Abstract
Background
Merosin-deficient congenital muscular dystrophy type 1A (MDC1A) is a rare autosomal recessive genetic condition caused by deleterious mutations in the LAMA2 gene encoding the laminin-α2 chain. It is the most frequent subtype of congenital muscular dystrophies (CMDs) characterized by total laminin-α2 deficiency with muscle weakness at birth or in the first six months of life. To the best of our knowledge, this study reports the first molecular diagnosis and genetic defect of this heterogeneous form of CMD performed in a Moroccan medical genetic center using next-generation sequencing (NGS). It allows us to expand the mutational spectrum of the LAMA2 gene.
Case presentation
We report the case of a female Moroccan child with clinical and paraclinical features in favor of a CMD. She has global congenital hypotonia with generalized muscle weakness, psychomotor retardation, increased serum creatine kinase, and normal brain scan at the age of six months. Targeted NGS leads to the identification of a novel homozygous nonsense mutation c.2217G > A, p.(Trp739*) in the exon 16 of LAMA2. Sanger sequencing confirmed this mutation in the affected patient and showed that her parents are heterozygous carriers.
Conclusions
A modern genetic analysis by NGS improves the genetic diagnosis pathway for adequate genetic counseling of affected families more precisely. An accession number from the National Center for Biotechnology Information (NCBI) ClinVar database was retrieved for this novel LAMA2 mutation.
Similar content being viewed by others
Background
Laminin α2-related muscular dystrophy (LAMA2 MD) is a rare autosomal recessive neuromuscular disorder caused by homozygous or compound heterozygous mutations in the LAMA2 gene (MIM*156225) on chromosome 6q22-q23, encoding the laminin-α2 chain [1]. A total loss of merosin function due to pathogenic LAMA2 mutations causes mainly a severe phenotype with early onset of merosin-deficient congenital muscular dystrophy type 1A (MDC1A). It is the most frequent form of congenital muscular dystrophy (CMD), responsible for 30% to 40% of all CMD cases in Europe [2]. MDC1A includes a broad range of phenotypes with different degrees of severity due to variability in the onset symptoms, clinical manifestations, and mutation effects on the laminin-α2 synthesis [3].
Herein, we report the case of a Moroccan female patient with a severe form of CMD caused by a novel homozygous nonsense mutation in the LAMA2 gene identified by next-generation sequencing (NGS).
Case presentation
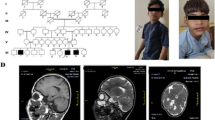
The proband (VI.3) is a female of 2 years and 7 months at the time of her genetic assessment. She is consanguineous, the third liveborn of a couple of half-first cousins (V.1 and V.8), sharing the same grand-mother (III.2). The grand-mother (III.2) was firstly married to her maternal cousin (III.1), grand-father of (V.1) and then to her paternal cousin (III.3), grand-father of (V.8). The proband’s siblings (VI.1 and VI.2) aged 10 and 6 respectively, were normal at the time of assessment. Grand-parents, parents, uncles, aunts and cousins of the proband (VI.3) were healthy with no particular disease history (Fig. 1a). The proband’s pregnancy was medically followed and presented without complication, but the baby was born prematurely at 33 weeks of gestation by vaginal delivery. Since the neonatal period, her parents have noticed a muscle weakness with hypotonia.
Pedigree of the Moroccan family with analysis of LAMA2 mutation. (a) Pedigree of the studied family. The filled symbol represents the affected patient and open symbols represent unaffected individuals. The genotypes for the mutation c.2217G > A in the LAMA2 gene are shown below the symbols V.1, V.8, and VI.3 (b) Electropherogram showing the c.2217G > A homozygous mutation in the proband. (c, d) Electropherograms showing the heterozygous mutation in both parents. Black arrows indicate homozygous and heterozygous mutation position detected in the proband and her parents, respectively
At 1 year of age, the neurologic evaluation showed a delay in motor development, tetraparesis with hypotonia, and an inability to sit. The tendon and bone reflexes were absent in lower limbs, and Babinski’s sign was negative. Her serum creatine kinase (CK) level was increased to 537 IU/l (normal range < 170 IU/l [4]). An electroneuromyographic analysis showed normal nerve conduction at the time of examination and the electromyogram (EMG) showed a myogenic pattern in muscles of upper and lower limbs. The computerized tomography (CT) scan at 6 months was normal and did not reveal any cerebral changes. Her intellectual development was normal and she never had seizures. She had no ocular signs and did not have cardiac nor respiratory dysfunction. She was unable to raise her head until 18 months and she was unable to stand up or stay up without support until now.
