Abstract
The purpose of this study was to determine the level of horizontal transmission of the blaCTX-M-65 gene and the role of its associated mobile genetic elements (MGEs) in the bovine-derived Escherichia coli. After PCR identification, two plasmids carrying blaCTX-M-65 were successfully transferred to the recipient E. coli J53 Azr through conjugation assays and subsequently selected for Whole-Genome sequencing (WGS) analysis. The resistance profiles of these two positive strains and their transconjugants were also determined through antimicrobial susceptibility tests. Whole genome data were acquired using both the PacBio sequencing platform and the Illumina data platform. The annotated results were then submitted to the Genbank database for accession number recording. For comparison, the genetic environment of plasmids carrying the resistance gene blaCTX-M-65 was mapped using the Easyfig software. WGS analysis revealed Tn3-like composite transposons bearing blaCTX-M-65, blaTEM-1, and blaOXA-10 in the IncHI2-type plasmids of these two E. coli ST1508 strains. A phylogenetic tree was generated from all 48 assembled E. coli isolates blaCTX-M-65, blaTEM-1, and blaOXA-10 from the NCBI Pathogen Detection database with our two isolates, showing the relationships and the contribution of SNPs to the diversity between genetic samples. This study suggests that the transmissibility of blaCTX-M-65 on Tn3-like composite transposons contributes to an increased risk of its transmission in E. coli derived from dairy cattle.
Similar content being viewed by others
Introduction
Since the 1980s, extended-spectrum β-lactamases (ESBL)-producing E. coli has spread globally, thus becoming the primary cause of nosocomial and community-acquired infections such as gastrointestinal and extra-intestinal infections which majorly includes urinary tract infections (UTIs) [1, 2]. Most of E. coli strains are extended beta-lactamase producing and identified as multidrug resistant (resistance to penicillin, cephalosporin’s, and monobactems) due to the hydrolyzaiton of beta-lactam ring. The most prevalent ESBL families include CTX-M, TEM, OXA and SHV, with CTX-M gradually emerging as the dominant variant [3,4,5]. The expression of CTX-M family genes is often associated with increase drug resistance to multiple beta-lactam antibiotics which restrict the more options for therapeutic purposes leading to the treatment options are severely limited [6]. In addition, CTX-M genes have been widely identified in E. coli isolated from multiple sources such as humans, livestock, companion animals, food products, and environment, raising significant concerns [1, 6].
The CTX-M genes are rarely found on chromosomes, instead encoded on mobile genetic elements (MGEs), usually on plasmids. Plasmid located CTX-M genes has a strong frequency of horizontal transfer and their increased dissemination within one health care (humans, animals, and environment) [7, 8]. MGEs located on plasmids can widely spread antibiotic resistance genes (ARGs) [9]. It follows that Enterobacteriaceae have adopted a crucial strategy to capture, stabilize and disseminate new genetic elements during the acquisition and evolution of ESBL enzymes [6]. The current epidemiological scenario suggests that the transmission pattern of the CTX-M enzyme may be allodemic rather than epidemic. In other words, the widespread prevalence of CTX-M enzyme is not due to the proliferation of specific clones, but rather to the proliferation of multiple specific clones or MGEs [10].
Co-transmission of CTX-M-65 with other ESBL-ARGs in E. coli have also been reported more frequently, ultimately leading to an increased risk of livestock-to-human transmission and global multidrug resistance (MDR) through horizontal gene transfer (HGT) [11,12,13,14]. Note that blaCTX-M-65 gene, with an elevated detection rate, has been reported in food animals [15, 16]. Food-producing chickens carrying extended-spectrum β-lactamase–producing Escherichia coli (ESBL-EC), including CTX-M-65–producing Escherichia coli have posed a potential threat to human and animal health [17]. As a result, there is an increased risk of transmission of these genes from food animals to humans. In this study, we characterized and investigated the horizontal transfer mechanism of blaCTX-M-65 from bovine origin through whole genome sequencing and conjugation assay.
Materials and methods
Strain isolation
73 non-repetitive fecal samples were collected from dairy cattle in the Xinjiang region of China in 2018. All samples were collected in sterile containers, transported to the laboratory at 6 °C ± 2 °C, and processed immediately for further assays. All isolates were selected on MacConkey agar plates (Huankai, China) and identified by MALDI-TOF–MS and 16 s rRNA sequencing using universal primers F (5’-GAGCGGATAACAATTTCACACAGG-3’) and R (5’-CGCCAGGGTTTTCCCAGTCACGAC-3’). All E. coli strains were stored in our laboratory after identification.
