Abstract
Background
The incidence of human infection and colonization with methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) has increased in the recent years. Environmental sources, including bird droppings, might play an important role as resistance reservoirs.
Results
Fresh fecal samples were collected from rooks and wild-living birds during the autumn-winter period of 2016/2017, and tested for the presence of bacteria associated with human diseases. Besides bacteria representing the genera Enterococcus, Campylobacter, Escherichia, and Staphylococcus, Enterobacter, Citrobacter, Proteus, Hafnia, and Pseudomonas were also identified. The susceptibility of S. aureus and Enterococcus spp. isolates to methicillin, and vancomycin and teicoplanin, respectively, was analyzed to assess the avian wildlife as a reservoir of MRSA and VRE strains. Twenty-two percent of all S. aureus isolates were methicillin-resistant. These strains were screened by polymerase chain reaction (PCR), using the most widely used primer sets specific for the mecA gene. Twenty percent of all Enterococcus strains were phenotypically vancomycin-resistant. The presence of van resistance genes in these strains was investigated by PCR using vanA and vanB gene-specific primers. A good correlation between mecA gene detection and disc diffusion data was observed, while some discrepancy was noted between the PCR data and the vancomycin/teicoplanin phenotypic resistance pattern. The incidence of strains resistant to methicillin and glycopeptide antibiotics in wild-living birds was twice that in rooks.
Conclusions
The study suggests that rooks from urban areas and passerine birds from the natural habitat carry antibiotic-resistant Enterococcus spp. and S. aureus strains, probably reflecting the presence of such isolates in the environmental food sources.
Similar content being viewed by others
Background
Wild-living birds are reservoirs of potentially pathogenic bacteria. They may play an important role in the epidemiology of human diseases, either directly or indirectly, by contaminating foodstuff, water, dust, or other environmental sources [1]. The risk to public health is most widely reported based on analyses of large numbers of feral pigeons whose populations greatly increased in the last few decades in European cities [2].
Free-living bird species that are frequently found in the human environment are naturally infected with viruses, bacteria, fungi, and protozoa that are also pathogenic to human [3,4,5]. Wild-bird populations attracted to urbanized areas can be colonized with strains of Enterococcus spp. [6], Escherichia coli, Salmonella spp. [7,8,9], and Staphylococcus that are all resistant to antibiotics [4, 5, 10]. Enterococcus, Campylobacter, E. coli, and coliforms are the predominating bacteria in feces of wild-living birds [2]. Although antimicrobial drug resistance is quite common in poultry, it has been also observed in bacteria isolated from wild birds [4].
Corvids are considered to be important environmental reservoirs and vectors of antibiotic-resistant bacterial strains [7, 8]. The rook (Corvus frugilegus) is an omnivore, with a Palearctic distribution. This bird leaves the roosting place during the day to search for food, usually within 0.5–2 km from the breeding colony, or even 10–40 km from the winter roost [6, 11, 12]. Currently, the rook is one of the most numerous species found in agroecosystems. Recently published evaluations of the current rook population in Poland vary, ranging from 150,000 to 200,000 pairs or from 250,000 to 310,000 [13]. Rook, on account of its increasing abundance and mobility, also plays a role in the dissemination of pathogenic microorganisms [8].
Enterococcus, considered a typical commensal of the intestinal tract, is fast emerging as a pathogen that causes a variety of infections, including urinary tract infections, bacteremia, endocarditis, and meningitis. It spreads to patients in the hospital or is transmitted through the food supply. Enterococci are intrinsically resistant or tolerant to many antibiotics, and can acquire drug resistance either via integrons, plasmid transfer, or transposon acquisition [14,15,16].
The first-line treatment of choice for systemic enterococcal infections are glycopeptide antibiotics, mainly vancomycin (Van) and teicoplanin (Tei). Enterococcus faecalis and E. faecium are most frequently described as etiological agents of nosocomial infections caused by vancomycin-resistant enterococci (VRE) [14]. Until now, nine distinct vancomycin resistance gene clusters have been described in enterococci (vanA, vanB, vanC, vanD, vanE, vanG, vanL, vanM, and vanN) [17]. The most predominant phenotype is VanA, characterized by high-level resistance to both vancomycin and teicoplanin. VanB-type phenotype is commonly observed among clinical isolates and in fecal strains of domestic animals. VanB strains are resistant to various concentrations of vancomycin but are susceptible to teicoplanin [14, 18].
