Abstract
Background
This study attempted to verify the potential of KCNJ14 as a biomarker in colorectal cancer (CRC).
Methods
Data on transcriptomics and DNA methylation and the clinical information of CRC patients were downloaded from The Cancer Genome Atlas and Gene Expression Omnibus databases. Biological information analysis methods were conducted to determine the role of KCNJ14 in the prognosis, diagnosis, immune cell infiltration, and regulation mechanism of CRC patients. The effect of KCNJ14 on the proliferation and migration of HCT116 and SW480 CRC cell lines was verified by in vitro experiments (MTT, colony-forming, wound healing, and transwell assays). Western blotting was performed to detect the effect of KCNJ14 on the levels of mTOR signalling pathway-related proteins.
Results
KCNJ14 expression was remarkably increased in CRC tissues and cell lines, which reduced the overall survival time of patients. KCNJ14 mRNA was negatively regulated by its methylation site cg17660703, which can also endanger the prognosis of patients with CRC. Functional enrichment analysis suggested that KCNJ14 is involved in the mTOR, NOD-like receptor, and VEGF signalling pathways. KCNJ14 expression was positively correlated with the number of CD4 + T cells and negatively correlated with that of CD8 + T cells in the immune microenvironment. KCNJ14 knockdown significantly reduced not only the proliferation and migration of CRC cell lines but also the levels of mTOR signalling pathway-related proteins.
Conclusions
This study not only increases the molecular understanding of KCNJ14 but also provides a potentially valuable biological target for the treatment of colorectal cancer.
Similar content being viewed by others
Background
Colorectal cancer is the fourth most lethal cancer worldwide, and its incidence is expected to increase to 2,500,000 cases in 2035 [1]. Evidence suggests that the development of CRC is due to hereditary and environmentally harmful factors and long-standing inflammatory bowel disease [2]. Owing to the complex pathogenesis of colorectal cancer, it is difficult to improve the prognosis of patients despite the comprehensive treatment by surgery and adjuvant radiotherapy [3]. Therefore, a treatment scheme that improves the prognosis of patients with colorectal cancer is urgently required. Previous studies have suggested that biologically targeted therapy may be ideal for the treatment of malignant tumours [4]. However, the basic premise of biologically targeted therapy is to identify key regulatory genes that affect the prognosis of patients with malignant tumours.
KCNJ14, also known as IRK4 and KIR2.4, has been mapped to the chromosomal locus 19q13, and the encoded protein has been identified to belong to a family of integral membrane proteins that act as ATP-sensitive inward rectifier potassium (K+) channels [5]. Central to the entire discipline of inward rectifier K+ channels is the concept of regulation of K+ flow into cells at potentials negative to the potassium equilibrium potential [6]. Hence, this class of proteins is a major interest of researches focusing on multiple biological processes, such as heart rate regulation, neurotransmitter release, epithelial electrolyte transport, and participation in immune regulation [7]. Recent advances in inward rectifier K+ channels have heightened the need for cancer research on lung cancer [8], neuroblastoma [9], and glioblastoma [10]. However, only few studies have explored on the regulatory effect of KCNJ14 on the pathological process of malignant tumours, especially in the prognosis of patients with colorectal cancer.
Therefore, this study attempted to investigate the effect of KCNJ14 on the prognosis of patients with colorectal cancer and evaluate its regulatory relationship with the complex pathological process of colorectal cancer. First, transcriptomic expression data, DNA methylation data, and the detailed clinical characteristics of patients with colorectal cancer were collected from the public database to explore the changes in KCNJ14 expression in colorectal cancer and the relationship between KCNJ14 expression and the clinical characteristics of patients. Subsequently, we verified that KCNJ14 knockdown could significantly reduce the proliferation and migration of colorectal cancer cell lines and revealed the regulatory mechanism of KCNJ14 leading to poor prognosis in colorectal cancer. To our knowledge, this is the first study to investigate the potential mechanism of KCNJ14 in colorectal cancer. From a genetic perspective, we identified a novel biological target for diagnosis, treatment, and prognosis of patients with CRC.
Methods
Data collection
TCGA transcriptome profiling data and the clinical information of 488 patients with colorectal adenocarcinomas and 42 adjacent tissues (workflow type: HTSeq-FPKM) were collected from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). Those with missing clinical data, such as age, sex, lymphatic invasion, pathological TNM classification, tumour stage, overall survival (OS), and survival status, were excluded. Thus, 339 patients with complete clinical information were included in the analysis (Table 1). DNA methylation data of colorectal cancer patients were downloaded to further explore the effects of KCNJ14 expression. We also downloaded microarray datasets for GSE50117 based on the GPL6480 platforms from the GEO database (http://www.ncbi.nlm.nih.gov/geo), which contained nine paired tumour-normal colorectal samples, to explore the changes in KCNJ14 expression in colorectal cancer [11]. GSE31595, based on the GPL570 platform, had 37 tissue samples of colorectal cancer and the survival status of patients [12]. The GSE31595 and TCGA transcriptomic data were used to perform a meta-analysis.
Meta-analysis
To date, no study has explored the prognostic value of KCNJ14 in colorectal cancer; thus, herein, we performed a meta-analysis to evaluate the importance of KCNJ14 expression in the prognosis of patients from both databases. OS was considered a prognostic outcome, and the prognostic significance of KCNJ14 expression is shown as hazard ratios (HRs) with 95% confidence interval (CIs). The Q test (I2 statistics) was used to evaluate heterogeneity between the two databases. A fixed effects model was chosen because there was no statistical heterogeneity (I2 < 50%, P ≥ 0.1). Otherwise, when there was statistical heterogeneity between the two databases (I2 > 50%, P < 0.1), a random-effects model was used [13].
