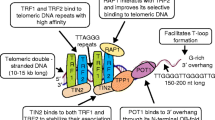
Telomeres are the nucleoprotein complexes located at the ends of eukaryotic chromosomes. Telomere structure was discovered by Muller and Meier in 1938. Telomeres consist of 5 to 20kb of repeating hexanucleotide DNA sequence TTAGGG (telomeric DNA) [4,5,6]. Telomeric DNA repeats are followed by a terminal 3´G-rich single-stranded overhang forming a telomeric loop (T-loop) that provides 3´end protection [7, 8]. Telomeric DNA is associated with the shelterin protein complex and together they protect chromosomal ends and maintain genomic and chromosomal integrity by preventing nucleolytic degradation, unnecessary recombination, and inter-chromosomal fusions [7, 9, 10]. The shelterin complex consists of a group of six telomere-specific proteins; telomeric repeat binding factor 1 and 2 (TERF1, TERF2) and protection of telomeres protein 1 (POT1) interact directly with TTAGGG repeats. These proteins are interconnected with three others: TERF1 Interacting Nuclear Factor 2 (TINF2), tripeptidyl-peptidase 1 (TPP1), and repressor activator protein 1 (RAP1) [7, 8, 11]. Telomeric DNA is masked with shelterin protective caps and these complexes enable DNA damage repair (DDR) machinery to distinguish telomeric DNA from genomic DNA damage [12, 13]. Throughout cellular lifespan, telomeric DNA is shortened after each replicative cycle due to the “end-replication problem”, oxidative damage, age, and lifestyle (including diet, smoking, professional environment and stress) [14,15,16]. Telomere shortening leads to a stage of cell growth arrest. At this stage (M1), DNA damage signalling and cellular senescence are triggered which constitutes a crucial protective mechanism that prevents progression to an oncogenic state [10, 17]. However, in some cases, cells surpass this senescence state (avoiding important cell cycle checkpoints provided by p16INK4a, TP53 and Rb) and enter a crisis state (M2) [17]. At this point, cells have very short telomeres and their chromosomal ends fuse, leading to chromosome bridge-breakage-fusion cycles, genomic instability, and eventually cell apoptosis [17]. However, in rare situations, cells may acquire the ability to continuously divide which may promote malignant transformation (Fig. 1). This process of unlimited self-renewal is mediated by telomerase that maintains or lengthen telomeres promoting cellular immortalization process [1, 3, 17].
Telomere length dynamics in different cells over time. Telomeres shorten over time. Germ cells and embryonic stem cells have long telomeres that are maintained by telomerase activity. Stem cells have shorter telomeres and somatic cells even shorter. After multiple cell divisions these cells achieve a senescence state (M1). At M2 stage cells enter crisis due to their short telomeres that lead to chromosomal and genomic instability resulting in apoptosis. Cancer cells escape from crisis through telomerase activation, reacquire longer telomeres and unlimited self-renewal capacity
Telomerase was discovered in 1985, as an enzyme capable of extending telomeric repeat sequences; and in 1989, telomerase activity was reported for the first time [18,19,20]. However, the protein component of telomerase was only identified and functionally characterized in 1997, more than a decade after its discovery [21]. This enzyme consists of a large ribonucleoprotein complex responsible for progressive synthesis of telomeric DNA repeats. Telomerase is a DNA polymerase that consists of two different subunits: a functional catalytic protein subunit called human telomerase reverse transcriptase (hTERT) encoded by the TERT gene, positioned at chromosome 5p15.33; and a RNA component known as human telomerase RNA component (hTERC or hTR), encoded by the TERC gene on chromosomal region 3q26 [22,23,24]. Other proteins including Pontin, Reptin, Gar1, Nhp2, and Tcab1 were shown to be associated with the telomerase core complex and required for proper telomerase assembly and recruitment to chromosomes [25, 26]. Dyskerin and telomerase protein component (TEP1) have an important role in stabilizing the telomerase complex [27, 28]. Es1p and Es3p are additional protein subunits (Ku heterodimer) involved in assembly and maturation, which also contribute to the telomerase enzymatic complex [29]. Despite extensive research on these proteins, the three-dimensional structure of human telomerase is yet to be fully understood [30]. Importantly, only hTERC and hTERT are necessary for the reestablishment of telomerase activity [31,32,33].
hTERT mRNA expression is strictly controlled and closely associated with telomerase activity, which suggests that hTERT is the primary determinant for the enzyme activity. Current knowledge proposes that the limiting factor for telomerase activity is hTERT expression which is tightly regulated at transcriptional level [34,35,36,37]. Experimental evidence suggests that telomerase activity shows strong association with hTERT expression [30]. hTERC acts as a template for the synthesis of telomeric DNA, and unlike hTERT, is ubiquitously expressed in all tissues. Therefore, it has been considered by some authors as a non-limiting factor of telomerase activity [38, 39]. However, another study performed in fibrosarcoma-derived HT1080 cells [40] revealed that hTERC is more abundant in tumors than in normal cells with its locus amplified, and is essential for telomerase activity and can be a limiting factor [40].
hTERT regulation is a multifarious process yet to be fully understood where both transcriptional and posttranscriptional mechanisms are involved [38]. These include pre-mRNA alternative splicing of the hTERT gene which was found to be involved in the regulation of telomerase activity [41,42,43] and has been associated with diagnosis, prognosis and clinical cancer parameters [43].
hTERT regulation in normal cells
Telomerase is constitutively activated in germline, hematopoietic, stem and also rapidly renewing cells [44, 45]. On the other hand, telomerase activity is very low or absent in somatic cells mainly due to tight hTERT regulation [46]. However, telomerase activity was found in normal human blood cells and other normal human cell types that are mitotically active, such as proliferative basal skin layer, endometrial tissue (during menstrual cycle), proliferative zone of intestinal crypts, and hair follicles [44, 45, 47,48,49,50,51,52].
Telomere length and telomerase activity diverge between normal and embryonic stem cells. While embryonic stem cells fully maintain their telomeres and exhibit telomerase activity, normal stem cells have progressive telomere shortening and minimal telomerase activity (Fig. 1). Since hTERT is not expressed in most normal human cells, it can be used as a potential cancer biomarker. In fact, there are studies suggesting that telomerase activity might be a useful marker for diagnosis (detecting cancer disease) and prognosis (associated with stage and disease outcome) in different cancers (e.g., prostate, bladder, thyroid, breast, colon, gastric and lung) [53,54,55,56,57,58,59,60,61,62,63,64,65].
hTERT regulation in Cancer
Cancer arises when normal cells accumulate genomic instability and acquires limitless proliferative capacity [66]. Cancer cells have acquired the ability to overcome their fate of senescence via telomere length maintenance mechanisms, mainly by telomerase activation or alternative mechanisms (alternative lengthening of telomeres – ALT) [3, 67,68,69]. In 1994, it was shown that telomerase is upregulated in up to 90% of malignancies, and is crucial for oncogenesis and disease progression [68, 70,71,72,73,74].
hTERT regulation mechanisms have been studied for the last 20 years, and recent advances mainly related to the discovery of hTERT promoter mutations have given new impetus to better understand the mechanisms involved in hTERT regulation [75].