CTX-M-65-positive strains were screened for the presence of blaCTX-M-9 group using PCR methods with primers obtained from the article [18]. PCR products were confirmed using Sanger sequencing.
Conjugation assay and determination of conjugation frequency
Conjugation assays were performed among 2 isolates. To determine the transferability of the resistance genes, filtered broth mating was performed using E. coli J53 Azr as the recipient [19]. The donor-recipient ratio was 1:1 using Mueller–Hinton medium (MHA, Huankai, Guangzhou, China) supplemented with cefotaxime (2 μg/mL) and sodium azide (200 μg/mL) as the selective medium. Positive clones of transconjugants were screened by PCR (blaCTX-M-9 primers, F: 5’-ATGGTGACA AAG AGAGTGCAAC-3’, R: 5’-TTACAGCCCTTCGGCGATG-3’), Sanger sequencing and antimicrobial susceptibility testing. Transfer frequencies were calculated as the number of transconjugants per recipient.
Antimicrobial susceptibility testing and resistant genes detecting
Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method according to Clinical & Laboratory Standards Institute (CLSI) M100 guidelines (http://clsi.org). Broth culture, equivalent to a 0.5 McFarland standard, was incubation for 16 h at 37℃. The following antibiotics were used for testing: cefotaxime (CTX), ceftazidime (CAZ), cefalotin (KF), tetracycline (TE), ampicillin (AMP), sulbactam-ampicillin (SAM), streptomycin (S), amoxicillin/clavulanic acid (AMC), amikacin (AK), ciprofloxacin (CIP), doxycycline (DO), fosfomycin (FOT), kanamycin (K), chloramphenicol (C), sulphamethoxazole-trimethoprim (SXT), gentamicin (GN), aztreonam (ATM). E. coli ATCC25922 was used as the quality control strain.
WGS sequencing and analysis
The total genomic DNA was extracted with the SDS method [20]. The harvested DNA was detected by the agarose gel electrophoresis and quantified by Qubit® 2.0 Fluorometer (Thermo Scientific). The whole genome of the isolate was sequenced on PacBio Sequel single-molecule real-time (SMRT) sequencing platform (Novogene Bioinformatics Technology Co., Beijing, China) with 400 bp paired-end libraries. It was preliminarily assembled with SMRT Link v5.0.1. The assembled data were optimized by Illumina reads using Burrows-Wheeler Aligner (BWA).
Gene prediction and annotation were performed using the RAST server (https://rast.nmpdr.org/) and the BLAST program of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The replicon of the plasmid was confirmed using PlasmidFinder 2.1 (with minimum identity 95% and minimum coverage 60%) (https://cge.cbs.dtu.dk/services/PlasmidFinder/). The antibiotic-resistant genes were identified using CARD data (https://card.mcmaster.ca/). Clonal analysis was assessed by MLST 2.0 (https://cge.food.dtu.dk/services/MLST/). The comparative analysis and plasmid map was generated with Easyfig2.2.3 [21]. The pathogenic and antibiotic-resistant information of strainswas supported by NCBI Pathogenic Detection (https://www.ncbi.nlm.nih.gov/pathogens/).
To confirm the phylogenetic relationship based on the single nucleotide polymorphisms (SNPs) present in the genomes carrying the blaCTX-M-65 gene, the two E. coli strains in this study and the 48 E. coli strains downloaded from the NCBI Pathogenic Detection were analyzed using CSI Phylogeny version 1.4 (10 reads of minimal depth at SNP positions, 10% minimal relative depth at SNP positions, 10 bp of minimal distance between SNPs, minimal SNP quality of 30, minimal read mapping quality of 25, and a minimal Z-score of 1.96 [22]. The phylogenetic tree was visualized using FigTree version v1.4.4.
Phylogenetic analysis
The phylogenetic analysis was performed using the method of Kaas [23]. A phylogenetic tree was generated using E. coli ATCC25922 as the reference genome for all assembled E. coli isolates carrying blaCTX-M-65, blaTEM-1, and blaOXA-10 from the NCBI Pathogenic Detection database. To be specific, a total of 50 E. coli isolates carrying these ARGs were collected from different regions of the world and isolated from various sources.