TIMER database analysis
The Tumour Immune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer) is an interactive database that provides comprehensive computation and visualisation of the inextricable relationship between tumour immunologic and genetic information, such as gene expression, mutation, and copy number variants [10]. In this study, we evaluated the association between KCNJ14 expression and the infiltration of six different immune cell types (B cells, CD4 + T cells, CD8 + T cells, macrophages, neutrophils, and dendritic cells).
GO and KEGG analysis
Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to explore the biological processes and signalling pathways associated with KCNJ14 expression in colorectal cancer. First, we divided the patients into two groups based on median KCNJ14 expression as follows: high and low KCNJ14 expression groups. We then selected the differentially expressed genes between the two groups, and R software (v.4.0.3) was used to analyse the significant biological functions and critical pathways related to KCNJ14. Results were considered significantly enriched when P < 0.05.
CMap analysis
The connectivity map (CMap, https://portals.broadinstitute.org/CMap/) is a public database that contains drug-induced gene expression profiles and reveals the biological connection between genes, drugs, and diseases [14]. We used CMap analysis to select candidate drugs for colorectal cancer by comparing the transcriptome data with database information. Genes with positive and negative relationship with KCNJ14 were selected to obtain information on alternative drugs by accessing the CMap database. Four candidate drugs were selected according to the most negative enrichment index, while setting P < 0.001 as the filter condition. The corresponding chemical structure formulas were obtained from the publicly available PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
Cell treatment and RT-PCR
The human normal colorectal mucosal cell line FHC and the colorectal cancer cell lines HCT116 and SW480 were purchased from the American Type Culture Collection (ATCC) and cultured in RPMI 1640 medium with 10% foetal bovine serum (FBS) in a 5% CO2 cell incubator at a constant temperature of 37 °C. Both HCT116 and SW480 cell lines were treated with S-adenosyl methionine at concentrations of 100 μM and 200 μM or decitabine at 5 μM and 10 μM, respectively, and expression of KCNJ14 was verified by RT-PCR. Total RNA was extracted from cells using TRIzol reagent (Invitrogen, US) and reverse-transcribed into cDNA. We performed RT-PCR to evaluate KCNJ14 expression using specific primers as follows: 5′-GGGGTCCCTCCGTCCAAT-3′ (sense) and 5′-CAGTGCCCGTCTTTCTTGAC-3′ (antisense). In addition, HCT116 and SW480 cells were transfected with the negative control shRNA (NC) or KCNJ14-targeted shRNA (shKCNJ14: 5′-GATCCGCCAGGATGTGGATGTGGGCTTTGATTTCAAGAGAATCAAAGCCCACATCCACATCCTGGTTTTTTGGAAG-3′) for 24 h and used for subsequent experiments.
Western blotting
The shRNA-treated HCT116 and SW480 cells were collected and lysed in RIPA buffer containing protease and phosphatase inhibitors. After incubation on ice for 30 min, cell lysates were centrifuged at 12,000 rpm at 4 ℃ for 15 min. An amount of 25 μL 4 × protein loading buffer was added to 75 μL supernatant after quantification of protein concentration. The mixed protein samples were analysed by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. Next, the membrane was sealed with skimmed milk at room temperature for 1 h and incubated with specific primary antibodies against mTOR signalling pathway-related proteins (KCNJ14, Cat 14-171-1-AP, Proteintech; phospho-AKT, Cat 66444-1-Ig, Proteintech; AKT, Cat 60203-2-Ig, Proteintech; Phospho-mTOR, Cat 67778-1-Ig, Proteintech; and mTOR, Cat 66888-1-Ig, Proteintech) at 4 °C overnight. After washing with TBST, the membranes were incubated with the corresponding secondary antibodies conjugated with horseradish peroxidase (HRP) at 25 °C in the dark for 1 h. Finally, band signals were detected using a chemiluminescence (ECL) detection system.
MTT and colony-forming experiment
The shRNA-treated HCT116 and SW480 cells were inoculated in 96 well plates at a density of 2000 cells per well, 20 μL MTT was added to each well, and cells were incubated at 37 ℃ for 4 h. After which, 150 μL DMSO was added to the wells, and further incubation for 15 min in the dark was performed to dissolve the formed formazan crystals. Finally, the absorbance of the treated SW480 cells was measured using a microplate reader at 490 nm. The number of HCT116 and SW480 cells transfected with shRNA was calculated to be 103 for culture in 60 mm dishes with complete medium. After 14 days, the cells were washed with PBS, fixed with paraformaldehyde, and stained with a crystal violet solution. Finally, the number of cell colonies was recorded and calculated.
Wound healing assay and Transwell assay
HCT116 and SW480 cells were cultured in six-well plates and treated with either NC or shKCNJ14. When the cells reached 95% confluence, a sterile pipette tip was used to wound them. After washing with PBS, the cells were continuously cultured for 48 h in serum-free medium. The relative distances between cells at the same location were recorded by photography. HCT116 and SW480 cells were treated with shRNA and resuspended in a medium containing 5% serum. A total of 105 cells were selected and cultured in the Transwell chamber, and 500 μL of medium with 20% serum was added to the lower chamber of the 24-well plate. Cells that invaded the bottom surface of the transwell chamber after 48 h were stained with crystal violet and photographed after fixation.