However, other alterations were recently reported, and hTERT expression is also up-regulated in tumors via multiple genetic and epigenetic mechanisms including hTERT amplifications (3%), hTERT structural variants (3%), hTERT promoter mutations (31%) and epigenetic modifications through hTERT promoter methylation (53%) [72, 76].
hTERT regulation in cancer: genetic mechanisms
hTERT amplifications
Gain or loss of genetic material occurs frequently in cancer where gene amplification is an important mechanism for the oncogenic process. Gene amplification results from a copy number increase associated with overexpression of the amplified gene. Different models have been proposed for the initiation of amplification including DNA replication errors, telomere dysfunction and the existence of chromosomal fragile sites [77]. Specifically, hTERT gene amplification can result from telomere dysfunction in addition to breakage at fragile sites and formation of chromosomal fusions [78]. In a large cohort made of 31 different types of cancer, it was demonstrated that 3% out of 95% of hTERT expressing tumours presented hTERT amplifications [76]. Therefore, hTERT might be a target for amplification during tumorigenesis, which contributes to the dysregulation of telomerase activity that usually occurs in human tumors [79].
hTERT amplifications: clinical relevance
Increased hTERT gene copy number is associated with up-regulation of hTERT expression, related to acquired drug resistance, and correlated with worse clinical outcomes in breast, skin and thyroid cancer [79,80,81,82]. However, in bladder cancer, no correlation was observed between increased hTERT gene copy number and hTERT mRNA, telomerase activity, or telomere length, suggesting that hTERT gene amplification may require another companion alteration for telomerase reactivation [83, 84].
TERT genomic rearrangements
Another potential mechanism of hTERT upregulation in tumors are the genomic rearrangements affecting the hTERT gene locus (5p15.33). Functionally, these rearrangements bring active enhancers in proximity to the hTERT gene, and the interaction between the promoter and these newly introduced enhancers drives hTERT expression [85, 86]. hTERT rearrangements were associated with increased hTERT expression, with poorer patient outcome, and found along with other telomere maintenance mechanisms including ALT and MYCN amplifications in neuroblastoma [87].
Further studies are essential to understand whether or not hTERT rearrangements are used by different types of cancers, and as well their clinical impact.
TERT promoter mutations
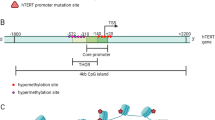
In 2013, two pivotal studies described two recurrent non-coding mutations within the hTERT promoter region in both familial and sporadic melanomas [88, 89]. These two mutations were located at -124 and -146 bp upstream from ATG (chr5:1,295,228 G>A and 1,295,250 G>A, C>T on opposite strand). After the initial discovery, hTERT promoter mutations (TERTpMut) have been identified in multiple and distinct tumor types, such as glioblastoma, bladder and thyroid cancer, with different prevalence according to cancer type and histology [90].
TERTpMut represent a frequent but unique genetic alteration that drives hTERT expression and telomerase activation. hTERT core promoter consists of 260 base pairs with multiple transcription-factors binding motifs that regulate gene transcription and telomerase activation [91]. The location of these mutations within the promoter creates additional binding sites for the E-twenty-six (ETS) transcription factor family, thus constituting a novel mechanism of genetic activation in cancer and a possible driver genomic alteration [92, 93].
The transcriptional controlling of hTERT gene is complex and includes regulation at multiple levels by various positive and negative factors or pathways. Recent knowledge has come from the cloning of hTERT promoter and identification of various transcription factor-binding motifs, involved in hTERT expression and telomerase regulation by TERTpMut [22, 30, 39, 94,95,96]. TERTpMut modulate transcriptional regulation without altering an encoding protein. Functionally, hTERT promoter mutations are associated with the formation of consensus binding sequence (CCGGAA) at the E-twenty-six/ternary complex (ETS/TCF) transcription factors (Fig. 2), providing a possible mechanism for cancer-specific upregulation of telomerase [88, 89]. Mechanistically, ETS transcription factor binding to the motifs (created by the mutations) causes a recruitment of a multimeric ETS family member, the GA-binding protein alpha subunit (GABPA) that activates hTERT transcription [88, 97, 98]. These findings were further explored through luciferase reporter assays showing increased telomerase activity in cells transfected with mutant constructs. [88, 89, 99] Moreover, there is an evidence of promoter mutations creating de novo transcription factor binding sites, as cells co-transfected with mutant promoter constructs and plasmids containing ETS1 cDNA display increased activity [100]. In cancer cells harboring TERTpMut, the mutant promoter recruits GABPA and exhibits H3K4m2/3, an active chromatin mark. On the other hand, wild type cell lines exhibit the H3K27me3, a mark of epigenetic silencing, suggesting that only the mutant promoters are transcriptionally active [98]. Despite both mutations are functionally active the TERTpMut, C228T is significantly more frequent than the C250T [101].
Mechanisms of hTERT regulation in cancer. Transcription factors and their binding sites, as well the positions of both hTERT promoter mutations, C228T and C250T, the hypermethylated region upstream to TSS (THOR) and TERT-miRNAs are shown. The cancer-specific mutations within the core promoter, at -124 and -146bp positions generate ETS binding motifs, leading to GABP transcription factor recruitment and consequently hTERT transcription. Binding of transcriptional activators (c-Myc) and repressors (WT1 and CTCF) to the hTERT promoter may be controlled by DNA methylation, in which methylated CpGs prevent their binding to the target sites, leading to hTERT activation (THOR region). MiRNAs targeting the 3’UTR promotes translation repression of hTERT. Black dots represent methylated CpG sites. ETS: E-twenty-six; TSS: transcription start site; ATG: start codon
The wide distribution across different tumors (urothelial cancer – bladder and upper urinary tract, melanoma, glioblastoma, thyroid cancer, hepatocellular carcinoma) and high frequency in some of them has created an important hub around these genetic alterations [90, 99, 102, 103]. Bladder, thyroid, cutaneous melanoma, basal cell and squamous carcinoma and oligodendrogliomas are examples of cancers where TERTpMut are widespread through different stages and grades of the disease, suggesting their role as an early tumorigenic event [102, 103]. Additionally, not all TERTpMut tumors display telomerase activation and some premalignant lesions also displayed these genetic alterations at the hTERT promoter region [104]. Together, these results support the fact that TERTpMut may act as early events in the oncogenic process [90, 105,106,107].
