Abstract
Background
Members of the Anaplasmataceae family, such as the Anaplasma and Ehrlichia species, cause economic losses and public health risks. However, the exact economic impact has not been comprehensively assessed in Mozambique due to limited data available on its basic epidemiology. Therefore, we investigated the molecular occurrence and identity of Anaplasma and Ehrlichia spp. infecting beef cattle in Maputo province, Mozambique.
Methods
A total of 200 whole blood samples were collected from apparently healthy beef cattle. Whole blood DNA was extracted and tested for presence of Anaplasma spp. and Ehrlichia ruminantium DNA through amplification of the 16S rRNA and map1 genes. Positive samples to Anaplasma spp. were subject to PCR assay targeting the A. marginale-msp5 gene. Amplicons obtained were purified, sequenced and subject to phylogenetic analyses.
Results
Anaplasma spp., A. marginale and E. ruminantium were detected in 153 (76.5%), 142 (71%) and 19 (9.5%) of all the samples analyzed, respectively. On this same sample group, 19 (9.5%) were co-infected with A. marginale and E. ruminantium. The 16S rRNA sequences of Anaplasma spp. obtained were phylogenetically related to A. marginale, A. centrale and A. platys. Phylogenetic analysis revealed that A. marginale-msp5 nucleotide sequences were grouped with sequences from Asia, Africa and Latin America, whereas E. ruminantium-map1 DNA nucleotide sequences were positioned in multiple clusters.
Conclusion
Cattle in Maputo Province are reservoirs for multiple Anaplasma species. A high positivity rate of infection by A. marginale was observed, as well as high genetic diversity of E. ruminantium. Furthermore, five new genotypes of E. ruminantium-map1 were identified.
Similar content being viewed by others
Introduction
Tick and tick-borne diseases are one of the most significant constraints on livestock production in Mozambique. Diseases such as East Coast fever (caused by Theileria parva), heartwater (caused by Ehrlichia ruminantium), anaplasmosis (caused by Anaplasma marginale) and babesiosis (caused by Babesia bigemina and Babesia bovis) cause significant economic losses for cattle in the country [1,2,3,4]. Infections by Anaplasmataceae agents produce significant economic losses in Africa, where roughly 150 million animals are susceptible to infection [5, 6]. Mozambique holds the highest pooled prevalence estimate of tick-borne pathogens in domestic animals across the Southern African Developing Community (SADC), which has warm subtropical and tropical climates [7].
Anaplasma spp. and Ehrlichia spp. are tick-borne obligate intracellular Gram-negative bacteria that infects hematopoietic cells and belongs to Anaplasmataceae family. These species are of veterinary and public health significance [8,9,10] and are maintained in nature through enzootic cycles that includes Ixodidae ticks and vertebrate hosts [11]. Animals that recover from infection act as long-lasting carriers, with a small number of infected erythrocytes. These carrier animals play a significant role in the transmission of these tick-borne infections [12].
The Anaplasmataceae family comprises four main genera, namely Anaplasma, Ehrlichia, Neorickettsia and Wolbachia [12]. The genus Anaplasma comprises seven species, namely Anaplasma marginale, A. bovis, A. centrale, A. ovis, A. phagocytophilum, A. capra and A. platys. The genus Ehrlichia consists of six species, namely Ehrlichia canis, E. chaffeensis, E. ewingii, E. muris, E. minasensis and E. ruminantium [13, 14].
Anaplasma species known to infect domestic ruminants including cattle are A. marginale, A. bovis, A. capra, A. centrale, A. ovis, A. phagocytophilum, and A. platys [15,16,17]. Additionally, three other putative novel species of Anaplasma were recently detected in cattle in Ethiopia, namely Anaplasma sp. Hadesa, Anaplasma sp. Dedessa, and Anaplasma sp. Saso [18]. When it comes to Ehrlichia genus, E. ruminantium is the most common species known to infect cattle [19]. A new Ehrlichia, namely Ehrlichia minasensis, was detected in Rhipicephalus microplus ticks and cattle in Brazil, Ethiopia and Kenya [18, 20, 21], causing clinical signs similar to those of canine ehrlichiosis in an experimentally infected calf [22].
Anaplasmosis is caused by several Anaplasma species and is responsible for significant challenges for animal breeders. Indeed, infection by Anaplasma spp. increases the costs for veterinary care since it causes a reduction in animal body weight, decrease in milk production, abortions, and often death [23, 24]. Anaplasma marginale is the main causative agent of bovine anaplasmosis worldwide. This species is biologically transmitted by approximately 20 tick species and mechanically transmitted by biting flies and blood-contaminated fomites [24, 25]. The disease is characterized by anemia, weight loss, abortion, and death, resulting in significant economic losses for the cattle industry [23, 25, 26].
Ehrlichia ruminantium is the etiological agent of heartwater disease in domestic ruminants and is transmitted by Amblyomma ticks [27]. The disease is limited to sub-Saharan Africa and some Caribbean islands [27, 28]. Heartwater is severe in exotic and malnourished or stressed local breeds of cattle, and high losses are also observed in naïve local small ruminants and cattle that have been moved to an area in which the disease is endemic [28].