Statistical analysis
R software (v.4.0.3) was used to analyse the raw data from the TCGA and GEO databases. The differential expression of KCNJ14 in colorectal and adjacent tissues was analysed using a t-test. Therein, 42 normal mucosae and their corresponding tumour tissues were further assessed using matched-pair analysis. Pearson’s correlation coefficient was used to identify co-expressed genes and the relationship between KCNJ14 expression and DNA methylation. Moreover, we used Kaplan–Meier curves to evaluate the association between patient survival and KCNJ14 expression, as well as KCNJ14 DNA methylation. Receiver operating characteristic (ROC) analysis was used to assess the diagnostic significance of KCNJ14. In addition, we utilised univariate and multivariate Cox regression models to estimate whether KCNJ14 is an independent prognostic factor for colorectal cancer. Finally, the results obtained from in vitro experiments are presented as mean ± SD, and a t-test and a one-way analysis of variance (ANOVA) test were used for statistical analysis. P-value less than 0.05 was considered statistically significant.
Results
KCNJ14 mRNA expression is abnormally increased in colorectal cancer
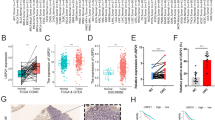
To explore the difference in KCNJ14 expression in colorectal cancer tissues [15], we obtained colorectal cancer and adjacent tissue samples from the TCGA (cancer: 488 cases; adjacent: 42 cases) and GSE50117 (cancer: 9 cases; adjacent: 9 cases) databases, respectively. The results showed that the mRNA expression level of KCNJ14 increased significantly in both the TCGA (P < 0.001) and GSE50117 (P = 0.019) datasets (Fig. 1a, c). In addition, we conducted a matched-pair analysis to process 42 pairs of tumour-normal samples and obtained a similar result (P < 0.001) (Fig. 1b).
KCNJ14 mRNA and protein levels in colorectal cancer cells and tissues and normal cells and adjacent tissues. a The mRNA level of KCNJ14 in colorectal cancer and adjacent tissue samples based on the TCGA database. b The mRNA level of KCNJ14 in 42 pairs of tumor-normal samples based on the matched-pair analysis. c The mRNA level of KCNJ14 in nine paired tumor-normal colorectal samples of GSE50117. d The mRNA level of KCNJ14 in normal human colonic epithelial cells (FHC) and colorectal cancer cell lines (HCT116 and SW480). e The protein level of KCNJ14 in three cell lines. f The correlation between KCNJ14 expression and tumour stage in CRC. ****P < 0.0001, compared with the FHC group
To verify the reliability of the results, we used RT-qPCR to evaluate the mRNA expression of KCNJ14. We found that HCT116 and SW480 CRC cells had a significantly upregulated KCNJ14 mRNA expression compared to the normal cell line (Fig. 1d). Western blotting revealed that the protein levels of KCNJ14 in HCT116 and SW480 CRC cells are significantly increased compared to those in the normal cell line (Fig. 1e). More importantly, the expression of KCNJ14 increased with an increase in tumour stage (P = 0.040) (Fig. 1f), and the increase in tumour stage of patients with colorectal cancer was a poor prognostic factor [16]. Collectively, these results suggest that KCNJ14 may have an important regulatory effect on the pathological process of colorectal cancer.
High KCNJ14 mRNA level is regulated by its DNA methylation in colorectal cancer
To determine why the expression of KCNJ14 is abnormally increased in colorectal cancer cells, we downloaded the DNA methylation data of patients with colorectal disease from the TCGA database. Based on the important regulatory effect of DNA methylation on the expression of downstream mRNA [17], from a large number of DNA methylation sites, we selected 11 CpG sites with regulatory effects on KCNJ14 expression (Additional file 1: Fig. S1a). Subsequently, we conducted a co-expression analysis and found that the methylation level of cg17660703 is positively correlated with KCNJ14 (Additional file 1: Fig. S1b). However, the methylation of the remaining 10 CpG sites showed no significant association. The high methylation status of cg17660703 was negatively correlated with overall survival (P = 0.006) (Additional file 1: Fig. S1c). Based on these results, the high expression of KCNJ14 in colorectal cancer may be positively regulated by the methylation site of cg17660703.
High expression of KCNJ14 can independently affect poor prognosis of patients
Based on the abnormally high expression of KCNJ14 in colorectal cancer, we further explored its influence on patient prognosis. First, we used Kaplan–Meier curves to assess the correlation between high expression of KCNJ14 and patient survival and found that high KCNJ14 expression is associated with shorter overall survival of patients (P = 0.012) (Fig. 2a). Subsequently, further subtype analysis showed that the high expression of KCNJ14 could significantly reduce the disease-free survival (stage I to III) (Additional file 2: Fig. S2a) and progression-free survival (stage IV) of patients (Additional file 2: Fig. S2b).