VRE intrinsically resistant to low levels of vancomycin and containing the vanC gene, such as Enterococcus gallinarum, E. casseliflavus, and E. flavescens, have been isolated from wild bird feces [17] and from a poultry farm [19]. The presence of VRE and/or high-level aminoglycoside-resistant enterococci in wild small mammals, rabbits, and birds has been documented [6, 10, 20]. Further, the same variants of the vanA gene cluster associated with Tn1546 [21] or located on a transferable broad host-range plasmid [15] have been detected in enterococci of both human and animal origin. This may indicate that resistance genes can spread among isolates from different sources [22].
The mecA gene is the major determinant of methicillin resistance in staphylococci, and is found in both Staphylococcus aureus and coagulase-negative staphylococci [23]. MRSA has been isolated from birds, including poultry, pigeons, parrots, and wild birds [4].
The aim of the current study was to evaluate and compare the presence of potentially pathogenic bacteria in the feces of rooks feeding in public sites and wild birds inhabiting places far from the urban areas, to verify whether feeding municipal food waste increases the occurrence of bacteria that could be harmful to human health. The fecal bacterial flora of rooks feeding in Lublin and the surrounding region, and Kraków (urban areas) was compared with that of wild birds that were caught and ringed in the Vistula river valley (outside urban areas). The identified potential pathogens Enterococcus spp. and S. aureus were tested for antibiotic resistance. Using antibiotic susceptibility assays, the occurrence of the mecA gene in S. aureus, and the vanA and vanB genes in enterococcal isolates from rooks and wild-living birds was determined.
Results
Locations of the sample collection sites and isolate culture analysis
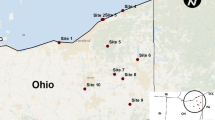
Samples collected from rooks and wild-living migratory birds from urban areas and the Vistula valley, respectively, were analyzed using bacteriological approaches. Overall, 190 samples were obtained: 163 from rooks (five locations) and 27 samples from migratory birds representing six genera (one location) (Table 1).
The results of microbiological examination and isolate identification in fecal samples from the specified areas are summarized in Table 2. The commonly identified genera Enterococcus spp., Campylobacter spp., E. coli, and S. aureus accounted for 55% (90/163), 57% (93/163), 56.4% (92/163), and 28.2% (46/163), respectively, of all isolates from rook feces; and 81.5% (22/27), 37% (10/27), 85% (23/27), and 29.6% (8/27), respectively, of all isolates from cloacal swabs of wild birds. The overall frequency of these genera suggested that they are normal constituents of the intestinal flora of the tested bird species. Except for the similar frequency of isolation of S. aureus from rooks and wild-living birds (28.2% vs. 29.6%, respectively), the differences in the relative abundance of dominant genera indicated host-dependence of the intestinal microflora composition and the effect of the bird feeding habits. Four species of Enterococcus (E. faecalis, E. faecium, E. casseliflavus, and E. raffinosus) were distinguished by biochemical tests (EN—COCCUStest).
Antimicrobial susceptibility testing and detection of methicillin and vancomycin resistance genes
Methicillin disc diffusion and oxacillin agar tests revealed that 19.5% (9/46) of S. aureus isolates from rook samples and 37.5% (3/8) from wild-living birds were resistant to methicillin. The incidence of MRSA isolation from rook samples was mainly related to locations B and D [8.3% (2/24) and 8% (2/25), respectively] (Tables 2 and 3). MRSA strains were isolated from the spotted flycatcher, robin, and common blackbird.