Eleven species of ixodid tick parasitize cattle in Maputo Province, namely Amblyomma hebraeum, Hyalomma rufipes, Ixodes cavipalpus, Rhipicephalus appendiculatus, Rhipicephalus evertsi evertsi, Rhipicephalus (Boophilus) microplus, Rhipicephalus simus, Rhipicephalus kochi, Rhipicephalus longus, Rhipicephalus pravus group, and Rhipicephalus turanicus [29]. Of these, A. hebraeum is the main transmission vector for E. ruminantium, while R. (B.) microplus is the main vector for A. marginale, B. bigemina, and B. bovis. Lastly, R. appendiculatus is the main transmission vector for T. parva, although other tick species can still transmit Anaplasma species, particularly A. marginale [21, 30].
Despite their economic importance, information about ticks and TBDs in the country remains fragmented and incomplete, making reasonable disease control methods difficult to implement. Concerning cattle, official records show that only one study detailing the genetic diversity of Anaplasma spp. [1] and another one on E. ruminantium [3] were carried out in the country. Therefore, the present study aims to contribute to a better knowledge of the molecular epidemiology of Anaplasma species and E. ruminantium that infect cattle in four districts of the northern region of Maputo Province.
Material and methods
Sampling
Between April and September 2022, 200 EDTA-blood samples were collected, by convenience, from apparently healthy adult cattle in four districts of Maputo province, Mozambique (Fig. 1). Fifty samples were collected in each of the four selected districts: Boane, Moamba, Marracuene and Manhiça. All the cattle sampled were Nguni and Nguni crossbreeds. Approximately 2–5 mL of blood was collected from the coccygeal vein into Ethylenediamine Tetra-Sodium Acetic Acid (EDTA)-buffered vacutainer tubes. The samples were kept on ice until they arrived at the laboratory, and then stored at -20 °C until analysis.
Ethical approval
All the procedures were carried out according to ethical guidelines for the use of animal samples permitted by the Institutional Animal Care and Use Committee (IACUC) of the Direcção Nacional de Desenvolvimento Pecuário, Maputo, Mozambique (License number: 161/MADER /DNDP/340/2023). The managers of the surveyed farms were informed about the study and gave their verbal approval prior to the cattle sampling.
Polymerase chain reaction
DNA extraction and molecular detection of Anaplasma spp. and Ehrlichia spp.
DNA was extracted from 200 μL of each blood sample using the DNeasy® Blood & Tissue Kit (Qiagen®, Valencia, CA), according to manufacturer’s instructions and stored at − 20 °C until its use in amplification reactions.
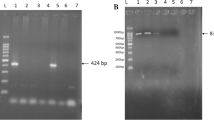
All DNA blood samples were subjected to a nested PCR targeting a 524 bp fragment of the 16S rRNA gene of Anaplasma spp. and Ehrlichia spp. as previously described by Rar et al. [11], using primers for initial reactions Ehr1 (5′-GAA CGA ACG CTG GCG GCA AGC-3′) and Ehr2 (5′-AGT A (T/C)C G(A/G)A CCA GAT AGC CGC-5′), and primers for nested reactions Ehr3 (5′-TGC ATA GGA ATC TAC CTA GTA G-3′) and Ehr4 (5′-CTA GGA ATT CCG CTA TCC TCT-3′).
All positive samples in the previously mentioned PCR assay were subjected to a semi-nested PCR targeting a 458 bp fragment of A. marginale major surface protein 5 (msp5) gene as previously described by Singh et at. [25], using Amar msp5 eF: GCATAGCCTCCGCGTCTTTC and Amar msp5 eR: TCCTCGCCTTGGCCCTCAGA as external primers and Amar msp5 iF: TACACGTGCCCTACCGAGTTA and Amar msp5 eR as internal primers.
In addition, all samples were subjected to semi-nested PCR assays targeting a fragment (720–738 bp) of the E. ruminantium Major Antigenic Protein 1 (map 1) gene as previously described by Matos et al. [3] using the following primers: External forward primer (ERF3) 5´- CCAGCAGGTAGTGTTTACATTAGCGCA-3´; External reverse (ERR1) 5´-CAAACCTTCCTCCAATTTCTATACC-3´; internal reverse (ERR3) 5´-GGCAAACATCAAGTGTTGCTGATGC-3´. Thus, the external forward primer (ERF3) in the first round of PCR was also maintained in the second round of amplification. Amplification reactions were performed in a conventional thermocycler (Gene Amp® PCR System Applied biosystems, Singapore).
Blood DNA samples positive to Anaplasma sp. (MH165337), Anaplasma marginale (MH124566), and E. ruminantium (KY860579), respectively, obtained from naturally infected cattle [1, 3], and ultra-pure sterile water were used as positive and negative controls, respectively.
PCR products were electrophoresed on 1.5% agarose gels to check the size of amplified fragments by comparison to a DNA molecular size marker (100 bp DNA ladder; Promega).