Important information came recently from a new study demonstrating that TERTpMut are necessary but not sufficient to maintain telomere length nor telomerase upregulation [108]. In fact these authors demonstrated that TERTpMut acquired at the transition from benign nevus to malignant melanoma do not support telomere maintenance suggesting that TERTpMut contribute to tumorigenesis in two distinct ways. Initially, TERTpMut do not prevent telomere shortening but act “healing” the shortest telomeres and later telomeres are critically short leading to genomic instability and telomerase reactivation [108].
These results might support the hypotheses that TERTpMut are not the unique event responsible to initiate an oncogenic process explaining their presence in premalignant lesions and non-hTERT expressing tumors. TERTpMut usually occur in cancers with low rate of self-renewal, such as brain tumors, liver, melanocytes and even low-grade bladder cancers suggesting a role in triggering telomerase activation [109, 110]. In adult gliomas, TERTpMut were found in 70% to 80% of glioblastomas, followed by oligodendrogliomas (60%-70%) and oligoastrocytomas (35%-55%). However, TERTpMut are rare events in ependymoma lesions [110, 111]. Urological malignancies have a different prevalence of TERTpMut varying from rare or absent in prostate cancer and testicular germ cell tumors to high frequency amongst urothelial cancers. In urothelial bladder cancer, mutations are present in up to 85% of the lesions, which rank these alterations as one of the most frequent genomic events in bladder cancer [112, 113]. However, the prevalence in renal cell carcinoma is low, at approximately 9% [106, 111,112,113]. Regarding thyroid cancer, the frequency of these genetic alterations varies according to histology. Papillary and follicular type lesions usually harbor 10-20% whereas in poorly undifferentiated and anaplastic lesions TERT mutations are found in 30-50% of the patients [107]. There is a stepwise increase in frequency of TERTpMut from well differentiated to poorly differentiated lesions in thyroid cancer being absent in medullary carcinomas [107, 114]. However, there are other cancers that do not harbor TERTpMut (testicular germ cell tumors; breast cancer, colorectal carcinoma, prostate cancer) but have telomerase activation [111]. These observations suggest that in hTERT-dependent tumors without TERTpMut, other mechanisms responsible for telomerase activation might be at play.
TERT promoter mutations: clinical relevance
Clinically, tumors carrying TERTpMut frequently express higher levels of hTERT mRNA and telomerase activity compared with those having a wild type promoter highlighting the prognostic potential of TERTpMut and their potential use as a clinical biomarker [90]. Several studies have looked at the role of TERTpMut in cancer diagnosis and prognosis. In urothelial bladder cancer patients, TERTpMut were detected in tissue and urine and has been proposed as a non-invasive diagnostic and prognostic marker, associated with decreased disease-free survival [102]. However, other studies did not find a clinical correlation with disease outcomes [84, 112]. Wu et al. [115] reported an important co-occurrence of TERTpMut and TP53/RB1 mutations and suggested that they might cooperatively contribute to the progression of bladder cancer.
In glioma, TERTpMut are distributed according to histology and are related to survival in combination with IDH1 mutations. Also, TERTpMut are not only prognostic factors for poor clinical outcomes, but also predictors of radiotherapy resistance [116,117,118,119]. Furthermore, BRAF/NRAS mutations are associated with decreased disease-free and melanoma-specific survival [120, 121]. In liver disease, TERTpMut are present in pre-malignant nodules and predict high risk for advanced disease and reduced disease-free and overall survival in hepatocellular carcinoma patients [122, 123]. Thyroid cancer patients with TERTpMut are associated with clinically aggressive and recurrent disease, with lower disease-free and overall survival when combined with BRAF mutations [124,125,126]. TERTpMut are a moderately prevalent genetic event in non-small cell lung cancer (NSCLC) associated with patient age, gender and distant metastasis [127]. These studies emphasize the hypothetical existence of a companion mechanism, necessary not only for telomerase activation but also to maintain the self-renewal capacity allowing cancer disease progression in TERTpMut patients [84, 112].
Current studies highlight the prognostic properties of TERTpMut and their potential use as a clinical biomarker. In general, these genetic alterations of the hTERT promoter are associated with adverse outcomes in several cancers. Nevertheless, recent studies show the presence of companion genetic alterations in patients with worse outcomes, suggesting the need for concomitant and possibly synergistic events resulting in not only telomerase activation but also disease progression.
Unanswered questions remain to be elucidated related to the diverse frequency of mutations amongst different cancers and histological types. Also, the coexistence of hTERT regulation mechanisms in the same tumor and the eventual collaborative effects between TERTpMut and other hTERT regulatory mechanisms resulting in differential telomerase activation is object for future studies.
hTERT regulation in Cancer: epigenetic mechanisms
hTERT promoter methylation
The epigenetic process of DNA methylation is crucial in gene expression regulation. DNA methylation occurs genome-wide at CpG sites usually located in non-coding regions. This process, mediated by DNA methyltransferases, occurs in the context of dinucleotide sequence 5’-CG-3’, often referred to as CpG methylation and consists of the addition of a methyl group (-CH3) on the 5-carbon of a cytosine (C) base followed by guanine (G) base. CpG dinucleotide sequences are spread throughout the genome, but there are specific regions known as CpG islands where high frequency of CpG dinucleotides is observed. 80% of CpG sites are methylated in intergenic regions while most sites in the promoter and exon 1 regions are unmethylated [128]. CpG islands are usually clustered near the gene promoters where transcription initiation occurs. About 70% of the human gene promoters contain CpG islands, and therefore DNA methylation has been thought to play an important role in gene expression [128, 129]. Promoter DNA methylation has been recognized as one of the most frequent and stable ways of gene expression controlling mechanisms. Hitherto, promoter DNA methylation is thought to promote gene silencing. In actively transcribed genes, the promoter tends to be unmethylated, since DNA methylation has been associated with gene silencing by hindering transcription factor binding or affecting chromatin architecture [130]. In fact, in most cases, genes with methylated promoters are usually silenced while genes with unmethylated promoters are actively transcribed, the pattern observed in oncogenes and tumor suppressors [131]. During cancer progression, there is a genome-wide hypomethylation of CpG sites along gene body and hypermethylation of CpG islands in gene promoter regions [132]. Thus, abnormal DNA methylation is a hallmark of cancer cells and is crucial in cancer development [133].
Despite the powerful role of recurrent hTERT promoter mutations in hTERT activation in cancers, there are several tumor types that exhibit low or no frequency of these mutations (e.g. prostate and breast cancer) [134]. Thus, the role of epigenetic mechanisms in cancer-specific hTERT regulation has been a topic of study for past decade, and several studies have shown contradicting effects of hTERT promoter methylation on hTERT expression.