Nucleotide sequence accession number
The data of the whole-genome sequence was uploaded and registered in the NCBI database with accession numbers: CP098227 (E. coli XJ6.3-chr1), CP098228 (E. coli XJ6.3-plas1), CP098231 (E. coli XJ34-chr1), CP098232 (E. coli XJ34-plas1), CP098233 (E. coli XJ34-plas2).
Availability of data and materials
The datasets generated and analyzed during the current study are available in the [NCBI] repository with accession numbers as mentioned above.
Results and discussion
A total of 108 E. coli strains were isolated on MacConkey agar from dairy cow and confirmed by 16 s rRNA and MALDI-TOF–MS. Among the 108 E. coli strains, three strains were identified positive for blaCTX-M-65 gene by PCR method in our previous study [24]. In two strains, blaCTX-M-65 gene was successfully transferred through conjugation which were further characterized for antimicrobial susceptibility testing, acquired ARGs, and related the genetic environment through WGS analysis. The information about the origin and source of the isolates is shown in Table 1.
The Kirby-Bauer disk diffusion method was used to evaluate the drug resistance profile against multiple antibiotics. The results showed that both of the strains were multidrug-resistant (MDR) and resistant to at least nine antibiotics. Both of the isolates showed resistance to cefalotin (KF), cefotaxime (CTX), tetracycline (TE), chloramphenicol (C), ampicillin (AMP), doxycycline (DO), sulbactam-ampicillin (SAM) and strimethoprim-sulfamethoxazole (SXT) (Table 1). A previous conducted by researchers also identified MDR E. coli strains isolated from dairy origin which is consistent to the current findings [25]. The ESBL-producing E. coli also displayed a wide spectrum antibiotic resistance to other commonly used antibiotics at clinical and livestock industry such as β-lactam, tetracycline and chloramphenicol and trimethoprim-sulfamethoxazole which results in limited treatment options and higher rate of therapeutic failures in infections caused by ESBL-producing E. coli.
The conjugation assay revealed that two plasmids carrying CTX-M-65 gene were successfully transferred to the recipient E. coli J53 (Sodium Azide resistance) indicating the presence of the target gene on a mobile plasmid. The conjugation frequencies of XJ6-3-J53 and XJ34-J53 carrying CTX-M-65 gene were noted to be 1.9 × 10–6 and 6.9 × 10–5 conjugants per recipient respectively which is in consistent with a previous study [26].
Both of the strains were sent for WGS and characterized for plasmid replicon typing, serotyping, MLST, acquired ARGs, and phylogenetic relationship using CGE website. Plasmid replicon typing revealed that blaCTX-M-65 gene was localized on IncHI2 plasmid type in both strains along with other replicon types such as IncI1, IncF, and IncA/C2 plasmids (Table 1) which are previously known as dominant plasmid replicon types [11, 27].
The MLST revealed that both CTX-M-65-positive strains belongs to ST1508. Surprisingly, after searching the Pathogen Detection database, none of the CTX-M-65 positive strain was ST1508 which indicate the emergence of a novel ST1508 in this study. Moreover, both XJ6.3 and XJ34 strains were categorized under O172:H45 serotype which belongs to toxin producing E. coli. E. coli O172 producing Shiga toxin type 2 has been reported from food and water sources commonly associated with illnesses [28, 29]. XJ6.3 and XJ34 have certain degree of pathogenicity and resistance, therefore their possible transmission need greater attention at the right time.
Two plasmids carrying CTX-M-65 gene were successfully transferred to the recipient E. coli J53 Azr according to conjugation assays, indicating the presence of the CTX-M-65 gene on a plasmid. The conjugation frequencies of J53XJ6-3 and J53XJ34 carrying CTX-M-65 gene were 1.9 × 10–6 and 6.9 × 10–5, respectively. The transconjugative frequencies of conjugants of harboring CTX-M-65 genes were 10–5 ~ 10–6 in consistent with the additional research of CTX-M-65 on IncHI2 [26].