Sequence and phylogenetic analyses
The amplicons obtained from Anaplasma spp. 16S rRNA, A. marginale msp5, and E. ruminantium map1-based PCR assays showing high intensity of the bands (strongly positive) of expected sizes were purified with Wizard® SV Gel and PCR Clean-Up System Ref A 9282 (Promega, United States) according to the manufacturer's recommendations. Purified amplified DNA fragments were submitted to LGC Genomics, Berlin, Germany for bidirectional DNA sequencing. Consensus sequences were obtained through the analysis of electropherograms using the Phred-Phrap program [31]. The Phred quality score (peaks around each base call) was established at ≥ 20 (99% accuracy of the base call). The Basic Local Alignment Search Tool [BLAST] (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search for homologous reference sequences using the BLASTn algorithm. Alignments of Anaplasma spp. 16S rRNA, A. marginale msp5 and E. ruminantium map 1 sequences were constructed, and manually edited using BioEdit (version 7.0. 2.5) program [32].
The phylogenetic analysis was performed using the Maximum Likelihood (ML) method, inferred with RAxML-HPC BlackBox (7.6.3.) [33] and performed in CIPRES Science Gateway [34]. The Akaike Information Criterion (AIC) available on MEGA v. 5 software [35] was applied to identify the most appropriate model of nucleotide substitution. The JC model was chosen as the most appropriate for the phylogenetic analysis of the 16S rRNA, TN93 for the phylogenetic analysis of the msp5, and GTR + G evolutionary model for the phylogenetic analysis of the map1 nucleotide sequence alignment.
Results
Detection of Anaplasma spp. and Ehrlichia spp. in cattle blood DNA samples
Among the 200 cattle blood DNA samples analyzed in the present study, 153 (76.5%) were positive in the PCR targeting the 16S rRNA of the Anaplasma genus. On the other hand, none of the samples were positive for Ehrlichia 16S rRNA. In addition, out of the 153 cattle DNA samples, 142 (71%) were positive for A. marginale as well as the 200 cattle DNA samples, 19 (9.5%) were positive for E. ruminantium, respectively. Co-infections with A. marginale and E. ruminantium were recorded in 19 (9.5%) samples (Table 1). Both tick-borne agents, A. marginale and E. ruminantium, were detected in all four districts investigated. The rate of infection of A. marginale and E. ruminantium varied among sampling locations, ranging from 66 to 76% for A. marginale, with an overall occurrence of 71%; for E. ruminantium, it ranged from 2 to 18%, with an overall occurrence of 9.5%.
A very low proportion of positivity for E. ruminantium was reported in Marracuene district (2%) (Table 1). Among the amplified fragments 17 representative sequences of Anaplasma sp. 16S rRNA, 19 A. marginale msp5 and 10 E. ruminantium map1 genes derived from this study were submitted to GenBank database and assigned accession numbers OP297676—OP297692, OQ282861—OQ282879 and OP271794—OP271803 respectively (Table 2).
Sequences analysis
Anaplasma spp-16S rRNA sequences
According to BLASTn analysis, the nucleotide sequences of Anaplasma spp. obtained in this study are divided into two main groups.
The first group was composed of six nucleotide sequences, two sequences (OP297676 and OP297679) from Boane district, two sequences (OP297683 and OP297685) from Manhiça district, one from Moamba district (OP297689), and one from Marracuene district (OP297691) respectively. These sequences shared 99.5% identity with published sequences of A. platys from Saint Kitts and Nevis (CP046391) and from Vietnam (MH686049). The same six sequences also shared 99.5% identity with two published nucleotide sequences of ‘Candidatus Anaplasma camelli’ from Iran (MK726038) and Saudi Arabia (KF843827), as well as 99.4% identity with ‘Candidatus Anaplasma cinensis’ from China (MH762079) and South Africa (MK814448).
The second group was composed of 11 nucleotide sequences (OP297677, OP297678, OP297680, OP297681, OP297682, OP297684, OP297686, OP297687, OP297688, OP297690 and OP297692), which shared identities ranging from 99.5 to 99.8% with different published sequences: A. marginale (MK804764) from Cuba, A. ovis (AF309865) from the USA, A. centrale (MF289480 and MH588232), from China and Iraq. Finally, six nucleotide sequences of this group, three from Boane district (OP297677, OP297678, and OP297680), one from Manhiça district (OP297682) and two from Moamba district (OP297687 and OP297688) shared 99.5% identity with A. phagocytophilum from India (DQ648489). The identity among these 17-nucleotide sequences of Anaplasma spp. ranged from 98 to 100%, with query coverage ranging from 99 to 100%.
Anaplasma marginale - msp5 sequences
Nineteen nucleotide sequences of msp5 obtained in this study shared identity ranging from 99.78 to 99.55% with sequences of A. marginale detected in Sri Lanka (LC467711) and Thailand (MK188829). These sequences showed query coverage ranging from 94 to 100%.
E. ruminantium - map1 sequences
The BLASTn analysis of E. ruminantium nucleotide sequences is summarized as follows: Two sequences from this study (OP271794 and OP271797) shared 96.1% identity with two published sequences (JX486794 and JX477668) from Cameroon. The other two sequences of this study (OP271799 and OP271802) shared 100% homology with two published sequences detected in animals from Mozambique (KY856827 and KY860588). One nucleotide sequence from Boane district (OP271795) showed 100% nucleotide sequence identity with one published sequence (AB818942) from Uganda.