Association between KCNJ14 expression and prognosis of CRC patients. a The overall survival of CRC patients with high or low expression of KCNJ14. b Results of univariate analysis of clinical characteristics related to CRC. c The relationship between of KCNJ14 expression and 1-year, 3-year, and 5-year survival rates of CRC patients. d Results of multivariate analysis of clinical characteristics related to CRC
ROC and AUC were used to estimate the diagnostic value of KCNJ14 (Fig. 2c). To further improve the reliability of our results, we used Cox regression models to predict the effect of KCNJ14 on the prognosis of patients. Univariate analysis showed that KCNJ14 and several clinical characteristics, such as lymphatic invasion, pathological TNM stage, and tumour stage (hazard ratio [HR] > 1; P < 0.05), are significantly related to OS (Fig. 2b). Multivariate analysis showed that KCNJ14, pathological T stage, and pathological M stage can independently affect the prognosis of colorectal patients (HR > 1; P < 0.05; Fig. 2d). Collectively, these results suggest that high KCNJ14 expression is an independent risk factor for the prognosis of patients and may play a pathogenic role in colorectal cancer.
KCNJ14 knockdown significantly inhibits the biological behaviour of colorectal cancer cell lines
We further verified the adverse effects of KCNJ14 expression on the prognosis of patients with colorectal cancer. A meta-analysis found no significant heterogeneity between the two databases (I2 = 0%, P = 0.98); thus, a fixed-effect model was applied. Because the pooled HR for the correlation between high KCNJ14 expression and patient OS was 2.24 (95% CI: 1.37–3.65), we can conclude that KCNJ14 high expression is an independent predictor of unfavourable OS in patients with colorectal cancer (Fig. 3a).
KCNJ14 knockdown remarkably reduced the proliferative and invasive capacities of CRC cells. a Forest plot of high KCNJ14 expression with unfavourable OS in CRC patients from two databases based on a meta-analysis. b Proliferation curves of HCT116 and SW480 cells treated with NC or shKCNJ14. c Staining results of cell colonies in NC or shKCNJ14 group of HCT116 and SW480 cells. d, e The relative distances of the wound healing assay of HCT116 and SW480 cells treated with NC or shKCNJ14. f Staining results of the Transwell assay in NC or shKCNJ14 group of HCT116 and SW480 cells. ***P < 0.001, ****P < 0.0001 vs NC group
To further verify the effect of KCNJ14 on the biological behaviour of colorectal cancer cells, we used gene interference technology to reduce the protein and mRNA levels of KCNJ14 in two colorectal cancer cell lines (Additional file 3: Fig. S3). We found that knocking down KCNJ14 significantly inhibits the proliferation of HCT116 and SW480 cells (Fig. 3b, c). In addition, wound healing assay results showed that knockdown of KCNJ14 significantly inhibits the migration of HCT116 (Fig. 3d) and SW480 (Fig. 3e) cell lines. Finally, Transwell assay results suggested that KCNJ14 knockdown can reduce the invasive ability of cancer cells (Fig. 3f). Collectively, these results suggest that knockdown of KCNJ14 expression can significantly inhibit the malignant behaviour of cancer cells in the pathological process of colorectal cancer.
Regulatory mechanism of KCNJ14 in the pathological process of colorectal cancer
To explore the possible mechanism behind KCNJ14, we collected the data of correlated genes from Pearson correlation analyses and presented the top five most relevant genes that were positively and negatively correlated with the expression of KCNJ14 (Additional file 4: Fig. S4a, b). Subsequently, we performed a gene annotation analysis on the genes with expression relationship of KCNJ14, and the results showed that there were neutrophil activation, neutrophil-mediated immunity, and Fc receptor-mediated stimulatory signalling pathways in biological processes, including immunoglobulin complex and ficolin-1-rich granules, and molecular functions were immunoglobulin receptor binding, cadherin binding, and cell adhesion molecule binding (Fig. 4a). In addition, KEGG pathway analyses were further applied to obtain more specific information about vital signaling pathways KCNJ14 participated in, such as the mTOR, NOD-like receptor, and VEGF signalling pathways (Fig. 4b). Finally, we verified the putative signalling pathway in KEGG results and found that knockdown of KCNJ14 remarkably inhibited the phosphorylation of AKT and mTOR in SW480 (Fig. 4c) and HCT116 (Fig. 4d) cell lines, thus blocking the mTOR signalling pathway, which greatly affected many biological activities of CRC cells, such as proliferation and migration. In addition, we also uploaded the genes with expression relationships with KCNJ14 to the CMap database to match the inhibitory drug discovery and found that four candidate drugs (thiostrepton, ivermectin, corticosterone, and indoprofen) may have potential value in the treatment of colorectal cancer (Additional file 4: Fig. S4c).
KCNJ14 knockdown significantly inhibited the mTOR signalling pathway. a GO annotation of KCNJ14. BP: biological process, CC: cell component, MF: molecular function. b KEGG analysis of the biological functions of KCNJ14. c The levels of mTOR signalling pathway-related proteins in NC and shKCNJ14 groups of SW480 cells. d The levels of mTOR signalling pathway-related proteins in NC and shKCNJ14 groups of HCT116 cells. **P < 0.01, ***P < 0.001 vs NC group
Relationship between KCNJ14 and immune cell infiltration in colorectal cancer
Due to the regulation of pathogenic genes in a variety of ways and the important impact of immune microenvironment on the prognosis of colorectal cancer [18, 19], Therefore, we explored the relationship between KCNJ14 expression and immune cell infiltration through the TIMER database to reveal the effect of KCNJ14 expression on immune microenvironment. As shown in Fig. 5, the expression level of KCNJ14 was positively correlated with CD4 + T cells in both COAD (colon adenocarcinoma) and READ (rectum adenocarcinoma) (P = 3.52e-06, COAD; P = 1.26e-05, READ) and negatively associated with CD8 + T cells in colorectal cancer (P = 2.18e-05, COAD; P = 1.58e-02, READ). In addition, KCNJ14 expression was found to be related to neutrophils and dendritic cells in COAD; however, there were no significant differences in READ, B cells, or macrophages in either COAD or READ. This demonstrates that KCNJ14 is significantly associated with the tumour immune microenvironment.