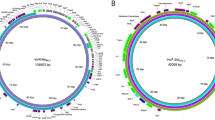
The genetic environment analysis of two blaCTX-M-65-positive isolates were done by WGS. For pXJ6.3-plas1, the complete genome contained a circular 181,896 bp plasmid with GC content of 46.0%, harboring blaCTX-M-65, blaTEM-1, tet(A), floR, dfrA14, aadA1, blaOXA-10, cmlA5, arr-2, and qnrS1 genes (Table 2). This plasmid was also harboring several mobile elements, including IS91-like, Tn3-like, IS26, IS2, IS6 and Int1. The genetic region of 14,251 bp around the blaCTX-M-65 gene was found 99.8% ~ 99.9% identical with 100% coverage to three plasmids, pA102-CTX-M-65 carried by E. coli (MN816370.1), p453-MDR carried by Salmonella sp. (CP060856.1) and pST95-32–1 carried by E. coli (CP043951.1), respectively (Fig. 1). The genome of E. coli XJ34 was carrying a circular 185,577-bp with GC content of 46.0%. The blaCTX-M-65 positive isolates harboring IncHI2 plasmid were acquiring same resistance genes with two additional mobile elements, IS903 and ΔIS1380 (Table 2). The genomic region of 6,747 bp was found 99.8% similar in sequence identity (coverage 100%) to the plasmid, p160070-CTXM carried by Klebsiella pneumonia (MG288677.1.1) (Fig. 1). These observation suggested that these multidrug-resistant region (MRR) on IncHI2-type plasmids carrying blaCTX-M-65 have a high degree of similarity, indicating Tn3-like–1S903-blaCTX-M-65–IS26 were conducive to spread between different species.
The results of genetic context showed that both strains were also carrying additional beta-lactam resistance genes, blaTEM-1 and blaOXA-10. More than 170 TEM-1 variants have been identified in E. coli from hospitals and clinical settings worldwide, which can hydrolyze beta-lactam drugs such as penicillin and cephalosporin’s [30]. The OXA family enzymes are mostly limited to penicillin group, but recently their resistance to last resort carbapenem was also observed which needs much attention because of horizontal transfer of genes as observed in this study in-vitro trial.
The mobile genetic elements such as IS903 and Tn3 are the most prevalent insertions and transposons which associated with blaCTX-M-65 which is consistent with our findings that blaCTX-M-65 gene was surrounded by IS903 and Tn3-like mobile elements [31]. The Tn3 belong to bacterial transposons group which are involved in transposition function and mediation of gene re-assortment, which make it conducive for horizontal gene transfer in gram-positive and negative bacteria [32]. In addition, the res site provides more clarity on the Tn3-like transposon formed by transduction and integration to transmit ARGs. The presence of these elements in the genome of blaCTX-M-65-positive strains mediates the mechanism of genes mobilization and increase the risk of horizontal transmission through multiple mobile elements [33].
The phylogenetic relationship of two strains harboring blaCTX-M-65 gene and other strains retrieved from NCBI database were done by CSIPhylogeny tool based on SNPs. Both strains of present study shared 14 SNPs with each other indicating much homology with each other. However, both strains shared lower homology with other 48 strains retrieved from NCBI based on SNP differences ranging from 26,834 to 35,699. In addition, SNP analysis showed that SAMD00532258 strain were having highest SNPs, between 58,737 and 53,612. The number of SNPs in the majority of the strains carrying three ARGs were ranged from 847 to 58,737. The phylogenetic tree distinguished the two strains of this study and 48 from NCBI into 4 major clades (Fig. 2). Clade 1 consist of 20 strains, one from USA and left from China isolated from chicken, swine, duck, Manis javancia, human and water sources. Clade 2 were containing 15 strains from four different countries and sources. Clade 3 were consisting of two strains characterized in present study harboring blaCTX-M-65 under separate sub-clade and 13 other strains isolated from humans and animals in China. The phylogenetic analysis revealed that there was a high degree of genetic homology between the two strains (XJ6.3 and XJ34), marked with green box of this study, and lower homology with other strains. The aforementioned genetic differences from phylogenetic analysis indicate that these genes are more likely to be transmitted horizontally by plasmids rather than clonal dissemination.
Conclusion
The WGS analysis obtained a complete map of the genetic environment of CTX-M-65 in both isolates through the Pacbio sequencing platform and Illumina data platform. The CTX-M-65 positive E. coli isolates derived from dairy cattle and was to show co-transmission of blaCTX-M-65 with other ESBLs genes (blaTEM-1, blaOXA-10) on the IncHI2 plasmid. A genetic environmental analysis of the CTX-M-65 gene revealed the presence of Tn3-like transposons and IS903 in both strains, possibly contributing to the mobilization of those ARGs. Genetic differences from phylogenetic analysis display showing the relationships and the contribution of SNPs to the diversity between genetic samples and co-transmission of these genes is more likely to occur through plasmids than through clones. Also, CTX-M-65 was derived from normal dairy cattle in this study, and it is still not absolutely certain that the source of CTX-M-65 is food dairy cattle. Finally, this study provides evidence of the need for appropriate measures to minimize the spread of MDR bacteria in dairy cattle samples.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the [NCBI] repository with accession numbers: CP098227 (E. coli XJ6.3-chr1), CP098228 (E. coli XJ6.3-plas1), CP098231 (E. coli XJ34-chr1), CP 098232 (E. coli XJ34-plas1), CP 098233 (E. coli XJ34-plas2).