Two-nucleotide sequences from Manhiça district (OP271796 and OP271798) shared identities ranging from 99.7% to 100% with sequences (CP063045 and CP040120) detected in South Africa. Three nucleotide sequences (OP271800, OP271801 and OP271803) obtained in this study shared identities ranging from 89.3 to 99.85% with published sequences (CP063043) from South Africa and two sequences (AB818944 and AB818943), both from Uganda. The identity among E. ruminantium-map1 nucleotide sequences obtained in the present study ranged from 85 to 100%, with query coverage of 95 to 100%.
Phylogenetic analysis
In the phylogenetic tree based on the 16S rRNA gene of Anaplasma spp., four nucleotide sequences (OP297679, OP297683, OP297689 and OP297691) detected in this study were positioned near to A. platys sequences. In addition, one amplified sequence (OP297685) was more closely related to Anaplasma sp. previously detected in cattle from Mozambique. The remaining 11 nucleotide sequences (OP297677, OP297678, OP297680, OP297681, OP297682, OP297684, OP297686, OP297687, OP297688, OP297690 and OP297692) grouped together with A. marginale and A. centrale. All clusters were supported by bootstrap values of 50% (Fig. 2).
Phylogenetic relationships within the Anaplasma genus based on the 16S rRNA region. The tree was inferred by using the Maximum Likelihood (ML) with the JC model. The sequences detected in the present study are highlighted. The numbers at the nodes correspond to bootstrap values higher than 50% accessed with 1,000 replicates. Ehrlichia caffeensis was used as an outgroup
The phylogenetic tree based on the A. marginale-msp5 gene positioned all the amplified sequences in the same main group and clustered with other A. marginale sequences from different countries (Fig. 3).
Phylogenetic relationships among A. marginale msp-5 sequences. The tree was inferred by using the Maximum Likelihood (ML) with the TN93 model. The sequences detected in the present study are highlighted. The numbers at the nodes correspond to bootstrap values higher than 60% accessed with 1,000 replicates. Anaplasma phagocytophilum was used as an outgroup
Finally, the phylogenetic tree based on E. ruminantium-map1 nucleotide sequences obtained in this study and those retrieved from GenBank clustered into nine clusters. The sequences obtained in this study were positioned in seven different clusters (#1, #3, #4, #5, #6, #7 and #9). In cluster #1, one Mozambican sequence (OP271796) is grouped with three sequences: one from Southern Africa (AF368011), one from Botswana (AF368015) and the last one from South Africa (AF368011). Cluster #3 was formed exclusively by two Mozambican nucleotide sequences (OP271794 and OP271797) obtained in the present study. Cluster #4, two Mozambican sequences, one obtained in this study (OP271801) is grouped with one sequence from Uganda (AB818944). Cluster #5 was formed by two nucleotide sequences: one Mozambican sequence (OP271798) obtained in this study and another one from South Africa (U50834). In the cluster #6, one Mozambican sequence (OP271795) is grouped with four sequences: one from Southern Africa (AF355202), one from Zambia (AF355201), one from Uganda (AB818942), and finally one sequence from Cameron (JX486796). Cluster #7 was formed by six nucleotide sequence. Among them, five from Mozambique, including three obtained in this study (OP271799, OP271800 and OP271802) and another one from South Africa (AF125274). In cluster #9, one Mozambican sequence (OP271803) is grouped with four sequences: one from Uganda (AB818943), one from Tanzania (AF368003), one from Cameroun (JX477671) and one nucleotide sequence from Namibia (HQ259910). All clusters were supported by bootstrap values of 69–100% (Fig. 4).
Phylogenetic relationships among the Ehrlichia ruminantium map1 sequences. The tree was inferred by using the Maximum Likelihood (ML) with the GTR + G model. The sequences detected in the present study are highlighted. The numbers at the nodes correspond to bootstrap values higher than 60% accessed with 1,000 replicates. Ehrlichia sp. was used as an outgroup
Discussion
In the present study, the occurrence and phylogenetic relationships of important tick-borne pathogens in four districts of Maputo province were determined and analyzed.
In this study, an infection rate of 76.5% (153/200) of Anaplasma spp. was obtained using a nested PCR protocol based on the 16S rRNA gene. Similarly, Fernandes et al. [1] reported an occurrence of 87.2% (191/219) among cattle in the south region of Maputo Province, while Machado et al. [36] recorded an infection rate of 67% (65/97) of Anaplasma spp. in African buffaloes (Syncerus caffer) from Sofala province, in the central region of Mozambique. In this study, Anaplasma spp. phylogenetically associated with A. marginale, A. centrale, and A. platys were detected. In a previous study carried out in the southern region of Maputo Province, the DNA sequences obtained in cattle were phylogenetically related to A. marginale, A. centrale, A. phagocytophilum, A. platys, A. ovis, and ‘Candidatus Anaplasma boleense’ [1].
Based on 16S rRNA sequences, different species of Anaplasma might be simultaneously infecting cattle sampled in the current study. Despite the 16S region being conserved, the utilization of this target was important to show the diversity of Anaplasma species occurring in cattle from Mozambique. These findings reinforce the relevance of using species-specific PCRs for the detection of Anaplasma species to better assist in the conclusion of a diagnosis and the conduction of epidemiological surveys [36].