Enterococcus strains resistant to vancomycin and teicoplanin represented E. faecalis (10 isolates) and E. faecium (13 isolates). Isolates of the other two species, E. casseliflavus and E. raffinosus, were sensitive to both glycopeptides. The minimal inhibitory values (MIC) of Van and Tei were determined (Table 4). The analysis confirmed the results of the disc diffusion test. According to the Clinical Laboratory Standards Institute (CLSI) criteria, six strains isolated from rooks and six strains isolated from wild birds exhibited the VanA phenotype (MIC ranges: Van ≥256 μg/ml and Tei 16–256 μg/ml). Of 23 VRE strains tested, five strains from rooks and one from a wild bird displayed the VanB phenotype (MIC ranges: Van > 256 μg/ml and Tei 1–4 μg/ml). Four strains exhibited intermediate susceptibility to vancomycin (MIC range 8–16 μg/ml) and sensitivity to teicoplanin (MIC 1–8 μg/ml); three of these strains were isolated from rooks (Table 4). Overall, combined vancomycin and teicoplanin resistance was more frequent among enterococcal isolates from wild-living birds than those from rooks. The VanA phenotype was predominant (2:1) in rooks feeding in the vicinity of a communal waste dump (location B) compared with those feeding in the municipal parks (locations A and C–E), (Table 1).
The primers used for polymerase chain reaction (PCR)-based detection of specific antibiotic resistance genes in S. aureus and Enterococcus spp. isolates resistant to methicillin or glycopeptides, respectively, are listed in Table 5. Using two primer pairs (MecA1/2 and MecA3/4), the presence of mecA gene was confirmed in all MRSA rook isolates (9) and wild-living bird isolates (3). Either 310-bp or 527-bp amplicons, or both, were detected by PCR (Table 3). Among the 12 MRSA isolates, both amplicons were detected in six isolates; only the small amplicon was detected in four isolates; and only the large amplicon was detected in two isolates. In general, there was a good agreement between the PCR-based mecA gene detection and the results of the disc agar diffusion test. Since the approach detected both mecA gene products in half of the isolates screened, the PCR analysis should be performed using at least two primer sets for MRSA confirmation.
The results of PCR used to screen glycopeptide-resistant isolates are shown in Table 4. Eight isolates tested positive for the vanA gene only (732-bp product); eight isolates tested positive for the vanB gene only (647-bp product); and four isolates displaying the VanA susceptibility pattern tested positive in both reactions. The vanA genotype was more prevalent in samples from wild-living birds. PCR products indicative of the presence of the vanA or vanB genes were detected in three different species of wild-living birds: the spotted flycatcher, common chiffchaff, and thrush nightingale. In three strains (E. faecium) exhibiting the intermediate level of resistance to vancomycin (MIC 8–16 μg/ml) and susceptibility to teicoplanin (MIC 1–8 μg/ml), neither vanA nor vanB gene products were detected.
Discussion
In the current study, we reported the isolation and characterization of bacteria from the feces of urban rooks and cloacal swabs of wild-living bird. From both rooks and wild-living birds, methicillin- and glycopeptide-resistant strains of S. aureus and Enterococcus spp., respectively, were isolated.
In Poland, in the years 2000–2014, a moderate decline in the rook breeding population was observed; however, in the eastern part of country, the rook population was regarded as stable or increasing [13]. The rooks frequently roam in the vicinity of inhabited areas, recreation areas, and parks, thus coming in close contact with human and animals. In fact, environmental (air and water) pollution by rook feces was reported [2, 26, 27]. Infection caused by feces of wild birds can be contracted in a number of ways: by direct contact or inhalation, or by food and water contamination. Although the risk of contracting an infection from bird droppings is relatively low, diseases caused by antibiotic-resistant staphylococci and enterococci may be severe. In the current study, four bacterial genera (Enterococcus spp., Campylobacter spp., E. coli, and S. aureus) were commonly identified in rook feces collected at five different localities and in samples from wild-living birds from one region.