Discussion
Colorectal cancer is responsible for almost 10% of newly diagnosed cancers and related deaths worldwide [20]. The current treatments for CRC have not achieving satisfactory prognosis. Because the mechanisms underlying CRC pathogenesis are not fully understood, targeted therapy is rapidly becoming a key instrument. In this study, we examined the relationship between KCNJ14 expression and CRC and investigated its potential diagnostic and therapeutic value.
We analysed the TCGA and GEO datasets and conducted a matched-pair analysis. We found that the expression of KCNJ14 was much higher in colorectal cancer tissues than in adjacent tissues. To verify the reliability of the results, RT-qPCR and western blotting were performed, and results showed that the mRNA and protein levels of KCNJ14 were significantly higher in colorectal cancer cells than in normal mucosal cells. In addition, the Kaplan–Meier curves showed that high KCNJ14 expression is correlated with shorter OS in patients with CRC, and multivariate Cox hazard analysis verified that KCNJ14 is an independent prognostic factor for CRC. Furthermore, we performed a meta-analysis and confirmed that high KCNJ14 expression is a critical prognostic factor for colorectal cancer. This study is the first to report the prognostic role of high expression of KCNJ14 in colorectal cancer.
Although the mechanism of action of KCNJ14 in cancer has not been elucidated, the roles of homologous proteins in various cancers have been extensively reported. For example, KCNJ2 regulates the expression of MRP1/ABCC1 to regulate cell growth and chemoresistance in small-cell lung cancer [21]. Kir2.2 can act as a constitutive activator to increase the phosphorylation of RelA, resulting in enhanced NF-kB activity and cell proliferation in cancer [22]. In this study, we demonstrated that high KCNJ14 expression is positively correlated with tumour stage, suggesting that KCNJ14 may be a pathogenic gene in colorectal cancer; therefore, we further verified the effect of KCNJ14 expression on the cellular behaviour of colorectal cancer cells. We knocked down KCNJ14 in two colorectal cancer cell lines and using in vitro experiments, found that the proliferation and migration ability of cancer cells decreased significantly, thereby verifying the carcinogenic effect of KCNJ14 in colorectal cancer. Hence, we infer that increased KCNJ14 expression in colorectal cancer can not only independently reduce the overall survival time of patients, but also enhance the malignant behaviour of colorectal cancer cells.
We next investigated whether increased KCNJ14 expression in the pathological process of colorectal cancer is regulated by DNA methylation. Previous studies have suggested that aberrant DNA methylation regulates the risk and prognosis of cancer by altering the expression of various genes [23]. Therefore, we detected the methylation level of 11 CpG sites of KCNJ14 in CRC and applied Pearson correlation analysis to screen out that methylation of the cg17660703 site might increase KCNJ14 expression. Generally, DNA methylation often leads to the silencing of gene expression; however, with the development of wide-scale analyses of gene expression profiles and DNA methylomes, the positive association between DNA methylation sites and gene expression has been confirmed [24]. The possible mechanisms included negative regulatory element methylation and gene-body DNA methylation [25, 26].
Our results showed that the cg17660703 high methylation status of KCNJ14 CpG sites is related to unfavourable OS in CRC patients. Similar conclusions have been published to support the theory that aberrant DNA methylation can regulate the activation of oncogenes, leading to malignant progression of cancers [27]. Taken together, our study is the first to demonstrate that the cg17660703 high methylation status of KCNJ14 CpG sites, along with KCNJ14 expression, can be a clear indicator of poor prognosis of colorectal cancer patients.
Previous studies have reported that tumour-infiltrating immune cells are recruited to generate a proinflammatory microenvironment, which benefits the progression of CRC and immune cells have become a prognostic marker for colorectal cancer [28, 29]. The TIMER database revealed that KCNJ14 expression is positively correlated with infiltration of CD4 + T cells and negatively correlatied with that of CD8 + T cells. Consistent with our expectations, CD8 + T cells are considered a preferable prognostic factor for relapse and overall survival in patients with CRC [30]. Increased CD4 + T cell infiltration can enhance the formation of a tumour inhibitory immune microenvironment and lead to a poor prognosis [31]. Our findings indicate that the mRNA level of KCNJ14 is positively correlated with CD4 + T cells, and that increased expression of KCNJ14 can lead to poor prognosis in colorectal patients. This suggests that KCNJ14 and CD4 + T cells may exert a synergistic effect to promote the formation of an inhibitory immune microenvironment in colorectal cancer. Overall, CD4 + T cells play a key role in regulating the cancer immune microenvironment, and our study demonstrated that KCNJ14 mainly regulates the infiltration of CD4 + and CD8 + cells to influence the development of CRC.