In Mozambique, particularly in the southern region of the country, cattle farmers also have a pack of dogs that accompany these ruminants to grazing areas, and those animals co-habit with each other, resulting in those cattle being infected by A. platys, the causative agent of infectious cyclic thrombocytopenia in dogs, so infection of cattle with this agent should not come as a surprise.
In this study, Anaplasma spp. phylogenetically associated with A. platys were detected. Anaplasma platys has been considered an emerging Anaplasma species whose clinical disease is yet to be described [37, 38]. Previous studies in Algeria [15], Mozambique [1], Senegal [38] and Tunisia [39] similarly itemized this pathogen in cattle. Yang et al. [40] suggested the possibility of domestic ruminants acting as alternative hosts or reservoirs for A. platys, which is typically a canine pathogen [41]. Therefore, the detection of this pathogen in cattle raises questions of host specificity, as earlier speculated [42]. Zobba et al. [37] noted that several domestic ruminants can harbor a number of strains of A. platys, although these strains have different cell tropisms compared to those infecting dogs. The ruminant strains infect neutrophils and are thought to be the ancestral pathogens that evolved to adopt to the canine platelets instead [37, 42]. Previous studies have recognized the zoonotic potential of A. platys, which can cause human disease characterized by headaches, intermittent edema, and muscle pains [43]. Consequently, more epidemiological studies are needed to determine the occurrence and clarify the zoonotic potential of A. platys in Maputo Province.
The MSP5 is a highly conserved 19-kDa protein and encoded by a single-copy 633 bp gene among A. marginale isolates, making it ideal for use in the molecular diagnosis of infection by this agent [25, 44, 45]. The A. marginale positivity rate of 71% (142 /153) based on the msp5 gene fragment detected in this study was lower than the recently reported 97.3% (213/219) based on a qPCR assay targeting the msp1β gene of A. marginale in Maputo Province [1]. This difference might be explained by the fact that the positivity rates of Anaplasma spp. might vary according to the diagnostic methods used [46]. On the other hand, reports indicate that the genus Anaplasma with causal agents of anaplasmosis in cattle had a higher prevalence in the SADC countries, and A. marginale was the most prevalent species of Anaplasma [7].
The high positivity rate of A. marginale observed in this present study and that reported in a study previously [1] warrants further investigation to evaluate the impact and diversity of this Anaplasmataceae agent on livestock production.
In the present study, E. ruminantium DNA was detected in all four searched districts with an overall proportion of infection of 9.5% (19/200), based on a map1-nested PCR assay. In a previous study, a positivity rate of 15% (31/210) was obtained according to the pCS20-nested PCR assay [3]. The positivity rate obtained here is relatively lower than that obtained in the previously performed study, and this variation in the positivity rate is probably due to the diagnostic methods used. The pCS20 gene is specific for E. ruminantium and is the most sensitive of the probes used for E. ruminantium detection, but it is not able to distinguish among the different genotypes. The map1 gene has also been used for the diagnosis and characterization of different genotypes of parasites [47]. The infection rate recorded in the present study is, however, sufficiently high to warrant the implementation of appropriate control strategies since clinical disease would be a risk if susceptible animals are present [21, 48]. We can assume that the E. ruminantium-map1 nucleotide sequences gained from the blood of cattle in the Maputo province were not conserved based on the results of the BLASTn and phylogenetic analyses.
The genetic diversity of E. ruminantium constitutes the main limitation for African countries to develop an efficient vaccine [3, 49,50,51]. Six DNA sequences, of which three from Moamba district were obtained in this study and three other nucleotide sequences obtained in a previous study [3] from three different localities in Maputo Province, shared identity or clustered with the Welgevonden sequence, one of the strains tested for vaccine development. Considering that five new genotypes were identified in this present study, these findings may help to improve current vaccine development and are also vital in understanding the epidemiology and control of heartwater disease.
Further research involving a large population of cattle, goats, and vectors in Mozambique is recommended in order to accurately determine the prevalence, geographic distribution, and genetic diversity of Anaplasma spp. and E. ruminantium throughout the country.
Conclusions
The present work indicates that cattle in Maputo Province are a reservoir for multiple Anaplasmataceae species. The 16S rRNA sequences of Anaplasma obtained were phylogenetically related to A. platys and A. marginale/A. centrale. The high positivity rate of infection by A. marginale in cattle observed in this present study warrants further investigation to evaluate the impact and diversity of this agent. High genetic diversity of E. ruminantium was observed, and five new genotypes of E. ruminantium-map1 were identified in cattle from Maputo province.
Availability of data and materials
The Anaplasma sp. 16S rRNA, Anaplasma marginale msp5 and E. ruminantium map1 sequences derived from this study were submitted to GenBank database and assigned accession numbers: OP297676-OP297692, OQ282861-OQ282879 and OP271794-OP271803, respectively.
References
Fernandes SJ, Matos CA, Freschi CR, Ramos IAS, Machado RZ, André MR. Diversity of Anaplasma species in cattle in Mozambique. Ticks Tick Borne Dis. 2019;10:651–64. https://doi.org/10.1016/j.ttbdis.2019.02.012.
