Abstract
Background
Ischemic stroke (IS) is a common and serious neurological condition that is highly fatal but so far no early diagnostic markers are available. Myocardial infarction-associated transcript (MIAT) is a long non-coding RNA (lncRNA) that could lead to IS by inducing autophagy and apoptosis in neuronal cells. However, there has been no report on the link between susceptibility to IS and the single-nucleotide polymorphisms (SNPs) of MIAT. This study aimed to investigate the association between MIAT gene polymorphisms and IS risk.
Methods
A total of 320 IS patients and 310 age-, sex- and race-matched controls were included in this study. Four polymorphisms (rs2157598, rs5761664, rs1894720, and rs9625066) were genotyped by using SNPscan technique.
Results
Among the 4 polymorphisms of MIAT, only rs9625066 was associated with IS risk (CA vs. CC: adjusted OR = 0.55, 95% CI, 0.37–0.85, P = 0.006; AA vs. CC: adjusted OR = 0.39, 95% CI, 0.16–0.94, P = 0.036; (AA + CA vs. CC: adjusted OR = 0.53, 95% CI, 0.35–0.80, P = 0.002; A vs. C adjusted OR = 0.59, 95% CI, 0.42–0.82, P = 0.002). Haplotype analysis showed a 1.32-fold increase (95% CI, 1.05–1.67, P = 0.017) in IS risk for rs2157598-rs5761664-rs1894720-rs9625066 (A-C-G-C). Logistic regression analysis identified some independent impact factors for IS including rs9625066 AA/AC, TC, TG, HDL-C (P < 0.05).
Conclusion
The rs9625066 polymorphism of MIAT might be associated with IS susceptibility in Chinese population, in which AA/CA plays a protective role in IS, whereas the CC genotype increases the risk of developing IS, suggesting it might be a marker predictive of IS risk.
Similar content being viewed by others
Introduction
Ischemic stroke (IS), also known clinically as cerebral infarction, represents a common and serious condition in which brain blood flow is reduced or interrupted due to stenosis or occlusion of arteries, resulting in ischemic necrosis of brain tissue in the affected area [1, 2]. IS is clinically characterized by sudden onset, rapid progression, and high mortality and disability rates, among others [3]. IS reportedly accounted for up to 80% of cardiovascular and cerebrovascular diseases and has been a subject of active studies. The risk factors associated with IS include age, gender, dietary habits, hyperglycemia, hyperlipidemia, and hypertension, etc. [4]. In recent years, the pathogenesis of IS has been found to be intimately linked to genetic factors, such as transcription, expression and variation of some genes [5, 6].
Long non-coding RNAs (lncRNAs), as an important class of non-coding RNAs, have been extensively studied in a wide array of biological processes and mounting evidence showed that they play a key role in the development of a multitude of diseases [7]. The lncRNA MIAT (Myocardial Infarction Associated Transcript) is located on chromosome 22q12.1 and was originally found to be a lncRNA associated with myocardial infarction [8]. However, recent studies revealed that the lncRNA MIAT was not only associated with cardiovascular diseases, but also bore close correlation with neurological conditions, including IS [9,10,11]. Guo et al. found that MIAT expression was increased in IS rats and in PC12 cells injured by oxygen-glucose deprivation/reoxygenation (OGD/R) and the MIAT over-expression exacerbated IS by promoting autophagy and apoptosis in neuronal cells through upregulating REDD1 (DNA damage responses 1) expression [12]. Li et al. demonstrated that MIAT overexpression increased infarct volume and induced neuronal apoptosis in the rat middle cerebral artery occlusion (MCAO) model [13]. These findings support that the MIAT might be implicated in the pathological process of IS.
At present, lncRNA-associated single-nucleotide polymorphisms (SNPs) have been shown to potentially influence the susceptibility to IS. For example, the rs2240183 CT/CC genotype and the C allele in the promoter of the lncRNA TUG1 gene were found to be associated with an increased risk of IS [14], whereas the rs1194338 AC/AA genotype in the promoter of the lncRNA MALAT1 might be a protective factor against IS [15]. In this study, we, by employing a case-control method, investigated the correlation between SNPs in the MIAT gene (rs2157598, rs5761664, rs1894720, and rs9625066) and the susceptibility to IS in Chinese population.