To determine the biological functions of KCNJ14, we performed GO annotation analysis. We found that KCNJ14 is mainly enriched in neutrophil -and immunoglobulin-mediated humoral immunity. Immune responses are believed to participate in the development of colorectal cancer. For example, neutrophil extracellular traps can interact with platelets and endothelial cells to mediate procoagulant activity and contribute to thrombogenesis in colorectal cancer [32]. Moreover, infiltrated neutrophils secrete metalloproteinases to activate latent TGFβ and suppress T-cells, leading to an immunosuppressive microenvironment in colorectal cancer [33]. Finally, immunoglobulin-related mechanisms were also investigated. Immunoglobulin-like receptors on killer cells, such as 3DS1 and 2SD1, contribute to a high risk of CRC [34]. In addition, studies have reported that engineered immunoglobulins with Fc regions can guide activated NK cells against CRC [35]. KEGG analysis also showed that KCNJ14 participates in cancer-related signalling pathways in colorectal cancer, such as the mTOR, NOD-like receptor, and VEGF signalling pathways. Activation of these pathways can lead to malignant progression of tumour cells [36, 37]; however, we could not verify the influence of KCNJ14 on these signalling pathways. Therefore, we only performed knockdown KCNJ14 experiments in two cell lines of colorectal cancer and found that the levels of mTOR signalling pathway-related proteins decreased significantly. This indicates that KCNJ14 regulates the activity of the mTOR signalling pathway in the pathological process of colorectal cancer.
To validate the possible molecular mechanism of KCNJ14, we probed the cancer-related functions of the most positively correlated co-expressed genes. Shadow of prion protein (SPRN) is the one which is accurately reported that the SPRN appears exclusively in leiomyoma in contrast to normal samples, and its overexpression can increase the migratory ability of bladder cancer cells [38]. As a potential splicing factor, SFSWAP can alternatively regulate gene expression, and the cg09170112 methylation site of SFSWAP has been verified to be significantly correlated with colon cancer prognosis [39]. AGAP4 and AGAP6 have not been previously reported; they belong to the GTPase-activating protein family that play a vital role in cancer progression. For example, the homologous protein AGAP1 mainly mediates the migration and invasion of breast cancer cells [40]. Considering its location and function, we speculate that as an ATP-sensitive inward rectifier potassium (K+) channel, KCNJ14 malfunctions in cancer-related microenvironment and consequently activates the following signalling pathways.
Finally, we expanded the treatment options for CRC using CMap analysis and identified four candidate drugs. Thiostrepton was previously considered a thiazole antibiotic and is currently identified as an effective therapeutic drug for colon cancer that targets the oncogenic transcription factor FoxM1 [41]. Ivermectin can reverse chemotherapy resistance in colorectal cancer and breast cancer cells by regulating the EGFR/ERK/Akt/NF-κB pathway [42]. Although the other two agents have not been used for the treatment of CRC, there are indications for their use in enteric diseases and cancers. Corticosterone can inhibit the invasion of bladder cancer cells, and its production in inflamed intestines is increased [43]. Indoprofen can function as a pyruvate kinase M2 (PKM2) inhibitor to facilitate the radiosensitivity of non-small cell lung cancer [44]. Our study expands drug indications and achieves the aim of drug repurposing for a more comprehensive treatment of CRC.
Conclusions
High expression of KCNJ14 in CRC can be used as an independent prognostic risk factor, resulting in the poor prognosis in patients with CRC. Therefore, it is expected to become a potential biological target for colorectal treatment. In addition, this study provides novels insights for future studies aiming to investigate the complex pathological process of colorectal cancer and broadens the molecular knowledge on the role of KCNJ14 in the pathological process of cancer.
Availability of data and materials
In this study, multiple data were obtained from web-based datasets. The gene expression profiles and clinical information of patients with colorectal cancer in the TCGA database were obtained from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). The GSE50117 and GSE31595 datasets were downloaded from the GEO dataset (http://www.ncbi.nlm.nih.gov/geo/). The correlation between KCNJ14 expression and immune cell infiltration was obtained from the TIMER database (https://cistrome.shinyapps.io/timer). Finally, candidate therapeutic drugs were screened for co-expressed genes using the CMap database (https://portals.broadinstitute.org/CMap/), and the chemical structure formulas of four drugs, namely corticosterone, indoprofen, ivermectin, and thiostrepton, were searched in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). All data generated from the analysis of this study are available from the corresponding author upon reasonable request.
Abbreviations
- CRC:
-
Colorectal cancer
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene Expression Omnibus
- OS:
-
Overall survival
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- TIMER:
-
Tumour Immune Estimation Resource
- CMap:
-
The connectivity map
- NSAIDs:
-
Non-steroidal anti-inflammatory drugs
- GDC:
-
Genomic data commons
- COAD:
-
Colon adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- SPRN:
-
Shadow of prion protein
- PKM2:
-
Pyruvate kinase M2
References
Arnold M, Sierra M, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–91.
Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2012;10(6):639–45.
Dekker E, Tanis P, Vleugels J, Kasi P, Wallace M. Colorectal cancer. Lancet (Lond, Engl). 2019;394(10207):1467–80.
Liu Z, Shen F, Wang H, Li A, Wang J, Du L, Liu B, Zhang B, Lian X, Pang B, et al. Abnormally high expression of HOXA2 as an independent factor for poor prognosis in glioma patients. Cell Cycle (Georget, Tex). 2020;19(13):1632–40.
Töpert C, Döring F, Derst C, Daut J, Grzeschik K, Karschin A. Cloning, structure and assignment to chromosome 19q13 of the human Kir2.4 inwardly rectifying potassium channel gene (KCNJ14). Mamm Genome Off J Int Mamm Genome Soc. 2000;11(3):247–9.