Martins TM, Neves L, Pedro OC, Fafetine JM, Rosário VE, Domingos A. Molecular detection of Babesia spp. and other haemoparasitic infections of cattle in Maputo Province, Mozambique. Parasit. 2010;1–8. https://doi.org/10.1017/S003118200999196X.
Matos CA, Gonçalves LR, Ramos IAS, Mendes NS, Zanatto DCS, André MR, Machado RZ. Molecular detection and characterization of Ehrlichia ruminantium from cattle in Mozambique. Acta Trop. 2019;191:198–203. https://doi.org/10.1016/j.actatropica.2019.01.007.
Tembue AAM, Silva JB, Silva FJM, Pires MS, Baldani CD, Soares CO, Massard CL, Fonseca AF. Seroprevalence of IgG antibodies against Anaplasma marginale in cattle from south Mozambique. Rev Bras Parasitol Vet. 2011;20:1–7. https://doi.org/10.1590/s1984-29612011000400011.
Minjauw B. and Mclead A. Veterinary Medicine Tick-Borne Diseases and poverty. The impact of ticks and Tick-Borne Diseases on the livelihood of small- Scale and marginal livestock owners in India and Eastern and Southern Africa. Research report DFID Animal Heaith programme, Centre for Tropical Veterinary Medicine, University of Edinburgh, UK. 2003. https://doi.org/10.5555/20063155090.
Kivaria FM. Estimated direct economic costs associated with tick-borne diseases on cattle in Tanzania. Trop Anim Health Prod. 2006;38:291–9. https://doi.org/10.1007/s11250-006-4181-2.
Tawana M, Onyiche TE, Ramatla T, Mtshali S, Thekisoe O. Epidemiology of Ticks and Tick-Borne Pathogens in Domestic Ruminants across Southern African Development Community (SADC) Region from 1980 until 2021: A Systematic Review and Meta-Analysis. Pathogens. 2022;11:929. https://doi.org/10.3390/pathogens11080929.
Eremeeva ME. and Dasch GA. Anaplasmataceae as Human pathogens: Biology. Ecology and Epidemiology. Revue Tunisienne d’Infectiologie Intracellular Bacteria: From Biology to Clinic. 2011;5 (S1):S7-S14: Revue Tunisienned’Infectiologie. http://www.rev-tun-infectiologie.org/detail_art.php?id_det=32.Dahmani M,.
Rymazewska A, Grenda S. Anaplasma-Characteristics of Anaplasma and their vector: A revie vet Med. 2008;11(11):573–84. https://doi.org/10.17221/1861-VETMED.
Welc-Falęciak R, Kowalec M, Karbowiak G, Bajer A, Behnk JM, Siński E. Rickettsiaceae and Anaplasmataceae infections in Ixodes ricinus ticks from urban and natural forested areas of Poland. Parasit Vector. 2014;7:121. https://doi.org/10.1186/1756-3305-7-121.
Rar VA, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, Ivanov LI. Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 2010;1:57–65. https://doi.org/10.1016/j.ttbdis.2010.01.002.
Dumler JS, Barbet AF, Bekker CPJ, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganisation of genera in the family’s Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia and Ehrlichia with Neorickettsia, description of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonymous of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–216. https://doi.org/10.1099/00207713-51-6-2145.
Aktas M. A Survey of Ixodid Ticks Species and Molecular Identification of Tick-Borne Pathogens. Vet Parasitol. 2014;200:3–4. https://doi.org/10.1016/j.vetpar.2013.12.008.
Dumler JS. Anaplasma and Ehrlichia infection. Ann N Y Acad Sci Washington. 2005;1063:361–73. https://doi.org/10.1196/annals.1355.069.
Davoust B, Benterki MS, Fenollar F, Raoult D, Mediannikov O. Development of a new PCR-Based Assay to detect Anaplasmataceae and the first report of Anaplasma Phagocytophilum and Anaplasma Platys in cattle from Algeria. Comp Immunol Microbiol Infect Dis. 2015;39:39–45. https://doi.org/10.1016/j.cimid.2015.02.002.
Park J, Han DG, Ryu JH, Chae JB, Chae JS, Yu DH, Park BK, Kim HC, Cho KS. Molecular detection of Anaplasma bovis in Holstein cattle in the Republic of Korea. Acta Vet Scand. 2018;60(1):1–5. https://doi.org/10.1186/s13028-018-0370-z.
Staji H, Yousefi M, Hamedani M A, Tamai I A, Khaligh S G. Genetic characterization and phylogenetic of Anaplasma capra in Persian onagers (Equus hemionus onager) Vet Microbiology 261 (2021) 109199. https://doi.org/10.1016/j.vetmic.2021.109199.
Hailemariam Z, Krücken J, Baumann M, Ahmed JS, Clausen PH, Nijhof AM. Molecular detection of tick-borne pathogens in cattle from Southwestern Ethiopia. PLoS ONE. 2017;12(11):1–16. https://doi.org/10.1371/journal.pone.0188248.
Allsopp BA. Heartwater-Ehrlichia ruminantium infection. Revue scientifique et technique (International office of Epizootics). 2015;34(2):557–68. https://doi.org/10.20506/rst.34.2.2379.