Abbreviations
- MGEs:
-
Mobile genetic elements
- E. coli :
-
Escherichia coli
- WGS:
-
Whole-Genome sequencing
- ESBL:
-
Extended-spectrum β-lactamases
- ARGs:
-
Antibiotic resistance genes
- MDR:
-
Multidrug resistance
- CTX:
-
Cefotaxime
- CAZ:
-
Ceftazidime
- KF:
-
Cefalotin
- TE:
-
Tetracycline
- AMP:
-
Ampicillin
- SAM:
-
Sulbactam-ampicillin
- S:
-
Streptomycin
- AMC:
-
Amoxicillin/clavulanic acid
- AK:
-
Amikacin
- CIP:
-
Ciprofloxacin
- DO:
-
Doxycycline
- FOT:
-
Fosfomycin
- K:
-
Kanamycin
- C:
-
Chloramphenicol
- SXT:
-
Sulphamethoxazole-trimethoprim
- GN:
-
Gentamicin
- ATM:
-
Aztreonam
- SMRT:
-
Single-molecule real-time
- ICE:
-
Integrative and conjugative element
- ST:
-
Sequence type
- SNPs:
-
Single nucleotide polymorphisms
References
Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrobial Resistance. 2021;3:1.
Moffat J, Chalmers G, Reid-Smith R, Mulvey MR, Agunos A, Calvert J, et al. Resistance to extended-spectrum cephalosporins in Escherichia coli and other Enterobacterales from Canadian turkeys. PLoS ONE. 2020;15(9):e0236442.
Dantas Palmeira J, Ferreira HMN. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production – a threat around the world. Heliyon. 2020;6:e03206.
Silva N, Carvalho I, Currie C, Sousa M, Igrejas G, Poeta P. Extended-spectrum-β-lactamase and carbapenemase-producing enterobacteriaceae in food-producing animals in Europe: an impact on public health? In: Antibiotic Drug Resistance. 2019. p. 261–73.
Rocha FR, Pinto VPT, Barbosa FCB. The Spread of CTX-M-Type Extended-Spectrum β-Lactamases in Brazil: A Systematic Review. Microb Drug Resist. 2016;22:301–11.
Naseer U, Sundsfjord A. The CTX-M conundrum: Dissemination of plasmids and Escherichia coli clones. Microb Drug Resist. 2011;17:83–97.
Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72:2145–55.
Shoaib M, He Z, Geng X, Tang M, Hao R, Wang S, et al. The emergence of multi-drug resistant and virulence gene carrying Escherichia coli strains in the dairy environment: a rising threat to the environment, animal, and public health. Front Microbiol. 2023;14:1197579.
Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–32.
Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–75.
Leão C, Clemente L, Moura L, Seyfarth AM, Hansen IM, Hendriksen RS, et al. Emergence and Clonal Spread of CTX-M-65-Producing Escherichia coli From Retail Meat in Portugal. Front Microbiol. 2021;12:653595.
Lü Y, Kang H, Fan J. A novel blactx-m-65-harboring inchi2 plasmid pe648ctx-m-65 isolated from a clinical extensively-drug-resistant escherichia coli st648. Infect Drug Resist. 2020;13:3383–91.
Hu Y, He Y, Nguyen SV, Liu C, Liu C, Gan X, et al. Antimicrobial resistance of Salmonella Indiana from retail chickens in China and emergence of an mcr-1-harboring isolate with concurrent resistance to ciprofloxacin, cefotaxime, and colistin. Front Microbiol. 2022;13:1–13.
Zhang X, Fang C, Zhang J, Hua W, He R, Zhou M. Carbapenemase- and Colistin Resistant Escherichia coli Strains from Children in China: High Genetic Diversity and First Report of blaNDM-5, blaCTX-M-65, blaOXA-10, blaTEM-1, and mcr-1.1 Genes Co-Occurrence in E. coli ST156. Infect Drug Resist. 2022;15:5315–20.