Clinical and paraclinical symptoms enable us to conclude that the patient’s phenotype may be compatible with a CMD. Thus, molecular genetic analysis was performed in the proband through a custom gene panel of 24 genes known to be related to inherited neuromuscular disorders in order to reach a precise molecular diagnosis (Table 1). The informed consent was obtained from the proband’s parents prior to molecular analysis. Samples of peripheral blood were collected from the affected child and her parents.
A DNA extraction was performed by the commercial PureLink™ Genomic DNA Mini Kit (Invitrogen-Thermo Fisher Scientific-USA). The quantity and quality of extracted genomic DNA (gDNA) were measured by a NanoDrop-2000 Spectrophotometer followed by Qubit 3.0 Fluorometer for more accurate DNA quantification using Qubit dsDNA HS (High Sensitivity) Assay Kit. In each sample, 10 ng of gDNA were used for multiplex PCR reactions using Ion Ampliseq™ On-Demand Primer Panel, which was designed via the Ion AmpliSeq™ Designer software (Thermo Fisher Scientific), available at (http://www.ampliseq.com). This allows highly multiplexed PCR amplification of 679 amplicons with superior coverage uniformity and specificity; in-silico covered 99% of regions of interest (ROI). Library construction was prepared using Ion AmpliSeq™ Library Kit v2.0 (Thermo Fisher Scientific), according to the manufacturer’s instructions. One of 16 barcodes of the Ion Xpress Barcode Adapters 1–16 Kit (Thermo Fisher Scientific) was added to each sample. The resulting DNA libraries were quantified with Qubit™ dsDNA BR Assay Kit on Qubit 3.0 Fluorometer (Molecular Probes) to get equimolar amounts of each library ready for downstream template preparation including clonal amplification by emulsion PCR that carried out on OneTouch™ 2 System (Thermo Fisher Scientific) with Ion PGM HI-Q™ view OT2 Kit (Thermo Fisher Scientific) coupled with the Ion OneTouch ES enrichment module (Thermo Fisher Scientific). Finally, the sequencing run was loaded on the 316™ Chip v2 (Thermo Fisher Scientific) using the Ion Torrent Personal Genome Machine (PGM, Thermo Fisher Scientific) through the Ion PGM HI-Q™ view Sequencing Kit as per the manufacturer’s protocol. Raw data were aligned to the hg19 human reference genome. Trimming, base calling, coverage analysis, and variant calling were performed on the Ion Torrent Server using the Torrent Suite software v.5.12.0 (Thermo Fisher Scientific), with default parameters.
Ion reporter software analysis v.5.10 (https://ionreporter.thermofisher.com/ir/) revealed a homozygous nonsense mutation in exon 16 of LAMA2, NM_000426.3(LAMA2):c.2217G > A (p.Trp739*). This mutation has never been reported in public human databases (accessed Nov 2020), including: ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), LOVD database (https://databases.lovd.nl/shared/genes/LAMA2), 1000 Genomes Project (https://www.internationalgenome.org/), gnomAD (https://gnomad.broadinstitute.org/), Exome Aggregation Consortium (http://exac.broadinstitute.org/), dbSNP Database (https://www.ncbi.nlm.nih.gov/snp/), and Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk/ac/). It was also not found in an in-house database of 100 Moroccan exomes (personal data).