Aguiar DM, Ziliani TF, Zhang X, Melo AL, Braga IA, Witter R, et al. A novel Ehrlichia genotype strain distinguished by the TRP36 gene naturally infects cattle in Brazil and causes clinical manifestations associated with ehrlichiosis. Ticks Tick Borne Dis. 2014;5(5):537–44. https://doi.org/10.1016/j.ttbdis.2014.03.010.
Peter SG, Gakuya DW, Maingi N, Mulei CM. Prevalence and risk factors associated with Ehrlichia infections in smallholder dairy cattle in Nairobi City County, Kenya. Vet World. 2019;12(10):1599–607. https://doi.org/10.14202/vetworld.2019.1599-1607.
Cruz AC, Zweygarth E, Ribeiro MF, da Silveira JA, de la Fuente J, Grubhoffer L, et al. New species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus shows an ortholog of the E. canis Major immunogenic glycoprotein gp36 with a new sequence of tandem repeats. Parasit Vectors. 2012;5:2. https://doi.org/10.1186/1756-3305-5-291.
Kocan KM, De la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. 2010;167:95–107. https://doi.org/10.1016/j.vetpar.2009.09.012.
Kocan KM, Blouin EF, Barbet AF. Anaplasmosis control. Past, present, and future. Ann N Y Acad Sci. 2000;916:501–9. https://doi.org/10.1111/j.1749-6632.2000.tb05329.x.
Singh H, Jyoti HM, Singh NK, Rath SS. Molecular detection of Anaplasma marginale infection in carrier cattle. Ticks Tick Borne Dis. 2012;3(1):55–8. https://doi.org/10.1016/j.ttbdis.2011.10.002.
Mccoske P. Global aspects of the management and control of ticks of veterinary importance. Med Environ Sci. 1979. https://doi.org/10.1016/B978-0-12-592202-9.50012-4.
Allsopp BA. Natural history of Ehrlichia ruminantium. Vet Parasit. 2010;167:2–4. https://doi.org/10.1016/j.vetpar.2009.09.014.
Bekker CPJ, deVos S, Taoufik A, Sparagano OAE, Jongejan F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantum in Amblyomma variegatum ticks by reverse line Blot hybridization. Vet Microbiol. 2002;89:223–38. https://doi.org/10.1016/s0378-1135(02)00179-7.
Horak IG, Nyangiwe N, Matos CA, Neves L. Species composition and geographic distribution of ticks infesting cattle, goats and dogs in a temperate and in a subtropical region of southeast Africa. Onderstepoort J Vet Res. 2009;76:263–76.
Walker JB. A review of the ixodid ticks (Acari, Ixodidae) occurring in southern Africa. Onderstepoort J Vet Res. 1991;58:81–105.
Ewing B, Hillier L, Wendl MC, Green P. Base calling of automated sequencer traces using phred. I Accuracy assessment Geno Res. 1998;8:175–85. https://doi.org/10.1101/gr.8.3.175.
Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Stamatakis A, Hoover P, Rougemont J, Renner S. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–71. https://doi.org/10.1080/10635150802429642.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In proceedings of the gateway computing environments workshop (GCE) (New Orleans. 2010;14:1–8. https://doi.org/10.1109/GCE.2010.5676129.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011;2. https://doi.org/10.1093/molbev/msr121.
Machado RZ, Teixeira MMG, Rodrigues AC, André MR, Gonçalves LR, Da Silva JB, et al. Molecular diagnosis and genetic diversity of tick-borne Anaplasmataceae agentes infecting the African buffalo Syncerus caffer from Marromeu Reserve in Mozambique. Parasit Vectors. 2016;9:454–462.29. https://doi.org/10.1186/s13071-016-1715-y.
Zobba R, Anfossi AG, Parpaglia MLP, Dore GM, Chessa B, Spezzigu A, et al. Molecular investigation and phylogeny of Anaplasma spp. in mediterranean ruminants reveal the presence of neutrophil-tropic strains closely related to A. platys. Appl Environ Microbiol. 2014;80:271–80.
Ben Said M, Belkahia H, El Mabrouk N, Saidani M, Alberti A, Zobba R, et al. Anaplasma platys-like strains in ruminants from Tunisia. Infect Genet Evol. 2017;49:226–33. https://doi.org/10.1016/j.meegid.2017.01.023.
Dahmani M, Davoust B, Sambou M, Bassene H, Scandola P, Ameur T, et al. Molecular investigation and phylogeny of species of the Anaplasmataceae infecting animals and ticks in Senegal. Parasites Vectors. 2019;12:1–15. https://doi.org/10.1186/s13071-019-3742-y.
Yang J, Han R, Niu Q, Liu Z, Guan G, Liu G, et al. Occurrence of four Anaplasma species with veterinary and public health significance in sheep, northwestern China. Ticks Tick Borne Dis. 2018;9:82–5. https://doi.org/10.1016/j.ttbdis.2017.10.005.
Ybañez AP, Ybañez RHD, Yokoyama N, Inokuma H. Multiple infections of Anaplasma platys variants in Philippine dogs. Vet World. 2016;9:1456–60.