Materials and methods
Study population
Three hundred twenty samples in the case group came from patients who presented at the Affiliated Hospital of Guilin Medical College in Guangxi, China, from June 2020 to June 2021. Their clinical symptoms, CT and MRI findings satisfied the diagnostic criteria of IS proposed by the 2014 Chinese Diagnostic Guidelines for Acute Ischemic Stroke, and the candidate subjects were excluded if their strokes were caused by traumas, brain tumors, cerebral parasitosis, or other diseases. Three hundred ten cases in the control group were sex- and age-matched healthy subjects recruited at the same period of time, and those with cardiovascular and cerebrovascular diseases, tumors, neurological diseases, and hereditary diseases were excluded. The following data were collected: sex, age, blood glucose level, triglycerides (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C).
Ethics
This study was conducted in accordance with the guidelines of the Helsinki Declaration and approved by the Ethics Review Committee of Affiliated Hospital of Guilin Medical University, and all subjects who participated in the study signed an informed consent form.
Screening of SNPs and the basis of selection
We searched for MIAT gene-related information in the NCBI database. Four SNPs (rs2157598, rs5761664, rs1894720 and rs9625066) were selected against the following criteria: (1) loci with minor allele frequency (MAF) > 5% in the Chinese Han population, (2) SNPs with potential functions in the promoter region of the MIAT gene as predicted by computerized analyses, and (3) loci that had been reported in the literature and have been studied in other diseases. Our selection criteria were to ensure that data were more in line with the genetic profile of the target population, with focus directed at SNPs that bore correlations with biological functions and potential disease, thus increasing the power of the study and the interpretability of the results.
DNA extraction and genotyping
3–5 ml of peripheral blood samples were harvested and genomic DNA was extracted using a DNA extraction kit (DP318, Qiagen, China). Genotyping was performed by using the SNPscan™ Multiple SNP typing kit on the ABI3730XL amplifier (PE Applied Biosystems, Foster City, CA, USA), and the data collected were analyzed using GeneMapper 4.1 (Applied Biosystems, USA). Testing was repeated on 10% of randomly selected DNA samples to verify the reproducibility of the experiments, and a consistency rate of > 99% was required.
Statistical analysis
All statistical analyses were performed by employing IBM SPSS 20.0 software package (SPSS Inc., Chicago, IL, USA). Continuous variables were expressed as mean ± standard deviation (SD) and analyzed using t-test. Hardy-Weinberg equilibrium (HWE) and categorical data, such as gender and diabetes, were analyzed using chi-square test. Logistic regression was utilized to analyze the association between SNP and IS. Risk was assessed by calculating odds ratio (OR) and 95% confidence intervals (95% CI). OR was adjusted for sex, age, presence of diabetes, TC, TG, HDL-C, and LDL-C. SHEsis software was employed for linkage disequilibrium (LD) and haplotype analysis. Difference was considered to be statistically significant when a P < 0.05.
Results
Characteristics of the study population
The features of the study population are shown in Table 1. There were no significant differences in the distribution of cases and controls in terms of age, sex and presence of diabetes mellitus (P > 0.05). Levels of TC, TG and LDL and lower levels of HDL were higher in IS patients than in controls (all P < 0.05).
Association between MIAT polymorphisms and IS risk
In this study, all four SNPs were found to have three genotypes as analyzed by SNPscan technique (Table 2). The distribution of genotypes in the case and control groups followed the HWE (P > 0.05). The A allele of rs9625066 was found to be associated with a lower risk of IS as compared with the C allele (A vs. C adjusted OR = 0.59, 95% CI, 0.42-0.82, P = 0.002), and that AA genotypes, CA genotypes, and the dominant model bore a correlation with the lower risk for IS (CA vs. CC: adjusted OR = 0.55, 95% CI, 0.37-0.85, P = 0.006; AA vs. CC: adjusted OR = 0. 39, 95% CI, 0.16–0.94, P = 0.036; AA + CA vs. CC: adjusted OR = 0.53, 95% CI, 0.35–0.80, P = 0.002). No significant associations were observed between the rs2157598, rs5761664, and rs1894720 polymorphisms and the risk of IS (P > 0.05). Subsequent correction for multiple comparisons revealed that the initially observed protective effect of the AA genotype on IS has lost statistical significance. Therefore, further studies with a larger sample size are needed to confirm these associations.