Confirmation of this mutation was performed by conventional Sanger sequencing in the patient and her parents, using the BigDye™ Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Thermo Fisher Scientific) and loaded on an ABI 3500 automated Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific). Exon 16 and its flanking regions were amplified using the set of primers (ex16F: 5′GCTTTGACAAGCACATTTGATG3′/ex16R: 5′GGTCCCCAGGGTAGTAGCTG3′). A direct nucleotide sequence analysis confirmed that the proband carried the mutation in a homozygous state (Fig. 1b). Both parents were heterozygous (Fig. 1c, d).
The accession number from ClinVar database, SCV001448189.1 (submitted December 07, 2020), was assigned to our novel mutation.
Discussion and conclusions
The CMDs are a group of inherited muscle diseases, genetically and clinically heterogeneous, mainly with an autosomal recessive type of inheritance. They are manifested in early life or infancy [5]. Some severe forms of CMD can be fatal in infancy, whereas others may have a milder course and can survive into adulthood [6]. To date, there are at least 32 genetic forms of CMD that have been associated with mutations in more than 30 genes coding for structural and functional proteins of skeletal muscle fibers [7, 8]. MDC1A, is the most common subtype of CMD in Caucasian populations due to laminin-α2 defects, accounting for 30 to 40 percent of total cases [2, 9].
We report here the clinical and molecular data of a patient with a severe phenotype of MDC1A. The patient manifested congenital hypotonia, muscle weakness, and had serum CK level which was three times the upper normal value. Similar values of CK levels were found in two unrelated Tunisian families with the same phenotype severity as our case, which had 3–5 times the normal range [10].
At the age of 6 months, our patient had a normal cerebral function without any specific change, observed by the CT scan. In contrast, Oliveira et al. reported a case of an MDC1A patient with white matter changes (WMC) observed in the CT scan [11].
In previous studies, patients with LAMA2 mutations, showed normal brain magnetic resonance imaging (MRI) when has been detected during the neonatal period or in the first 6 months of life [11,12,13]. However, white matter abnormalities have been shown in the later period of life except for the case of an Italian patient reported by Saredi et al. [14], who carried a compound heterozygous nonsense mutation with an atypical phenotype and nearly normal brain MRI performed at 16 years of age. Thus, in our case, we could not completely exclude the absence of WMC, as well as the first detection by CT, which was done at an early age, it was considered as a period in which the changes are not always visible even with MRI observation in some patients.
Because of clinical heterogeneity and overlap between the different forms of CMD, NGS technology may be more efficient to increase the molecular diagnostic success rate, bypassing immunohistochemistry, which is not always available like in the Moroccan context [15, 16].
To the best of our knowledge, we report here the first molecular study performed on a Moroccan female proband underlining a novel nonsense mutation in the LAMA2 gene using an Ion Torrent PGM platform. Our mutation c.2217G > A may cause a complete deficit in laminin-α2 function due to a premature termination codon (PTC) at 739 amino acid residue. The current mutation has never been reported in public human databases.
To date, more than 400 different pathogenic mutations in the LAMA2 gene have been reported in HGMD professional release 2019.4 (accessed Nov 2020), (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=LAMA2), with no noticeable mutational hotspot. The most frequently reported genotype in the literature for patients with MDC1A were mutations that create a PTC associated with a total absence of laminin-α2 and detected in a homozygous state [11, 12, 17].
We have illustrated in (Fig. 2), all pathogenic LAMA2 nonsense mutations reported in the public version of HGMD in patients with severe merosin-deficient CMD or milder myopathy according to zygosity state. We observed that the effect of mutations on phenotype development and its severity differs between patients regardless of their mode of inheritance, which makes the understanding of the physiopathological mechanisms so far unknown. Interestingly, in the same exon 16, a nonsense mutation c.2230C > T (p.Arg744*) was previously reported at a homozygous state in two adult siblings with a mild MD phenotype [18]. Whereas the same mutation at compound heterozygous state together with another nonsense mutation c.4048C > T (p.Arg1350*) in exon 27 is responsible for causing a severe CMD [17].