The occurrence of bacteria and fungi in urban rook populations in Zagreb (Croatia) during the breeding period in 2010 was described by Vlahović et al. [26]. Microbiological examination of fresh fecal samples revealed the presence of E. coli, Bacillus spp., Staphylococcus spp., Streptococcus spp., Agrobacterium radiobacter, and Acinetobacter spp. [3, 26, 28]. In the current study, no Salmonella isolates were detected, although S. Enteritidis was found in the feces of rooks wintering in Czechia in another study [7]. Further, one study of Salmonella prevalence in free-living birds in southern Poland reported no Salmonella isolation from the feces or tissue of Corvus cornix; by contrast, 16.6% of samples from C. frugilegus were positive [29].
The high frequency of detection of Campylobacter species (57% of all rook isolates) noted herein was not described in other reports. For example, in a study conducted in Zagreb, only one rook sample out of 57 fecal samples was positive for C. jejuni [26]. Nonetheless, other studies suggest that C. jejuni may be a typical component of the intestinal flora of at least some bird species [4]. Campylobacter spp. that could potentially be transmitted to human have been isolated from migrating ducks, pigeons, passerine birds, and crows (Corvus) [1]. Phylogenetic analyses of free-ranging American crows confirmed the notion that some C. jejuni strains potentially associated with disease might be shared by crows, human, poultry, and livestock [30].
Rooks could be important vectors for the dissemination of antibiotic-resistant enterococci [6] and MRSA [28] in Europe because they are scavengers and cover relatively long distances. Corvids and gulls that feed on garbage in urbanized areas are also considered to be environmental reservoirs and vectors of resistant isolates [8]. In the current study, we noted a relatively high frequency of isolation of Enterococcus spp. in rook samples (55%) and in wild-living birds (81.5%) in the examined localities in south-east Poland (Table 2).
The prevalence of S. aureus in fecal samples from rooks and wild birds was comparable (Table 2), but the average incidence of methicillin-resistant isolates was two times higher among isolates from wild-living birds than those from rooks (Table 3). Loncaric et al. [28] observed significant difference in the occurrence of MRSA isolates between migratory population compared with resident population of C. frugilegus from eastern Europe.
In the current study, glycopeptide-resistant strains were detected relatively more frequently in samples from rook feces collected in the vicinity of communal waste dumps (location B) than in city (locations A and E) or village (locations C and D) parks. Samples from wild-living birds contained two times more glycopeptide-resistant enterococci than fecal samples from rooks (36.4% vs. 16.6%, respectively). Since it was not possible to obtain the feces of wild bird species, the only available material was cloacal swabs. It is possible that the sampling site may have slightly affected the final results. The prevalence of VRE in rooks in individual countries may differ significantly, with Poland (14% in Gdynia and 8% in Jaroslaw) and Czechia (10%) as countries with the most frequent VRE isolation [6].
The results of the disc agar diffusion test and PCR determination of the presence of resistance genes were in good agreement. Specifically, the presence of the mecA gene was confirmed in all phenotypically resistant S. aureus isolates, whereas the vanA and/or vanB genes were identified in 83% of phenotypically vancomycin- and teicoplanin-resistant, or intermediate-resistant Enterococcus spp. isolates. In the PCR analysis, most strains yielded only one amplicon, which was consistent with the phenotypic resistance pattern. The amplicons from both, vanA and vanB genes, in strains with the VanA resistance phenotype were mainly detected in enterococci from wild-living birds. The amplicons from these two genes were also detected in VanA phenotypically resistant clinical isolates of E. faecium and E. faecalis [31], and in VanB phenotype E. faecium [32].
Examination of VRE presence in fecal samples of rooks wintering in Europe resulted in the identification of 6% of 1073 enterococci growing on a selective medium as E. faecium, of which 12.5% harbored the vanA and ermB (macrolide resistance) genes [6]. Enterococci with plasmid-encoded glycopeptide-resistance genes vanA, vanB, and vanM are reservoirs for the transmission of these genes by conjugation to other enterococci and to other Gram-positive cocci, including S. aureus [6, 33, 34].
Based on the findings of the current study, bacterial colonization of the host bird is associated with its feeding and breeding place. The obtained data may be used to map the dissemination of antibiotic-resistant strains by urban rooks and other wild birds, and to track the possible consequences of transferring the resistance determinants to other animals or to human.