Haplotype analysis of MIAT polymorphisms and IS risk
We assessed the haplotype frequencies of the four SNPs in the MIAT gene in IS patients and controls by using the online SHEsis software (Table 3). The results revealed that rs2157598-rs5761664-rs1894720-rs9625066 (A-C-G-C) were the predominant haplotypes in IS patients and healthy controls. We also found that the elevated risk of IS was associated with the presence of haplotypes (A-C-G-C) (OR = 1.32,95% Cl, 1.05–1.67, P = 0.017). The presence of linkage disequilibrium (LD) indicated there existed a correlation between specific genotypes. We observed linkage disequilibrium in the four loci, among which a strong linkage disequilibrium was found between rs2157598 and rs5761664 (D' = 0.99, r2 = 0.71) (Fig. 1).
Analysis of risk factors for IS
Logistic regression was applied to analyze the risk factors for IS, and the results are listed in Table 4. The risk factors included: TG (OR = 1.49, 95%CI, 1.26-1.77), TC (OR = 1.65, 95%CI, 1.28–2.11), HDL-C (OR = 0.02; 95%CI, 0.01-0.04), LDL-C (OR = 0.98, 95% CI, 0.76-1.25) and rs9625066AA/AC (OR = 0.55; 95% CI, 0.37-0.83) (P < 0.05). Logistic regression analysis exhibited that the TC, TG and HDL-C were still associated with the risk of IS.
Correlation between MIAT gene SNPs and tissue-specific expression
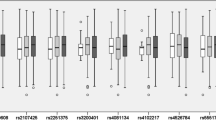
We used GTEx data (https://www.gtexportal.org/home/) to determine the correlation between MIAT gene SNPs and tissue-specific expressions. Expression quantitative trait loci (eQTL) analysis, or expression quantitative trait loci analysis, revealed how genetic variation at a locus can impact the expression levels of specific genes across different tissues. The eQTL analysis showed that the rs9625066 polymorphism was correlated with MIAT expression in tissues, and CC carriers of rs9625066 had elevated MIAT expression in brain tissues, such as cerebral-hippocampus, and cerebral-cerebellar hemispheres (P < 0.001) (Fig. 2).
Discussion
In this study, we examined the association between four single nucleotide polymorphisms (rs2157598, rs5761664, rs1894720, and rs9625066) in the MIAT gene and the risk for the development of IS. We found that individuals carrying the MIAT gene rs9625066 AA/CA genotype and A allele had significantly reduced risk for IS development. However, we failed to found any significant association between rs2157598, rs5761664, rs1894720 and risk for IS. Haplotype analysis revealed that the (A-C-G-A) haplotype increased the risk of IS development by 1.32-fold. Logistic regression analysis identified some independent impact factors for IS including rs9625066 AA/AC, TC, TG, HDL-C. These findings suggest that MIAT rs9625066 might etiologically contributed to the pathogenesis of IS.
Initially identified as a lncRNA in 2006, MIAT is a polyadenylated transcript, measuring roughly 10 kb and containing seven exons [16, 17]. MIAT is highly conserved in mammals, where it accumulates in the nucleus and is expressed in the central nervous system, heart, lungs and spleen [18]. The role of MIAT in neurological diseases is reportedly regulated by miRNAs. Specifically, upregulated miR-204-5p could effectively mitigate the injury of cerebral microvascular endothelial cells by silencing MIAT expression, while inducing neovascularization and significantly increasing the number of surviving neurons [19]. Overexpression of MIAT exacerbated the impaired behavioral activity and neurological function in mice by competitively binding to miR-874-3p, causing neuronal apoptosis and up-regulating the expression of inflammatory factors [20]. In addition, MIAT promoted the proliferation of human carotid smooth muscle cells through the ERK-ELK1-EGR1 pathway, thereby playing a pivotal role in the development and destabilization of atherosclerotic plaques [21]. It has also been found that MIAT expression was significantly elevated in atherosclerotic plaques, and this plaque buildup led to lumen narrowing, which, in turn, contributed to the development of IS [22]. A study by Zhu et al. also confirmed that MIAT was significantly upregulated in IS patients, further supporting that MIAT might be involved in the development and progression of IS [23]. Overall, MIAT might act as an important regulator in a variety of neurological diseases, especially in the development of IS.