Although some authors have reported hypomethylation in the CpG islands covering hTERT promoter, others identified increased DNA methylation in hTERT expressing cancer cells [135,136,137,138]. In fact, hTERT was one of the first genes in which methylation of its promoter sequence was positively correlated with gene expression [135]. This correlation among hTERT promoter methylation with hTERT mRNA and telomerase activity suggests that methylation of hTERT promoter may be implicated in hTERT regulation, but in a different manner from other genes regulated by promoter methylation [135].
As mentioned above, promoter methylation is often associated with gene silencing. However, several studies have shown that methylation of specific regions within hTERT promoter, particularly, upstream of the hTERT core promoter, is associated with gene activation [72].
The precise mechanisms by which the methylation pattern of hTERT promoter results in hTERT activation is still under investigation (Fig. 2). Recently, the possible role of hTERT promoter methylation on activation of hTERT expression has been functionally shown [72, 139].
There are several explanations as to how hTERT promoter methylation can result in hTERT activation: first possibility is based on the prevention of repressive elements binding caused by DNA methylation at the repressive region. If hTERT promoter is hypomethylated (unmethylated), the transcriptional repressors would bind to the promoter and block the transcriptional machinery (Fig. 2). However, if methylated, hTERT would prevent this binding and therefore would allow the promoter to be activated by appropriate transcriptional factors. An interesting observation from these results is that proximal hTERT core promoter – allowing essential drivers of gene expression to access the promoter is almost always hypomethylated, and the region upstream of core promoter is often hypermethylated [140, 141]. Whether coincidental or reasonable, recurrent hTERT mutations seem to occur in the unmethylated region, which supports the hypothesis stating ETS family factors binding to these sites activate hTERT expression. Evidence has been also given by demethylation of repressor binding sites by 5-aza-2-deoxycytidine, globally reducing DNA methylation, and consequently resulting in reduced levels of hTERT transcription [142]. Also, factors such as CTCF, which interact with hTERT promoter, are known for organizing global chromosomal architecture, and methylation-sensitive binding of CTCF may be changing not only the accessibility but also chromosomal conformation and possible interactions with enhancers or silencers far away in distance. CTCF binds adjacent to transcriptional start site (TSS) and represses hTERT transcription, but DNA methylation prevents CTCF binding and consequently allows for the activation of telomerase [143].
Wilms tumor protein (WT1) is another repressor of TERT expression [144]. WT1 exhibits methylation-sensitive binding to DNA sequence, with reduced binding when one or more methylated bases are present in the binding sequence. WT1 binding sites exhibit increased CpG methylation in cancer, which results in the blocking of repressive effects and consequently hTERT expression [135, 136]. MYC proto-oncogene encodes a ubiquitous transcription factor (c-Myc) involved in the control of cell proliferation and differentiation. c-Myc has a direct role in induction of telomerase activity [145]. As CTCF and WT1, c-Myc binding is also methylation-sensitive and its binding is absent or reduced when binding site is methylated, resulting in reduced hTERT expression [146].
Another possible explanation is a more complex mechanism involving DNA methylation and chromosome structural changes [147]. DNA methylation can contribute to changes in chromatin conformation influencing gene expression by affecting DNA exposure to transcription factor binding [148]. DNA methylation is often linked to histone modifications and might control the accessibility of transcription factors to the promoter. Specific conformational changes caused by methylation of hTERT promoter may be causing differential recruitment and binding of factors that can drive hTERT expression in cancer [94]. There are several histone post-translational modifications, such as histone acetylation and methylation, that can affect the compaction state of chromatin, which influences the folding, position and organization of DNA, thereby affecting gene expression [149]. Generally, high levels of H3K4me3 and H3K27ac marks are associated with active chromatin while the gain of H3K9me and H3K27me3 marks has been linked to transcriptional repression [150].
hTERT promoter methylation: clinical relevance
Several tumor types including malignant tumors of brain, prostate, urothelium, colon, and blood have shown high frequency of hypermethylation signature in a specific region upstream of hTERT core promoter. More interestingly, even in melanomas – where hTERT promoter mutations were first identified and is known to be a mechanism of hTERT activation – hTERT promoter methylation was associated with hTERT upregulation [151]. Despite high prevalence of this tumor-specific signature across various tumor types, there has been little effort put into translating these findings to apply in clinical settings. Methylation of a specific region in the hTERT promoter was identified as potential biomarker of tumor progression and survival in pediatric gliomas [72]. This region termed THOR (TERT Hypermethylated Oncological Region) is hypermethylated in malignant tumours and hypomethylated in normal tissues and stem cells [72]. THOR is 100% specific and 96% sensitive for detection of hTERT expressing malignant neoplasms. THOR methylation showed prognostic properties as well, and identified which low-grade tumours would progress to high-grade ones and predicted survival in a subset of paediatric cancers [72]. THOR was further explored in prostate cancer and has shown its role as a potential marker with diagnostic and prognostic properties [139]. These findings have been expanded upon by multiple groups implicating hTERT promoter methylation in hTERT upregulation, and further demonstrating not only its diagnostic but, importantly, its clinical significance in cancer prognostic including thyroid cancer, acute myeloid leukemia/myelodisplastic syndrome, esophageal carcinoma, meningioma, pituitary adenomas, colorectal cancer and hepatocellular carcinoma) [72, 82, 139, 152,153,154,155,156,157]. In these studies, hTERT promoter hypermethylation was positively correlated with high hTERT expression, telomerase reactivation and in the vast majority of the cases correlated with worse clinical outcomes.
MicroRNAs
MicroRNAs (miRNAs) are short (20-23nucleotides) endogenous non-coding RNA molecules that have a crucial role in gene expression regulation [158, 159].
The biological importance of miRNAs has been recognized and associated with the pathogenesis of cancer and mechanisms that govern metastatic spread [160]. miRNAs are implicated in genome instability, acting as tumour suppressors or oncogenic drivers. Specifically, miRNAs have been reported to play critical roles in fundamental pathophysiological processes, such as cell proliferation, apoptosis, differentiation and metabolism and present in several human diseases, including cancer [158, 161,162,163,164,165].
Alterations in miRNA patterns in cancer are often associated with genomic events such as mutations, deletions, amplifications and transcriptional changes or due to defects in enzymes involved in miRNA biogenesis. More recent studies however report that epigenetic alterations are crucial regulators of miRNAs in cancer [166, 167]. Functionally, miRNAs mediate the post-transcriptional gene silencing of their target genes, inducing translation repression or mRNA degradation [166]. Downregulation of miRNAs in tumor tissue suggests a tumor suppressor function (suppressor-miRNAs), since a decrease in their expression levels normally contributes to oncogenesis. On the other hand, overexpression of miRNAs that target tumor suppressor genes have been associated with oncogenic activity (onco-miRNAs) [167, 168]. Therefore, depending on their target genes, miRNAs can act as tumor suppressors or oncogenes.