Conclusions
Bacteria from the genera Campylobacter, Enterococcus, Escherichia, and Staphylococcus are predominant in the intestinal flora of rooks and in eight species of other wild-living migratory birds. Other genera, e.g., Enterobacter, Citrobacter, Hafnia, Cronobacter, Kluyvera, Proteus, Morganella, and Pseudomonas, are identified much less frequently. Among S. aureus isolates, 19.6% from rooks and 37.5% from wild-living birds were resistant to methicillin. Enterococcus isolates carrying van resistance genes were identified with a frequency of 14.4% (13/90) and 31.8% (7/22) in samples from rooks and wild birds, respectively. The overall frequency of isolation of strains resistant to methicillin and glycopeptide antibiotics in wild-living bird samples was approximately two times higher than that in rook samples.
Methods
Bird sampling
To obtain rook samples, swabs were taken from the top surface of fresh feces. Cloacal swabs were collected from eight different species of wild-living migratory birds of the order Passeriformes at the time of ringing, between 1 September 2016 and 31 October 2017.
The ornithologist ringed the birds with the consent of the General Directorate of Environmental Protection, Poland (nos. 291/2016 and 291/2017) in accordance with decisions DZP-WG.6401.03.98.2016.km, DZP-WG.6401.03.97.2017.jro, and Minister of Environment DLP-VIII-6713-21/29762/14/RN. In accordance with the Polish law, the Act of January 15th 2015 on the Protection of Animals Used for Scientific or Educational Purposes (Dz.U.2015.266), fecal and cloacal sampling does not require the consent of the local ethics committee (Article 1.2, point 5).
The samples (n = 190) were collected at six localities in south-east Poland (Table 1). Samples were collected using Amies clear gel collection and transport swabs (BioMerieux, Marcy-l’Etoile, France), and stored on ice (4–7 °C) for 1–7 h prior to culture. Individual samples were placed overnight in buffered peptone water (Biomaxima, Gdańsk, Poland) at 35–37 °C, and then inoculated onto selective media.
Hektoen enteric agar, MacConkey agar, and triple sugar iron agar (BioMerieux) were used for the recovery of Gram-negative gastrointestinal pathogens (Salmonella and Shigella) and other intestinal genera (E. coli, Klebsiella, and Enterobacter). All isolates were processed using API 20E strips (BioMerieux), following the manufacturer’s instructions. Supplementary investigation of cytochrome oxidase was performed using Oxidase Test (Merck, Darmstadt, Germany). β-glucuronidase test with the substrate 4-methylumbelliferyl-beta-d-glucuronide (BD Difco), Rapid Enterococci ChromoSelect agar (Sigma, St Louis, MO), Enterococcus-selective agar (Slanetz-Bartley) (BTL, Łódź, Poland), bile-esculin agar (Merck), and EN-COCCUStest (Pliva-Lachema, Brno, Czechia) based on acid production from sorbitol, l-arabinose, mannitol, sorbitol, mellibiose, raffinose, melezitose, and production of arginine dihydrolase, to isolate and identify enterococci. Further, mannitol salt agar (BTL) was used for the isolation of pathogenic staphylococci. S. aureus isolates were identified using colony morphology, catalase test, API Staph biochemical tests (BioMerieux), the ability to coagulate rabbit plasma, and based on clumping factor production. Campylosel agar (BioMerieux), a selective medium for the isolation of intestinal Campylobacter (mainly C. jejuni and C. coli) from stools, was used. Campylobacter isolation was performed following standard conditions, after a 24-h enrichment period at 42 °C under micro-aerobic conditions in a CampyPak Plus microaerophilic system with a palladium catalyst (BD Diagnostic Systems, Sparks, MD).