In recent years, the association between lncRNA-related polymorphisms and disease risk has become a topic of active investigations. Since IncRNA is functionally-diverse, MIAT-associated SNPs have attracted the attention of researchers [24, 25]. For example, Ma et al. explored the relationship between nine MIAT SNPs and acute myocardial infarction (AMI) in the Chinese Han population and confirmed that patients carrying the rs5752375 TT genotype had a 3.91-fold increased risk of developing AMI compared to the carriers of CC genotype [26], and rs1894720 of MIAT was significantly associated with paranoid schizophrenia, with AA carriers exhibiting an increased risk for this disorder [27]. Furthermore, Li and his colleagues and others found that subjects with the GT and TT genotypes of rs1894720 were at a higher risk of age-related cataracts [28]. These studies implied that functional SNPs in MIAT could serve as potential indicators of related diseases. Globally, IS has become a major public health concern due to the unavailability of early diagnostic markers, and a high mortality rate. In recent years, with the development of molecular biology and the completion of the Human Genome Project, some advances have been made in the regulation of the expression of SNPs of lncRNAs in diseases [29,30,31]. For example, the rs145204276 del allele of lncRNA- GAS5 was found to raise the risk of IS by increasing the transcriptional activity and the expression of rs145204276 of lncRNA-GAS [32]. Rezaei et al. found that the rs217727 recessive model of lncRNA H19 increased the risk of IS by 2.80-fold [33]. Moreover, the lncRNA-MALAT1 rs619586 AA and rs3200401 CT, TT genotypes were associated with an elevated risk of IS, whereas the lncRNA-ANRIL rs10965215 GG genotype was found to be protective against IS [34]. These studies provided new insights into the genetic mechanisms of IS. We further confirmed the hypothesis that SNPs of MIAT are associated with the risk of IS, and found that individuals with rs9625066 AA/CA genotypes and the A allele significantly lowered the risk for the development of IS. The A-C-G-A haplotype was associated with a reduced risk of IS development. Logistic regression also showed that the rs9625066 AA/CA exerted an effect on risk for IS. Additionally, the GTEx database search exhibited that subjects with the rs9625066 CC genotype had higher levels of MIAT expression in brain-hippocampus and brain-cerebellar hemisphere. The exact mechanisms underlying the findings need to be confirmed by further studies. In conclusion, the rs9625066 in MIAT might serve as a new target for the treatment of IS.
This study revealed a correlation between MIAT gene polymorphisms and IS. However, some limitations of this study have to be acknowledged. Firstly, we included samples from hospitals in the same region, which rendered it difficult to completely eliminate the possibility of selection bias. Secondly, due to the limited clinical data of the patients, we were unable to obtain relevant information on smoking and alcohol consumption, which prevented the further analysis of gene-environment interactions. In addition, the relatively small sample size might affect the power of our findings. Therefore, a larger sample size is needed to further validate the role of MIAT SNPs in IS susceptibility.
Conclusions
In conclusion, we demonstrated a significant association between lncRNA-MIAT rs9625066 and IS in the Chinese population. The rs9625066 A allele contributed to a reduced risk of IS, whereas the C allele increased the risk of developing IS. These findings suggest that rs9625066 may be a potential biomarker for the occurrence and development of IS.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Feske SK. Ischemic Stroke. Am J Med. 2021;134(12):1457–64.
Coupland AP, Thapar A, Qureshi MI, Jenkins H, Davies AH. The definition of stroke. J R Soc Med. 2017;110(1):9–12.