Schematic representation of the LAMA2 gene structure and localization of identified LAMA2 nonsense mutations that are listed in the public version of HGMD. Two classifications, homozygous and heterozygous state, of LAMA2 nonsense mutations scattered along the coding sequence containing 65 exons (only those containing nonsense mutations are described and marked in black) are shown in patients with severe or milder phenotype of CMD (in dark blue) and patients with mild myopathy (in light blue). Nonsense mutations listed in HGMD are shown in black. Mutations of the second allele are illustrated in the following manner: nonsense (in red), frameshift (in green), missense (in yellow), splice site (in purple), and in-frame deletion (in grey). [?] refers to an unknown second mutation from the other allele due to using classical tests that could not identify the second mutation in the proband. The asterisk (*) refers to the novel mutation identified in our Moroccan patient. cDNA reference sequence that has been used to identify these mutations was NM_000426.3
In conclusion, NGS has revolutionized molecular genetic research and has become an essential genetic tool for molecular diagnosis of heterogeneous disorders as CMDs. It allows us to improve our understanding of the origin of MDC1A-causing by the identification of a mutation in the LAMA2 gene. The novel mutation identified here provides an appropriate course of management to the patient to offer genetic counseling to the family, to expand the genetic spectrum of LAMA2-related CMDs, and to raise awareness to the pediatric neurologists on the morbidity of this severe form of CMD that is so far under-diagnosed in the Moroccan population, and to show them the added value of NGS technology in reducing diagnostic wandering.
Availability of data and materials
The principal data generated and/or analyzed during the current study are included in the published article. The datasets used in this study are available from the corresponding author on reasonable request.
Abbreviations
- MDC1A:
-
Merosin-deficient congenital muscular dystrophy type 1A
- CMD:
-
Congenital muscular dystrophy
- CK:
-
Creatine kinase
- NGS:
-
Next-generation sequencing
- MD:
-
Muscular dystrophy
- EMG:
-
Electromyogram
- CT:
-
Computerized tomography
- WMC:
-
White matter changes
- MRI:
-
Magnetic resonance imaging
- PTC:
-
Premature termination codon
- HGMD:
-
Human gene mutation database
References
Helbling-Leclerc A, Zhang X, Topaloglu H, Cruaud C, Tesson F, Weissenbach J, Tome FM, Schwartz K, Fardeau M, Tryggvason K, et al. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat Genet. 1995;11(2):216–8.
Durbeej M. Laminin-alpha2 chain-deficient congenital muscular dystrophy: pathophysiology and development of treatment. Curr Top Membr. 2015;76:31–60.
Beytia Mde L, Dekomien G, Hoffjan S, Haug V, Anastasopoulos C, Kirschner J. High creatine kinase levels and white matter changes: clinical and genetic spectrum of congenital muscular dystrophies with laminin alpha-2 deficiency. Mol Cell Probes. 2014;28(4):118–22.
Tran KT, Le VS, Vu CD, Nguyen LT. A novel de novo variant of LAMA2 contributes to merosin deficient congenital muscular dystrophy type 1A: case report. Biomed Rep. 2020;12(2):46–50.
Mah JK, Korngut L, Fiest KM, Dykeman J, Day LJ, Pringsheim T, Jette N. A systematic review and meta-analysis on the epidemiology of the muscular dystrophies. Can J Neurol Sci. 2016;43(1):163–77.
Bertini E, D’Amico A, Gualandi F, Petrini S. Congenital muscular dystrophies: a brief review. Semin Pediatr Neurol. 2011;18(4):277–88.
Kaplan JC. Gene table of neuromuscular disorders. http://www.musclegenetable.fr. Accessed 30 Nov 2020.
Kang PB, Morrison L, Iannaccone ST, Graham RJ, Bonnemann CG, Rutkowski A, Hornyak J, Wang CH, North K, Oskoui M, et al. Evidence-based guideline summary: evaluation, diagnosis, and management of congenital muscular dystrophy: Report of the Guideline Development Subcommittee of the American Academy of Neurology and the Practice Issues Review Panel of the American Association of Neuromuscular & Electrodiagnostic Medicine. Neurology. 2015;84(13):1369–78.
Allamand V, Guicheney P. Merosin-deficient congenital muscular dystrophy, autosomal recessive (MDC1A, MIM#156225, LAMA2 gene coding for alpha2 chain of laminin). Eur J Hum Genet. 2002;10(2):91–4.