Tinker A, Aziz Q, Li Y, Specterman M. ATP-sensitive potassium channels and their physiological and pathophysiological roles. Compr Physiol. 2018;8(4):1463–511.
Conti M. Targeting K+ channels for cancer therapy. J Exp Ther Oncol. 2004;4(2):161–6.
Sakai H, Shimizu T, Hori K, Ikari A, Asano S, Takeguchi N. Molecular and pharmacological properties of inwardly rectifying K+ channels of human lung cancer cells. Eur J Pharmacol. 2002;435:125–33.
Zschüntzsch J, Schütze S, Hülsmann S, Dibaj P, Neusch C. Heterologous expression of a glial Kir channel (KCNJ10) in a neuroblastoma spinal cord (NSC-34) cell line. Physiol Res. 2013;62(1):95–105.
Thuringer D, Chanteloup G, Boucher J, Pernet N, Boudesco C, Jego G, Chatelier A, Bois P, Gobbo J, Cronier L, Solary E, Garrido C. Modulation of the inwardly rectifying potassium channel Kir4.1 by the pro-invasive miR-5096 in glioblastoma cells. Oncotarget. 2017;8(23):37681–93.
Pesson M, Volant A, Uguen A, Trillet K, De La Grange P, Aubry M, Daoulas M, Robaszkiewicz M, Le Gac G, Morel A, et al. A gene expression and pre-mRNA splicing signature that marks the adenoma-adenocarcinoma progression in colorectal cancer. PLoS ONE. 2014;9(2):e87761.
Thorsteinsson M, Kirkeby L, Hansen R, Lund L, Sørensen L, Gerds T, Jess P, Olsen J. Gene expression profiles in stages II and III colon cancers: application of a 128-gene signature. Int J Colorectal Dis. 2012;27(12):1579–86.
Tan Y, Zhang S, Xiao Q, Wang J, Zhao K, Liu W, Huang K, Tian W, Niu H, Lei T, et al. Prognostic significance of ARL9 and its methylation in low-grade glioma. Genomics. 2020;112(6):4808–16.
Lamb J, Crawford E, Peck D, Modell J, Blat I, Wrobel M, Lerner J, Brunet J, Subramanian A, Ross K, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35.
Huang C, Zhao J, Zhu Z. Prognostic Nomogram of Prognosis-Related Genes and Clinicopathological Characteristics to Predict the 5-Year Survival Rate of Colon Cancer Patients. Front Surg. 2021;8:681721.
Bryan S, Masoud H, Weir H, Woods R, Lockwood G, Smith L, Brierley J, Gospodarowicz M, Badets N. Cancer in Canada: Stage at diagnosis. Health Rep. 2018;29(12):21–5.
Pinho R, Maga E. DNA methylation as a regulator of intestinal gene expression. Br J Nutr. 2021;126(11):1611–25.
Yang J, Qi M, Fei X, Wang X, Wang K. LncRNA H19: a novel oncogene in multiple cancers. Int J Biol Sci. 2021;17(12):3188–208.
Ganesh K, Stadler Z, Cercek A, Mendelsohn R, Shia J, Segal N, Diaz L. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–75.
Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Liu H, Huang J, Peng J, Wu X, Zhang Y, Zhu W, Guo L. Upregulation of the inwardly rectifying potassium channel Kir2.1 (KCNJ2) modulates multidrug resistance of small-cell lung cancer under the regulation of miR-7 and the Ras/MAPK pathway. Mol Cancer. 2015;14:59.
Lee I, Lee S, Kang T, Kang W, Park C. Unconventional role of the inwardly rectifying potassium channel Kir2.2 as a constitutive activator of RelA in cancer. Cancer Res. 2013;73(3):1056–62.
Guo Y, Mao X, Qiao Z, Chen B, Jin F. A novel promoter CpG-based signature for long-term survival prediction of breast cancer patients. Front Oncol. 2020;10:579692.
Irizarry R, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–86.
Dejeux E, Olaso R, Dousset B, Audebourg A, Gut I, Terris B, Tost J. Hypermethylation of the IGF2 differentially methylated region 2 is a specific event in insulinomas leading to loss-of-imprinting and overexpression. Endocr Relat Cancer. 2009;16(3):939–52.
Jjingo D, Conley A, Yi S, Lunyak V, Jordan I. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3(4):462–74.
Robertson K. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610.
Kostic A, Chun E, Robertson L, Glickman J, Gallini C, Michaud M, Clancy T, Chung D, Lochhead P, Hold G, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15.
Ye L, Zhang T, Kang Z, Guo G, Sun Y, Lin K, Huang Q, Shi X, Ni Z, Ding N, et al. Tumor-infiltrating immune cells act as a marker for prognosis in colorectal cancer. Front Immunol. 2019;10:2368.
Hu W, Sun R, Chen L, Zheng X, Jiang J. Prognostic significance of resident CD103CD8T cells in human colorectal cancer tissues. Acta Histochem. 2019;121(5):657–63.
Toor S, Murshed K, Al-Dhaheri M, Khawar M, Abu Nada M, Elkord E. Immune checkpoints in circulating and tumor-infiltrating CD4 T cell subsets in colorectal cancer patients. Front Immunol. 2019;10:2936.