Different miRNAs have been described as important regulators of hTERT in multiple types of cancer. hTERT-targeting miRNAs regulate negatively its expression, inhibiting tumorigenesis and are frequently downregulated in cancer [167, 169]. hTERT-targeting miRNAs biology have been widely studied and their function elucidated through pre-clinical in vivo model-based validation studies [164, 170,171,172]. MiRNAs can regulate hTERT in either direct or indirect manner. MiRNAs may directly bind to hTERT 3’ untranslated region (3’UTR), and interfere with hTERT protein production in cancer cell lines [169, 170, 172, 173]. For example, downregulation of mir-138 was shown to be associated with hTERT overexpression in anaplastic thyroid carcinoma cells, and the enforced overexpression of mir-138 induced a significant reduction in hTERT expression through interaction with hTERT 3’UTR [173]. Additionally, let-7g*, miR-133a, miR-342-5p and miR-491-5p downregulate telomerase activity and inhibit cell proliferation [169]. These miRNAs co-regulate hTERT and Wnt pathway-genes and importantly, might regulate other genes involved in oncogenesis, suggesting the presence of an oncogenic miRNA regulatory network involving telomerase activation [169, 174,175,176]. MiR-1182 is other hTERT 3’UTR modulator that is downregulated in bladder cell lines and tumor tissues, and whose overexpression was able to inhibit cell proliferation, colony formation, and invasion [171].
MicroRNAs can also regulate hTERT indirectly by targeting transcription factors involved in hTERT regulation [94]. For example, mir-494 and mir-1294 were reported to downregulate c-Myc, which is a known transcriptional activator of hTERT, in pancreatic cancer and esophageal squamous cell carcinoma [94, 177]. Further, miR-34a, a known tumor suppressor in multiple types of cancer, was reported to induce cellular senescence by targeting c-Myc and FoxM1 in the telomere pathway [176].
MiRNAs: Clinical relevance
MiRNAs are highly stable in a wide range of tissues, including formalin-fixed paraffin embedded (FFPE) tissues and body fluids. These characteristics highlight their use as potential diagnostic and prognostic biomarkers, as well as therapeutic targets [94, 164, 170,171,172, 178,179,180]. hTERT miRNAs are aberrantly expressed in cancer, and thus constitute a rich source of biological information with high diagnostic and prognostic value. Specifically, miR-1182, miR-1207-5p, miR-1266, miR-532 and miR-3064, which bind within the hTERT 3’UTR, are downregulated and associated with a poor clinical outcome in bladder, gastric and ovarian cancer [169,170,171]. Furthermore, miR-1182 induced chemosensitivity to cisplatin in bladder cancer, and thus, might eventually contribute for a better patient’s response to cisplatin-based chemotherapy [171].
miRNA targeting of genes involved in telomere pathway, might enable telomerase activity suppression and cellular senescence and eventually allow the modulation of other relevant cancer gene pathways, contributing more effectively to inhibit cancer cells self-renewal [181, 182]. Specifically, ongoing clinical research (Phase I, NCT01829971) are testing miR-34a mimics in multiple solid malignancies [182].
Although there is still much to understand about the complexity of telomerase regulation, the discovery of miRNAs that target hTERT appears to be a promising approach to prevent and treat cancers that are telomerase-dependent. However, further research is needed in order to provide a more comprehensive view of miRNA-based therapies in terms of delivery systems and toxicity effects and this way promote their translation into clinical reality.
Future research
Telomerase activation is crucial for cancer development, and was initially thought to be an attractive target for the development of a novel biomarker and anti-cancer therapeutics target [46]. Nonetheless, attempts to inhibit telomerase was devoted to disappointment from the beginning, with the inability of compounds to effectively repress hTERT expression and the risk of long-term toxicity to normal stem cells and their self-renewal capacity. Future approaches might be centred on mechanisms responsible for hTERT upregulation, as markers for clinical outcomes in cancer. So far, hTERT promoter mutations and hTERT promoter methylation are strong regulatory alterations that affect telomerase activation and might become useful as potential biomarkers in a wide range of tumors. Moreover, recent studies on ependymomas revealed that the CpG island methylator phenotype (CIMP) tumors, which are associated with poor prognosis, are responsive to drugs that target either DNA or H3K27 methylation [183].
Overall, further research is needed to confirm the potential of these mechanisms as drug-actionable biomarkers, and establish them as non-invasive tools (circulating tumor DNA or circulating tumor cells) with clinical application.
Conclusion
Cellular self-renewal is a hallmark of cancer which is regulated by telomerase activation, and current studies have shown different mechanisms involved in telomerase regulation. Until recently, telomerase regulation was thought to be controlled uniquely by transcriptional mechanisms. However, different genetic and epigenetic mechanisms have been showing a strong association with telomerase reactivation in different cancers, and importantly showing interesting properties as biomarkers – with diagnostic and prognostic abilities. Particularly, hTERT promoter mutations, hTERT promoter methylation and miRNAs targeting hTERT have gained special attention as mechanisms associated with hTERT reactivation. hTERT promoter mutations have been frequently identified as early events in tumors with low self-renewal capacity and related to worse clinical outcome. However, several important questions remain to be clarified regarding their role as a tumor initiating mechanism or a long-standing process crucial for oncogenesis and cancer progression. At an epigenetic level, hTERT promoter hypermethylation have been positively correlated with telomerase reactivation acting as a predictive marker for oncological outcomes in different cancers. miRNAs targeting hTERT have also been considered potentially useful clinical biomarkers, and as more are identified, further avenues for the development of effective cancer therapies are open.
These recent findings generate a spark of hope in advancing our understanding of telomere biology. However, more studies are needed in order to completely understand the complex telomerase regulatory mechanisms and the possible interplay between these mechanisms. Future research should be centred on the discovery of mechanisms responsible for hTERT upregulation specifically in cancers, establishing correlations of these biological findings with clinical outcomes and founding these mechanisms as relevant biomarkers. Moreover, hTERT regulation remains a very attractive therapeutic target. Understanding the mechanisms responsible for hTERT activation might unveil possible means to prevent the acquisition of aberrant self-renewal capacity in cancer cells.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015;40(9):504–15.
Shay JW, Wright WE, Werbin H. Defining the molecular mechanisms of human cell immortalization. Biochim Biophys Acta. 1991;1072(1):1–7.
Meier R, Muller R. A new arrangement for the registration of diaphragm movements. J Physiol. 1938;94(2):227–31.
Blackburn EH. Structure and function of telomeres. Nature. 1991;350(6319):569–73.
Moyzis RK, Buckingham JM, Cram LS, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6.
de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–10.
Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155(2):345–56.
de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol. 2010;75:167–77.