Feigin VL, Brainin M, Norrving B, Martins S, Sacco RL, Hacke W, et al. World Stroke Organization (WSO): global stroke fact sheet 2022. Int J Stroke. 2022;17(1):18–29.
Barthels D, Das H. Current advances in ischemic stroke research and therapies. Biochim Biophys Acta Mol Basis Dis. 2020;1866(4):165260.
Hassan A, Markus HS. Genetics and ischaemic stroke. Brain. 2000;123(Pt 9):1784–812.
Dichgans M, Pulit SL, Rosand J. Stroke genetics: discovery, biology, and clinical applications. Lancet Neurol. 2019;18(6):587–99.
Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–16.
Ghafouri-Fard S, Azimi T, Taheri M. Myocardial Infarction Associated Transcript (MIAT): review of its impact in the tumorigenesis. Biomed Pharmacother. 2021;133:111040.
He X, Zhang J, Guo Y, Yang X, Huang Y, Hao D. LncRNA MIAT promotes spinal cord injury recovery in rats by regulating RBFOX2-mediated alternative splicing of MCL-1. Mol Neurobiol. 2022;59(8):4854–68.
Bountali A, Tonge DP, Mourtada-Maarabouni M. RNA sequencing reveals a key role for the long non-coding RNA MIAT in regulating neuroblastoma and glioblastoma cell fate. Int J Biol Macromol. 2019;130:878–91.
Li EY, Zhao PJ, Jian J, Yin BQ, Sun ZY, Xu CX, et al. LncRNA MIAT overexpression reduced neuron apoptosis in a neonatal rat model of hypoxic-ischemic injury through miR-211/GDNF. Cell Cycle. 2019;18(2):156–66.
Guo X, Wang Y, Zheng D, Cheng X, Sun Y. LncRNA-MIAT promotes neural cell autophagy and apoptosis in ischemic stroke by up-regulating REDD1. Brain Res. 2021;1763:147436.
Li S, Fu J, Wang Y, Hu C, Xu F. LncRNA MIAT enhances cerebral ischaemia/reperfusion injury in rat model via interacting with EGLN2 and reduces its ubiquitin-mediated degradation. J Cell Mol Med. 2021;25(21):10140–51.
Wei YS, Yang J, He YL, Shi X, Zeng ZN. A functional polymorphism in the promoter of TUG1 is associated with an increased risk of ischaemic stroke. J Cell Mol Med. 2019;23(9):6173–81.
Wang Y, Gu XX, Huang HT, Liu CH, Wei YS. A genetic variant in the promoter of lncRNA MALAT1 is related to susceptibility of ischemic stroke. Lipids Health Dis. 2020;19(1):57.
Liao J, He Q, Li M, Chen Y, Liu Y, Wang J. LncRNA MIAT: myocardial infarction associated and more. Gene. 2016;578(2):158–61.
Yang C, Zhang Y, Yang B. MIAT, a potent CVD-promoting lncRNA. Cell Mol Life Sci. 2021;79(1):43.
Yang L, Deng J, Ma W, Qiao A, Xu S, Yu Y, et al. Ablation of lncRNA Miat attenuates pathological hypertrophy and heart failure. Theranostics. 2021;11(16):7995–8007.
Deng W, Fan C, Shen R, Wu Y, Du R, Teng J. Long noncoding MIAT acting as a ceRNA to sponge microRNA-204-5p to participate in cerebral microvascular endothelial cell injury after cerebral ischemia through regulating HMGB1. J Cell Physiol. 2020;235(5):4571–86.
Zhang S, Zhang Y, Wang N, Wang Y, Nie H, Zhang Y, et al. Long non-coding RNA MIAT impairs neurological function in ischemic stroke via up-regulating microRNA-874-3p-targeted IL1B. Brain Res Bull. 2021;175:81–9.
Fasolo F, Jin H, Winski G, Chernogubova E, Pauli J, Winter H, et al. Long noncoding RNA MIAT controls advanced atherosclerotic lesion formation and plaque destabilization. Circulation. 2021;144(19):1567–83.
Arslan S, Berkan Ö, Lalem T, Özbilüm N, Göksel S, Korkmaz Ö, et al. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis. 2017;266:176–81.