Park H, Kim J, Ryu S, Jeon B. Predominance of bla CTX-M-65 and bla CTX-M-55 in extended-spectrum β-lactamase-producing Escherichia coli from raw retail chicken in South Korea. J Glob Antimicrob Resist. 2019;17:216–20.
Zheng H, Zeng Z, Chen S, Liu Y, Yao Q, Deng Y, et al. Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J Antimicrob Agents. 2012;39(4):305–10.
Li S, Zhao M, Liu J, Zhou Y, Miao Z. Prevalence and antibiotic resistance profiles of extended-spectrum β-lactamase-producing Escherichia coli isolated from healthy broilers in Shandong Province, China. J Food Prot. 2016;79:1169–73.
Batchelor M, Hopkins K, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH, et al. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother. 2005;49:1319–22.
Liu C, Qin S, Xu H, Xu L, Zhao D, Liu X, et al. New Delhi Metallo-β-Lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant enterobacter cloacae from Henan Province, China. PLoS ONE. 2015;10:e0135044.
Lim HJ, Lee EH, Yoon Y, Chua B, Son A. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J Appl Microbiol. 2016;120:379–87.
Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–10.
Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–52.
Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:e104984.
Wei X, Wang W, Lu N, Wu L, Dong Z, Li B, et al. Prevalence of Multidrug-Resistant CTX-M Extended Spectrum Beta-Lactamase-Producing Escherichia coli From Different Bovine Faeces in China. Front Vet Sci. 2022;9:1–12.
Anwar MA, Aqib AI, Ashfaq K, Deeba F, Khan MK, Khan SR, et al. Antimicrobial resistance modulation of MDR E. coli by antibiotic coated ZnO nanoparticles. Microb Pathog. 2020;148:104450.
Yang X, Liu W, Liu Y, Wang J, Lv L, Chen X, et al. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front Microbiol. 2014;5:688.
Jing W, ZhenLing Z, XinYi H, ZhenBao M, ZeWen G, LuChao L, et al. Evolution and comparative genomics of F33:A-:B- plasmids carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. mSphere. 2018;3:e00137-18.
Leung PHM, Yam WC, Ng WWS, Peiris JSM. The prevalence and characterization of verotoxin-producing Eschericha coli isolated from cattle and pigs in an abattoir in Hong Kong. Epidemiol Infect. 2001;126:173–9.
Vettorato MP, Leomil L, Guth BEC, Irino K, Pestana De Castro AF. Properties of Shiga toxin-producing Escherichia coli (STEC) isolates from sheep in the State of São Paulo, Brazil. Vet Microbiol. 2003;95:103–9.
Salverda MLM, de Visser JAGM, Barlow M. Natural evolution of TEM-1 β-lactamase: Experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34:1015–36.
Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: Origin and diffusion. Front Microbiol. 2012;3:110.
Nicolas E, Lambin M, Dandoy D, Galloy C, Nguyen N, Oger CA, et al. The Tn3-family of replicative transposons. In: Mobile DNA III. 2015. p. 693–726.
Lei CW, Chen YP, Kang ZZ, Kong LH, Wang HN. Characterization of a novel SXT/R391 integrative and conjugative element carrying cfr, bla CTX-M-65, fosA3, and aac(6=)-Ib-cr in Proteus mirabilis. Antimicrob Agents Chemother. 2018;62:e00849-18.
Acknowledgements
The authors thank the anonymous reviewers for their insightful suggestions and comments.
Funding
The funding bodies played no role in the design of the study and collection, analysis, interpretation of data, and in writing the manuscript. This work was supported by the research on the National Natural Science Foundation of China (grant number 32273068 & 31872520) and the earmarked fund for China Agriculture Research System (CARS) [grant number CARS-37].
Author information
Authors and Affiliations
Contributions
JY and WW conceived and designed this study. LY, QQ and CY performed the experiment. WW, Z and YB analyzed the data. XJ, S and Q interpreted the reference and provided the background. WW and JY wrote and revised the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
We clarify that in our study no permission was necessary to collect stool samples from dairy cows. All animal experiments were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of the People’s Republic of China, Policy No. 2006398).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, W., Wei, X., Zhu, Z. et al. Tn3-like structures co-harboring of blaCTX-M-65, blaTEM-1 and blaOXA-10 in the plasmids of two Escherichia coli ST1508 strains originating from dairy cattle in China. BMC Vet Res 19, 279 (2023). https://doi.org/10.1186/s12917-023-03847-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-023-03847-2