Shay JW. Telomerase therapeutics: telomeres recognized as a DNA damage signal: commentary re: K. Kraemer et al., antisense-mediated hTERT inhibition specifically reduces the growth of human bladder cancer cells. Clin Cancer Res. 2003;9:3794–800. Clin eCancer Res. 2003;9(10 Pt 1):3521-3525
Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28(22):2477–91.
van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–13.
Griffith JD, Comeau L, Rosenfield S, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–14.
Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp Gerontol. 1992;27(4):383–9.
Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256(2-6):271–82.
Shammas MA. Telomeres, lifestyle, cancer, and aging. Curr Opin Clin Nutr Metab Care. 2011;14(1):28–34.
Wright WE, Pereira-Smith OM, Shay JW. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol Cell Biol. 1989;9(7):3088–92.
Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337(6205):331–7.
Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–13.
Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59(3):521–9.
Harrington L, Zhou W, McPhail T, et al. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11(23):3109–15.
Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8(1):137–42.
MacNeil DE, Bensoussan HJ, Autexier C. Telomerase Regulation from Beginning to the End. Genes (Basel). 2016;7(9)
Feng J, Funk WD, Wang SS, et al. The RNA component of human telomerase. Science. 1995;269(5228):1236–41.
Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132(6):945–57.
Vulliamy T, Beswick R, Kirwan M, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci U S A. 2008;105(23):8073–8.
Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315(5820):1850–3.
Saito T, Matsuda Y, Suzuki T, et al. Comparative gene mapping of the human and mouse TEP1 genes, which encode one protein component of telomerases. Genomics. 1997;46(1):46–50.
Liu L, Lai S, Andrews LG, Tollefsbol TO. Genetic and epigenetic modulation of telomerase activity in development and disease. Gene. 2004;340(1):1–10.
Akincilar SC, Unal B, Tergaonkar V. Reactivation of telomerase in cancer. Cell Mol Life Sci. 2016;73(8):1659–70.
Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8(3):177–80.
Weinrich SL, Pruzan R, Ma L, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17(4):498–502.
Ishikawa F. Regulation mechanisms of mammalian telomerase. A review. Biochemistry (Mosc). 1997;62(11):1332–7.
Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S, Greider CW. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56(3):645–50.
Yi XJ, Jiang HY, Lee KK, WS O, Tang PL, Chow PH. Expression of vascular endothelial growth factor (VEGF) and its receptors during embryonic implantation in the golden hamster (Mesocricetus auratus). Cell Tissue Res. 1999;296(2):339–49.
Morales CP, Holt SE, Ouellette M, et al. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet. 1999;21(1):115–8.
Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–52.
Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66(3):407–25. table of contents
Kyo S, Inoue M. Complex regulatory mechanisms of telomerase activity in normal and cancer cells: how can we apply them for cancer therapy? Oncogene. 2002;21(4):688–97.
Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25(3):565–74.
Nakamura TM, Morin GB, Chapman KB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277(5328):955–9.
Kilian A, Bowtell DD, Abud HE, et al. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6(12):2011–9.
Liu X, Wang Y, Chang G, Wang F, Wang F, Geng X. Alternative Splicing of hTERT Pre-mRNA: A Potential Strategy for the Regulation of Telomerase Activity. Int J Mol Sci. 2017;18(3)
Counter CM, Gupta J, Harley CB, Leber B, Bacchetti S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood. 1995;85(9):2315–20.
Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci U S A. 1995;92(20):9082–6.
Cifuentes-Rojas C, Shippen DE. Telomerase regulation. Mutat Res. 2012;730(1-2):20–7.
Harle-Bachor C, Boukamp P. Telomerase activity in the regenerative basal layer of the epidermis inhuman skin and in immortal and carcinoma-derived skin keratinocytes. Proc Natl Acad Sci U S A. 1996;93(13):6476–81.
Kyo S, Takakura M, Kohama T, Inoue M. Telomerase activity in human endometrium. Cancer Res. 1997;57(4):610–4.
Saito T, Schneider A, Martel N, et al. Proliferation-associated regulation of telomerase activity in human endometrium and its potential implication in early cancer diagnosis. Biochem Biophys Res Commun. 1997;231(3):610–4.
Brien TP, Kallakury BV, Lowry CV, et al. Telomerase activity in benign endometrium and endometrial carcinoma. Cancer Res. 1997;57(13):2760–4.
Hiyama K, Hirai Y, Kyoizumi S, et al. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J Immunol. 1995;155(8):3711–5.
Ramirez RD, Wright WE, Shay JW, Taylor RS. Telomerase activity concentrates in the mitotically active segments of human hair follicles. J Invest Dermatol. 1997;108(1):113–7.
Glybochko PV, Zezerov EG, Glukhov AI, et al. Telomerase as a tumor marker in diagnosis of prostatic intraepithelial neoplasia and prostate cancer. Prostate. 2014;74(10):1043–51.
Breslow RA, Shay JW, Gazdar AF, Srivastava S. Telomerase and early detection of cancer: a National Cancer Institute workshop. J Natl Cancer Inst. 1997;89(9):618–23.
Yoshida K, Sugino T, Goodison S, et al. Detection of telomerase activity in exfoliated cancer cells in colonic luminal washings and its related clinical implications. Br J Cancer. 1997;75(4):548–53.
Yoshida K, Sugino T, Tahara H, et al. Telomerase activity in bladder carcinoma and its implication for noninvasive diagnosis by detection of exfoliated cancer cells in urine. Cancer. 1997;79(2):362–9.
Umbricht CB, Saji M, Westra WH, Udelsman R, Zeiger MA, Sukumar S. Telomerase activity: a marker to distinguish follicular thyroid adenoma from carcinoma. Cancer Res. 1997;57(11):2144–7.
Lin Y, Miyamoto H, Fujinami K, et al. Telomerase activity in human bladder cancer. Clin Cancer Res. 1996;2(6):929–32.
Kulic A, Plavetic ND, Gamulin S, Jakic-Razumovic J, Vrbanec D, Sirotkovic-Skerlev M. Telomerase activity in breast cancer patients: association with poor prognosis and more aggressive phenotype. Med Oncol. 2016;33(3):23.
Carey LA, Kim NW, Goodman S, et al. Telomerase activity and prognosis in primary breast cancers. J Clin Oncol. 1999;17(10):3075–81.
Tahara H, Kuniyasu H, Yokozaki H, et al. Telomerase activity in preneoplastic and neoplastic gastric and colorectal lesions. Clin Cancer Res. 1995;1(11):1245–51.
Wu Y, Tang Y, Jiang ZQ. Diagnosis of human bladder cancer by detecting the telomerase activity in exfoliated urothelial cells. Hunan Yi Ke Da Xue Xue Bao. 2000;25(6):599–600.
Ahn MJ, Noh YH, Lee YS, et al. Telomerase activity and its clinicopathological significance in gastric cancer. Eur J Cancer. 1997;33(8):1309–13.