Louhichi N, Richard P, Triki CH, Meziou M, Ayadi H, Guicheney P, Fakhfakh F. Novel mutations in LAMA2 gene responsible for a severe phenotype of congenital muscular dystrophy in two Tunisian families. Arch Inst Pasteur Tunis. 2006;83(1–4):19–23.
Oliveira J, Santos R, Soares-Silva I, Jorge P, Vieira E, Oliveira ME, Moreira A, Coelho T, Ferreira JC, Fonseca MJ, et al. LAMA2 gene analysis in a cohort of 26 congenital muscular dystrophy patients. Clin Genet. 2008;74(6):502–12.
Geranmayeh F, Clement E, Feng LH, Sewry C, Pagan J, Mein R, Abbs S, Brueton L, Childs AM, Jungbluth H, et al. Genotype-phenotype correlation in a large population of muscular dystrophy patients with LAMA2 mutations. Neuromuscul Disord. 2010;20(4):241–50.
Hashemi-Gorji F, Yassaee VR, Dashti P, Miryounesi M. Novel LAMA2 Gene Mutations Associated with Merosin-Deficient Congenital Muscular Dystrophy. Iran Biomed J. 2018;22(6):408–14.
Saredi S, Gibertini S, Matalonga L, Farina L, Ardissone A, Moroni I, Mora M. Exome sequencing detects compound heterozygous nonsense LAMA2 mutations in two siblings with atypical phenotype and nearly normal brain MRI. Neuromuscul Disord. 2019;29(5):376–80.
Fattahi Z, Kalhor Z, Fadaee M, Vazehan R, Parsimehr E, Abolhassani A, Beheshtian M, Zamani G, Nafissi S, Nilipour Y, et al. Improved diagnostic yield of neuromuscular disorders applying clinical exome sequencing in patients arising from a consanguineous population. Clin Genet. 2017;91(3):386–402.
Oliveira J, Gruber A, Cardoso M, Taipa R, Fineza I, Goncalves A, Laner A, Winder TL, Schroeder J, Rath J, et al. LAMA2 gene mutation update: Toward a more comprehensive picture of the laminin-alpha2 variome and its related phenotypes. Hum Mutat. 2018;39(10):1314–37.
Xiong H, Tan D, Wang S, Song S, Yang H, Gao K, Liu A, Jiao H, Mao B, Ding J, et al. Genotype/phenotype analysis in Chinese laminin-alpha2 deficient congenital muscular dystrophy patients. Clin Genet. 2015;87(3):233–43.
Di Blasi C, He Y, Morandi L, Cornelio F, Guicheney P, Mora M. Mild muscular dystrophy due to a nonsense mutation in the LAMA2 gene resulting in exon skipping. Brain. 2001;124(Pt 4):698–704.
Acknowledgments
We thank the family for its participation in this study.
Y. EL KADIRI was supported by the excellence research grant from the National Center for Scientific and Technical Research (CNRST) in Rabat, Morocco.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
YEK collected the genetic data and literature search, performed the molecular genetics study by NGS, analyzed and interpreted the data, and wrote the original draft. IR performed the clinical genetic diagnosis and reviewed the manuscript. FZL helped in the Sanger sequencing. YK performed the clinical neurological evaluation. AS conceptualized and supervised the study, and edited the final version. JL performed NGS data analysis, reviewed the manuscript, and supervised the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All authors state that they have obtained written consent from the parents/legal guardians for genetic studies.
Consent for publication
Written consent has been obtained from the parents/legal guardians for publication of clinical details, radiological and biological data.
Competing interests
The authors declare that they have no financial or other conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
El Kadiri, Y., Ratbi, I., Laarabi, F.Z. et al. Identification of a novel LAMA2 c.2217G > A, p.(Trp739*) mutation in a Moroccan patient with congenital muscular dystrophy: a case report. BMC Med Genomics 14, 113 (2021). https://doi.org/10.1186/s12920-021-00959-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-021-00959-2