Zhu M, Li N, Luo P, Jing W, Wen X, Liang C, et al. Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J Stroke Cerebrovasc Dis. 2018;27(2):326–37.
Mohammad HMF, Abdelghany AA, Al Ageeli E, Kattan SW, Hassan R, Toraih EA, et al. Long non-coding RNAs gene variants as molecular markers for diabetic retinopathy risk and response to anti-VEGF therapy. Pharmacogenomics Pers Med. 2021;14:997–1014.
Ishii N, Ozaki K, Sato H, Mizuno H, Susumu S, Takahashi A, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–99.
Ma R, He X, Zhu X, Pang S, Yan B. Promoter polymorphisms in the lncRNA-MIAT gene associated with acute myocardial infarction in Chinese Han population: a case-control study. Biosci Rep. 2020;40(2):BSR20191203.
Rao SQ, Hu HL, Ye N, Shen Y, Xu Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr Res. 2015;166(1–3):125–30.
Li Y, Zhang W, Ke H, Wang Y, Duan C, Zhu Q, et al. Rs1894720 polymorphism is associated with the risk of age-related cataract by regulating the proliferation of epithelial cells in the lens via the signalling pathway of MIAT/miR-26b/BCL2L2. Arch Med Sci. 2022;18(1):223–36.
Yang J, Gu L, Guo X, Huang J, Chen Z, Huang G, et al. LncRNA ANRIL expression and ANRIL gene polymorphisms contribute to the risk of ischemic stroke in the Chinese Han population. Cell Mol Neurobiol. 2018;38(6):1253–69.
Zhu R, Liu X, He Z. Long non-coding RNA H19 and MALAT1 gene variants in patients with ischemic stroke in a northern Chinese Han population. Mol Brain. 2018;11(1):58.
Ahmed Elsabagh DT, Shaker OG, Elgendy HH, Mona MM, AbdElRahimBadr AM. Long noncoding Rna GAS5 And Mir-137 and two of their genetic polymorphisms contribute to acute ischaemic stroke risk in an Egyptian population. J Pak Med Assoc. 2023;73(Suppl 4)(4):S184-s90.
Zheng Z, Liu S, Wang C, Han X. A functional polymorphism rs145204276 in the promoter of long noncoding RNA GAS5 is associated with an increased risk of ischemic stroke. J Stroke Cerebrovasc Dis. 2018;27(12):3535–41.
Rezaei M, Mokhtari MJ, Bayat M, Safari A, Dianatpuor M, Tabrizi R, et al. Long non-coding RNA H19 expression and functional polymorphism rs217727 are linked to increased ischemic stroke risk. BMC Neurol. 2021;21(1):54.
Fathy N, Kortam MA, Shaker OG, Sayed NH. Long noncoding RNAs MALAT1 and ANRIL gene variants and the risk of cerebral ischemic stroke: an association study. ACS Chem Neurosci. 2021;12(8):1351–62.
Acknowledgements
The authors are indebted to all participants who agreed to take part in the study.
Funding
The study was supported by the National Natural Science Foundation of China (NOs.82160313); Guangxi Natural Science Foundation (NOs.2020GXNSFDA297027), Guangxi Medical and Health Key Discipline Construction Project and Graduate Research Program of Guilin Medical University (Nos. GYYK2023015).
Author information
Authors and Affiliations
Contributions
Hong-Bo Liu and Jun-Yang were involved in the conceptualization and revision of the paper. Yin-Hua Weng and Jie Chen were responsible for laboratory tests and drafting of the manuscript; Wen-Tao Yu, Yan-Ping Luo and Chao Liu collected, analyzed, and interpreted the data. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Review Board of the Affiliated Hospital of Guilin Medical University (2022YJSLL-79). All subjects signed informed consent to participate in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Weng, YH., Chen, J., Yu, WT. et al. lncRNA-MIAT rs9625066 polymorphism could be a potential biomarker for ischemic stroke. BMC Med Genomics 17, 58 (2024). https://doi.org/10.1186/s12920-024-01830-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-024-01830-w