Fernandez-Marcelo T, Gomez A, Pascua I, et al. Telomere length and telomerase activity in non-small cell lung cancer prognosis: clinical usefulness of a specific telomere status. J Exp Clin Cancer Res. 2015;34:78.
Graham MK, Meeker A. Telomeres and telomerase in prostate cancer development and therapy. Nature Reviews Urology. 2017;14(10):607–19.
Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8(1):69.
Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3(11):1271–4.
Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–5.
Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11(5):319–30.
Koziel JE, Fox MJ, Steding CE, Sprouse AA, Herbert BS. Medical genetics and epigenetics of telomerase. J Cell Mol Med. 2011;15(3):457–67.
Umbricht CB, Sherman ME, Dome J, et al. Telomerase activity in ductal carcinoma in situ and invasive breast cancer. Oncogene. 1999;18(22):3407–14.
Castelo-Branco P, Choufani S, Mack S, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14(6):534–42.
Shay JW, Gazdar AF. Telomerase in the early detection of cancer. J Clin Pathol. 1997;50(2):106–9.
Shay JW, Wright WE. The reactivation of telomerase activity in cancer progression. Trends Genet. 1996;12(4):129–31.
Naderlinger E, Holzmann K. Epigenetic Regulation of Telomere Maintenance for Therapeutic Interventions in Gliomas. Genes (Basel). 2017;8:5.
Barthel FP, Wei W, Tang M, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;
Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22(8):447–55.
McClintock B. The Fusion of Broken Ends of Chromosomes Following Nuclear Fusion. Proc Natl Acad Sci U S A. 1942;28(11):458–63.
Zhang A, Zheng C, Lindvall C, et al. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000;60(22):6230–5.
Piscuoglio S, Ng CK, Murray M, et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J Pathol. 2016;238(4):508–18.
Xie H, Liu T, Wang N, et al. TERT promoter mutations and gene amplification: promoting TERT expression in Merkel cell carcinoma. Oncotarget. 2014;5(20):10048–57.
Wang N, Kjellin H, Sofiadis A, et al. Genetic and epigenetic background and protein expression profiles in relation to telomerase activation in medullary thyroid carcinoma. Oncotarget. 2016;7(16):21332–46.
Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44.
Borah S, Xi L, Zaug AJ, et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347(6225):1006–10.
Valentijn LJ, Koster J, Zwijnenburg DA, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47(12):1411–4.
Peifer M, Hertwig F, Roels F, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526(75):700–4.
Kawashima M, Kojima M, Ueda Y, Kurihara S, Hiyama E. Telomere biology including TERT rearrangements in neuroblastoma: a useful indicator for surgical treatments. J Pediatr Surg. 2016;51(12):2080–5.
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–9.
Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61.
Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185.
Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99(8):1528–38.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46(12):1258–63.
Lewis KA, Tollefsbol TO. Regulation of the Telomerase Reverse Transcriptase Subunit through Epigenetic Mechanisms. Front Genet. 2016;7:83.
Aisner DL, Wright WE, Shay JW. Telomerase regulation: not just flipping the switch. Curr Opin Genet Dev. 2002;12(1):80–5.
Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene. 1999;232(1):97–106.
Bell RJ, Rube HT, Kreig A, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348(6238):1036–9.
Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015;29(21):2219–24.
Huang DS, Wang Z, He XJ, et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur J Cancer. 2015;51(8):969–76.
Vallarelli AF, Rachakonda PS, Andre J, et al. TERT promoter mutations in melanoma render TERT expression dependent on MAPK pathway activation. Oncotarget. 2016;7(33):53127–36.
Hurst CD, Platt FM, Knowles MA. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur Urol. 2014;65(2):367–9.
Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73(24):7162–7.
Wang N, Liu T, Sofiadis A, et al. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer. 2014;120(19):2965–79.
Shain AH, Garrido M, Botton T, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47(10):1194–9.
Populo H, Boaventura P, Vinagre J, et al. TERT promoter mutations in skin cancer: the effects of sun exposure and X-irradiation. J Invest Dermatol. 2014;134(8):2251–7.
Liu X, Wu G, Shan Y, Hartmann C, von Deimling A, Xing M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle. 2013;12(10):1637–8.
Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23(3):R143–55.
Chiba K, Lorbeer FK, Shain AH, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. 2017;357(6358):1416–20.
Cheng L, Montironi R, Lopez-Beltran A. TERT Promoter Mutations Occur Frequently in Urothelial Papilloma and Papillary Urothelial Neoplasm of Low Malignant Potential. Eur Urol. 2017;71(3):497–8.
Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6.
Vinagre J, Pinto V, Celestino R, et al. Telomerase promoter mutations in cancer: an emerging molecular biomarker? Virchows Arch. 2014;465(2):119–33.
Allory Y, Beukers W, Sagrera A, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65(2):360–6.
Rachakonda PS, Hosen I, de Verdier PJ, et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc Natl Acad Sci U S A. 2013;110(43):17426–31.
Liu X, Bishop J, Shan Y, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20(4):603–10.
Wu S, Huang P, Li C, et al. Telomerase reverse transcriptase gene promoter mutations help discern the origin of urogenital tumors: a genomic and molecular study. Eur Urol. 2014;65(2):274–7.
Chan AK, Yao Y, Zhang Z, et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol. 2015;28(2):177–86.
Gao K, Li G, Qu Y, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7(8):8712–25.
Killela PJ, Pirozzi CJ, Healy P, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–25.
Zhang ZY, Chan AK, Ding XJ, et al. TERT promoter mutations contribute to IDH mutations in predicting differential responses to adjuvant therapies in WHO grade II and III diffuse gliomas. Oncotarget. 2015;6(28):24871–83.
Griewank KG, Murali R, Puig-Butille JA, et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014;106(9)
Nagore E, Heidenreich B, Rachakonda S, et al. TERT promoter mutations in melanoma survival. Int J Cancer. 2016;139(1):75–84.
Nault JC, Calderaro J, Di Tommaso L, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014;60(6):1983–92.
Pinyol R, Tovar V, Llovet JM. TERT promoter mutations: gatekeeper and driver of hepatocellular carcinoma. J Hepatol. 2014;61(3):685–7.
Xing M, Liu R, Liu X, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014;32(25):2718–26.
Liu R, Bishop J, Zhu G, Zhang T, Ladenson PW, Xing M. Mortality Risk Stratification by Combining BRAF V600E and TERT Promoter Mutations in Papillary Thyroid Cancer: Genetic Duet of BRAF and TERT Promoter Mutations in Thyroid Cancer Mortality. JAMA Oncol. 2016;
Xing M, Alzahrani AS, Carson KA, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol. 2015;33(1):42–50.