Bastos ADS, Mohammed OB, Bennett NC, Petevinos C, Alagaili AN. Molecular detection of novel Anaplasmataceae closely related to Anaplasma platys and Ehrlichia canis in the dromedary camel (Camelus dromedarius). Vet Microbiol. 2015;179:310–4. https://doi.org/10.1016/j.vetmic.2015.06.001.
Breitschwerdt EB, Hegarty BC, Qurollo BA, Saito TB, Maggi RG, Blanton LS, et al. Intravascular persistence of Anaplasma platys, Ehrlichia chaffeensis, and Ehrlichia ewingii DNA in the blood of a dog and two family members. Para-sites Vectors. 2014;7:1–7.
Torioni de Echaide S, Knowles DP, McGuire TC, Palmer GH, Suarez CE, McElwain FF. Detection of cattle naturally infected with Anaplasma marginale in a region of endemicity by nested PCR and a competitive enzyme-linked immunosorbent assay using recombinant major surface protein 5. J Clin Microbiol. 1998;36:777–82. https://doi.org/10.1128/JCM.36.3.777-782.1998.
Ybañez AP, Sivakumar T, Battsetseg B, Battur B, Altangerel K, Matsumoto K, Yokoyama N, Inokuma H. Specific Molecular Detection and Characterization of Anaplasma marginale in Mongolian Cattle. J Vet Med Sci. 2013;75(4):399–406. https://doi.org/10.1292/jvms.12-0361.
Chaisi ME, Baxter JR, Hove P, Choopa CN, Oosthuizen MC, Brayton KA, Khumalo ZTH, Mutshembele AM, Mtshali MS, Collins NE. Comparison of three nucleic acid-based tests for detecting Anaplasma marginale and Anaplasma centrale in cattle. Onderstepoort J Vet Res. 2017;84(1): a1262. https://doi.org/10.1186/s13071-017-2595-5.
Collins NE, Allsopp MTEP, Allsopp BA, Molecular diagnosis of theileriosis and heartwater in bovines in Africa. Trans. R. Soc. Trop. Med. Hyg. 2002;96 (supplement 1), s1/217-s1/224. https://doi.org/10.1016/s0035-9203(02)90079-9.
Swai ES, Mtui PF, Chang’a AK, Machange GE. The prevalence of serum Antibodies to Ehrlichia ruminantium infection in Ranch cattle in Tanzania: A cross-sectional study. J S Afr Vet Association. 2008;79(2):71–5. https://doi.org/10.4102/jsava.v79i2.247.
Martinez D, Vachi´ery N, Stachurski F, Kandassamy Y, Raliniaina M, Aprelon R, et al. Nested PCR for detection and genotyping of Ehrlichia ruminantium; use in genetic diversity analysis. Ann N Y Acad Sci. 2005;1026:106–13. https://doi.org/10.1196/annals.1307.014.
Mnisi SS, Mphuthi MBN, Ramatla T, Mofokeng LS, Thekisoe O, Syakalima M. Molecular detection and genetic characterization of Ehrlichia ruminantium harbored by Amblyomma hebraeum ticks of domestic ruminants in North West Province, South Africa. Animals. 2022;12(19):2511. https://doi.org/10.3390/ani12192511.
Vachiéry N, Jeffery H, Pegram R, Rosalie AR, Pinarello V, Kandassamy RLY, Raliniaina M, Sophie MS, Savage H, Alexander R, Frebling M, Martinez D, Lefrancois T. Amblyomma variegatum ticks and Heartwater on three caribbean islands tick infection and Ehrlichia ruminantium genetic diversity in bovine herds. Anim Biodivers Emerg Dis Ann N Y Acad Sci. 2008;1149:191–5. https://doi.org/10.1196/annals.1428.081.
Acknowledgements
The authors would like to thank all the staff at the farms and Direçcão de Ciências Animais, Maputo, Mozambique for the support for sample collections. The support in terms of equipment, reagents and consumables given by IAEA is highly appreciated. MRA was supported by CNPq (National Council for Scientific and Technological Development; Productivity Grant Process #303701/2021-8]).
Institutional review board statement
Not applicable.
Conflict of interest
The authors have no any conflict of interest and no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript. Similarly, this manuscript has not been submitted to, nor is under review at, another journal or other publishing method.
Funding
The authors declare no financial support for the research, authorship, or publication of this article.
Author information
Authors and Affiliations
Contributions
CAM conceived, designed the study, and conducted the sequences and phylogenetic analysis and wrote the paper; CFN conducted the molecular studies; LRG conducted the phylogenetic analysis, revised the manuscript and submission; ACC, CFS, APRV and CMM analyzed the samples; MCC and MRA revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Animal Care and Use Committee (IACUC) of the Direcção Nacional de Desenvolvimento Pecuário, Maputo, Mozambique (License number: 161/MADER /DNDP/340/2023). The managers of surveyed farms were informed about the study and gave their approval for the sampling of cattle. All the procedures were carried out according to ethical guidelines for the use of animal samples.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Matos, C.A., Nomboro, C.F., Gonçalves, L.R. et al. Molecular diagnosis and characterization of Anaplasma marginale and Ehrlichia ruminantium infecting beef cattle of Maputo Province, Mozambique. BMC Vet Res 20, 185 (2024). https://doi.org/10.1186/s12917-024-04045-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-024-04045-4