Zhang Y, Wang C, Yu M, Zhao X, Du J, Li Y, Jing H, Dong Z, Kou J, Bi Y, et al. Neutrophil extracellular traps induced by activated platelets contribute to procoagulant activity in patients with colorectal cancer. Thromb Res. 2019;180:87–97.
Germann M, Zangger N, Sauvain M, Sempoux C, Bowler A, Wirapati P, Kandalaft L, Delorenzi M, Tejpar S, Coukos G, et al. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGFβ. EMBO Mol Med. 2020;12(1):e10681.
Al Omar S, Mansour L, Dar J, Alwasel S, Alkhuriji A, Arafah M, Al Obeed O, Christmas S. The relationship between killer cell immunoglobulin-like receptors and HLA-C polymorphisms in colorectal cancer in a Saudi population. Genet Test Mol Biomarkers. 2015;19(11):617–22.
Schmied B, Riegg F, Zekri L, Grosse-Hovest L, Bühring H, Jung G, Salih H. An Fc-optimized CD133 antibody for induction of natural killer cell reactivity against colorectal cancer. Cancers. 2019;11(6):789.
Narayanankutty A. PI3K/ Akt/ mTOR pathway as a therapeutic target for colorectal cancer: a review of preclinical and clinical evidence. Curr Drug Targets. 2019;20(12):1217–26.
Liu S, Fan W, Gao X, Huang K, Ding C, Ma G, Yan L, Song S. Estrogen receptor alpha regulates the Wnt/β-catenin signaling pathway in colon cancer by targeting the NOD-like receptors. Cell Signal. 2019;61:86–92.
Sahar T, Nigam A, Anjum S, Waziri F, Biswas S, Jain S, Wajid S. Interactome analysis of the differentially expressed proteins in uterine leiomyoma. Anticancer Agents Med Chem. 2019;19(10):1293–312.
Yang H, Jin W, Liu H, Wang X, Wu J, Gan D, Cui C, Han Y, Han C, Wang Z. A novel prognostic model based on multi-omics features predicts the prognosis of colon cancer patients. Mol Genet Genomic Med. 2020;8(7):e1255.
Tsutsumi K, Nakamura Y, Kitagawa Y, Suzuki Y, Shibagaki Y, Hattori S, Ohta Y. AGAP1 regulates subcellular localization of FilGAP and control cancer cell invasion. Biochem Biophys Res Commun. 2020;522(3):676–83.
Ju S, Huang C, Huang W, Su Y. Identification of thiostrepton as a novel therapeutic agent that targets human colon cancer stem cells. Cell Death Dis. 2015;6:e1801.
Jiang L, Wang P, Sun Y, Wu Y. Ivermectin reverses the drug resistance in cancer cells through EGFR/ERK/Akt/NF-κB pathway. J Exp Clin Cancer Res CR. 2019;38(1):265.
Salmenkari H, Issakainen T, Vapaatalo H, Korpela R. Local corticosterone production and angiotensin-I converting enzyme shedding in a mouse model of intestinal inflammation. World J Gastroenterol. 2015;21(35):10072–9.
Ye X, Sun Y, Xu Y, Chen Z, Lu S. Integrated in silico-in vitro discovery of lung cancer-related tumor pyruvate kinase M2 (PKM2) inhibitors. Med Chem. 2016;12(7):613–20.
Acknowledgements
We thank all the staff who contributed to this study.
Funding
This study did not receive any financial grants.
Author information
Authors and Affiliations
Contributions
LSP designed and managed the entire study; LB performed the main experiments, analysed the data, and drafted the original manuscript; GN, PZP, and HCF analysed the data and wrote the manuscript; XK, WDF, and LJW analysed the data; WJ and DFH revised the manuscript; and LMY provided professional advice on the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The surgical tissue samples used in this study were all obtained from public databases; thus, there were no ethical issues involved. The sample information stored in the public database complied with the Declaration of Helsinki (revised in 2013).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1
The methylation regulation of KCNJ14 expression. (a) The methylation status of 11 CpG sites of KCNJ14 in colorectal cancer tissue samples based on the TCGA database. (b) The relationship between cg17660703 methylation and KCNJ14 expression. (c) The overall survival of CRC patients with high or low methylation status of cg17660703.
Additional file 2: Fig. S2
Association between KCNJ14 expression and survival of patients at different stages of CRC. (a) Overall survival of stages I–III CRC patients with high or low expression of KCNJ14. (b) Overall survival of stage IV CRC patients with high or low expression of KCNJ14.
Additional file 3: Fig. S3
KCNJ14 expression in CRC cell lines treated with NC or shKCNJ14. (a) mRNA levels of KCNJ14 in both HCT116 and SW480 cells treated with NC or shKCNJ14. (b) Protein levels of KCNJ14 in both HCT116 and SW480 cells treated with NC or shKCNJ14.
Additional file 4: Fig. S4
Pearson correlation analysis and CMap analysis of KCNJ14 in CRC. (a)-(b) The 10 most related co-expressed genes of KCNJ14, including five positively associated genes (SPRN, AGAP4, BRICD5, SFSWAP and AGAP6) and five negatively associated genes (COPS4, RHOA, CASP3, PAFAH2 and GSKIP). (c)-(f) Pubchem information of four candidate drugs for CRC based on CMap analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, B., Ge, N., Pan, Z. et al. KCNJ14 knockdown significantly inhibited the proliferation and migration of colorectal cells. BMC Med Genomics 15, 194 (2022). https://doi.org/10.1186/s12920-022-01351-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-022-01351-4