Yuan P, Cao JL, Abuduwufuer A, et al. Clinical Characteristics and Prognostic Significance of TERT Promoter Mutations in Cancer: A Cohort Study and a Meta-Analysis. PLoS One. 2016;11(1):e0146803.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22.
Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103(5):1412–7.
Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34.
Hatada I, Fukasawa M, Kimura M, et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25(21):3059–64.
Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–54.
Bartlett TE, Zaikin A, Olhede SC, West J, Teschendorff AE, Widschwendter M. Corruption of the intra-gene DNA methylation architecture is a hallmark of cancer. PLoS One. 2013;8(7):e68285.
Stoehr R, Taubert H, Zinnall U, et al. Frequency of TERT Promoter Mutations in Prostate Cancer. Pathobiology. 2015;82(2):53–7.
Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101(4):335–41.
Shin KH, Kang MK, Dicterow E, Park NH. Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br J Cancer. 2003;89(8):1473–8.
Dessain SK, Yu H, Reddel RR, Beijersbergen RL, Weinberg RA. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60(3):537–41.
Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59(24):6087–90.
Castelo-Branco P, Leao R, Lipman T, et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: a retrospective cohort study. Oncotarget. 2016;7(36):57726–36.
Azouz A, Wu YL, Hillion J, et al. Epigenetic plasticity of hTERT gene promoter determines retinoid capacity to repress telomerase in maturation-resistant acute promyelocytic leukemia cells. Leukemia. 2010;24(3):613–22.
Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67(1):194–201.
Tsujioka T, Yokoi A, Itano Y, et al. Five-aza-2'-deoxycytidine-induced hypomethylation of cholesterol 25-hydroxylase gene is responsible for cell death of myelodysplasia/leukemia cells. Sci Rep. 2015;5:16709.
Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33(21):6850–60.
Lopatina NG, Poole JC, Saldanha SN, et al. Control mechanisms in the regulation of telomerase reverse transcriptase expression in differentiating human teratocarcinoma cells. Biochem Biophys Res Commun. 2003;306(3):650–9.
Wu KJ, Grandori C, Amacker M, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21(2):220–4.
Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251(4990):186–9.
Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9(2):158–63.
Bert SA, Robinson MD, Strbenac D, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23(1):9–22.
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95.
Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–68.
Seynnaeve B, Lee S, Borah S, et al. Genetic and Epigenetic Alterations of TERT Are Associated with Inferior Outcome in Adolescent and Young Adult Patients with Melanoma. Sci Rep. 2017;7:45704.
Zhao X, Tian X, Kajigaya S, et al. Epigenetic landscape of the TERT promoter: a potential biomarker for high risk AML/MDS. Br J Haematol. 2016;175(3):427–39.
Deng J, Zhou D, Zhang J, et al. Aberrant methylation of the TERT promoter in esophageal squamous cell carcinoma. Cancer Gene Ther. 2015;208(12):602–9.
Kochling M, Ewelt C, Furtjes G, et al. hTERT promoter methylation in pituitary adenomas. Brain Tumor Pathol. 2016;33(1):27–34.
Choi JH, Park SH, Park J, et al. Site-specific methylation of CpG nucleotides in the hTERT promoter region can control the expression of hTERT during malignant progression of colorectal carcinoma. Biochem Biophys Res Commun. 2007;361(3):615–20.
Zhang H, Weng X, Ye J, He L, Zhou D, Liu Y. Promoter hypermethylation of TERT is associated with hepatocellular carcinoma in the Han Chinese population. Clin Res Hepatol Gastroenterol. 2015;39(5):600–9.
Furtjes G, Kochling M, Peetz-Dienhart S, et al. hTERT promoter methylation in meningiomas and central nervous hemangiopericytomas. J Neuro-Oncol. 2016;130(1):79–87.
Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–59.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34.
Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7.
Huppi K, Volfovsky N, Mackiewicz M, et al. MicroRNAs and genomic instability. Semin Cancer Biol. 2007;17(1):65–73.
Vincent K, Pichler M, Lee GW, Ling H. MicroRNAs, genomic instability and cancer. Int J Mol Sci. 2014;15(8):14475–91.
Di Leva G, Calin GA, Croce CM. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res C Embryo Today. 2006;78(2):180–9.
Li J, Lei H, Xu Y, Tao ZZ. miR-512-5p suppresses tumor growth by targeting hTERT in telomerase positive head and neck squamous cell carcinoma in vitro and in vivo. PLoS One. 2015;10(8):e0135265.
Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–38.
Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258.
Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60.
Perez-Rivas LG, Jerez JM, Carmona R, et al. A microRNA signature associated with early recurrence in breast cancer. PLoS One. 2014;9(3):e91884.
Hrdlickova R, Nehyba J, Bargmann W, Bose HR Jr. Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS One. 2014;9(2):e86990.
Bai L, Wang H, Wang AH, Zhang LY, Bai J. MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. PLoS One. 2017;12(3):e0173912.
Zhou J, Dai W, Song J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun. 2016;470(2):445–52.
Chen L, Lu MH, Zhang D, et al. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5:e1034.
Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99(2):280–6.
Cittelly DM, Das PM, Spoelstra NS, et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317.
Ostling P, Leivonen SK, Aakula A, et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71(5):1956–67.
Xu X, Chen W, Miao R, et al. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget. 2015;6(6):3988–4004.
Liu K, Li L, Rusidanmu A, Wang Y, Lv X. Down-Regulation of MiR-1294 is Related to Dismal Prognosis of Patients with Esophageal Squamous Cell Carcinoma through Elevating C-MYC Expression. Cell Physiol Biochem. 2015;36(1):100–10.
Catto JW, Alcaraz A, Bjartell AS, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59(5):671–81.
Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–41.
Peng F, Zhang Y, Wang R, et al. Identification of differentially expressed miRNAs in individual breast cancer patient and application in personalized medicine. Oncogene. 2016;5:e194.
Cimino-Reale G, Gandellini P, Santambrogio F, Recagni M, Zaffaroni N, Folini M. miR-380-5p-mediated repression of TEP1 and TSPYL5 interferes with telomerase activity and favours the emergence of an “ALT-like” phenotype in diffuse malignant peritoneal mesothelioma cells. J Hematol Oncol. 2017;10(1):140.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22.
Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–50.
Acknowledgements
Not applicable.
Funding
This work was supported by the research grant UID/BIM/04773/2013 CBMR from FCT. RL is supported by FCT Doctoral Grant SFRH/BD/102232/2014 and JDA by a PD/BD/105899/2014 FCT fellowship.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
RL, JDA and DL equally contributed to this manuscript – literature review, manuscript conception and design; drafting and writing. AF, UT and PCB provided scientific guidance, critical revision and manuscript writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Leão, R., Apolónio, J.D., Lee, D. et al. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J Biomed Sci 25, 22 (2018). https://doi.org/10.1186/s12929-018-0422-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12929-018-0422-8