Antibiotic susceptibility testing
Susceptibility to antibiotics was tested on Mueller Hinton agar (Biomaxima) by using the Kirby-Bauer disc diffusion method, and commercially prepared discs of 30 μg vancomycin, 30 μg teicoplanin, and 5 μg methicillin (Liofilchem, Italy), according to CLSI guidelines [35]. Plates were incubated at 35 °C for 24 h. S. aureus strains identified as methicillin resistant by disc diffusion assay were then tested by oxacillin agar screening. Specifically, a colony suspension was spotted on MH agar supplemented with 4% NaCl and oxacillin at 6 μg/ml (Sigma), according to CLSI recommendations. After incubation for 24 h, any growth was interpreted as a positive result for oxacillin resistance [36]. MIC values for vancomycin and teicoplanin (Sigma) were tested by broth microdilution method, according to CLSI guidelines [37]. The final concentrations of vancomycin and teicoplanin ranged from 0.125 to 256 μg/ml.
Primers
The mecA gene was PCR-amplified using primers MecA1 and MecA2 [24], and MecA3 and MecA4 [25]. The vanA gene was PCR-amplified using primers VanA(+) and VanA(−). The vanB gene was PCR-amplified using primers VanB(+) and VanB(−) [18] (Table 5).
PCR
The presence of the mecA gene, and vanA and vanB genes was determined by PCR in MRSA and Enterococcus spp. isolates exhibiting phenotypical vancomycin resistance, respectively. The template DNA was isolated and purified using the Wizard genomic DNA purification kit (Promega, Madison, WI) supplemented with either lysostaphin (5 mg/ml; Sigma), for MRSA, or lysozyme (10 mg/ml, Sigma), for enterococci, as described elsewhere [18]. PCR was performed in a 25-μl reaction mixture containing template DNA (10–200 ng), 200 μg of each dNTP, 1.5 mM MgCl2, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 200 nM (each) primer, and 2.5 U of Taq polymerase (Sigma). Thermal cycling for vanA detection was conducted using the following program: an initial denaturation step at 96 °C for 180 s; followed by 35 cycles of 94 °C for 45 s, 46 °C for 45 s, and 72 °C for 60 s; and a final extension step at 72 °C for 360 s. The vanB and mecA genes were amplified using the same protocol, except that the annealing temperature used was 50 °C and 54 °C, respectively. PCR products were visualized on 1% agarose gel.
The following strains were used as positive and negative controls: S. aureus ATCC 43300, methicillin R, mecA gene positive control; S. aureus ATCC 25923, methicillin and vancomycin S, negative control; E. faecium ATCC 51559, vancomycin and teicoplanin R, vanA gene positive control; E. faecalis ATCC 51299, vancomycin R and teicoplanin S, vanB gene positive control; E. faecalis ATCC 29212, vancomycin S, negative control.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CLSI:
-
Clinical and Laboratory Standards Institute
- MH:
-
Mueller Hinton
- MIC:
-
Minimal inhibitory concentration
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- PCR:
-
Polymerase chain reaction
- Tei:
-
Teicoplanin
- Van:
-
Vancomycin
- VRE:
-
Vancomycin-resistant enterococcus
References
Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, Falagas ME. Human infections associated with wild birds. J Inf Secur. 2008;56:83–98.
Casanovas L, de Simon M, Ferrer MD, Arques J, Monzon G. Intestinal carriage of campylobacters, salmonellas, yersinias and listerias in pigeons in the city of Barcelona. J Appl Bacteriol. 1995;78:11–3.
Vlahović K, Matica B, Bata I, Pavlak M, Pavičić Ž, Popović M, Nejedli S, Dovč A. Campylobacter, Salmonella and Chlamydia in free-living birds of Croatia. Eur J Wildl Res. 2004;50:127–32.
Benskin CM, Wilson K, Jones K, Hartle IR. Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biol Rev Camb Philos Soc. 2009;84:349–73.
Wang J, Ma ZB, Zeng ZL, Yang XW, Huang Y, Liu JH. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool Res. 2017;38:55–80.
Oravcova V, Ghosh A, Zurek L, Bardon J, Guenther S, Cižek A, Literák I. Vancomycin-resistant enterococci in rooks (Corvus frugilegus) wintering throughout Europe. Environ Microbiol. 2013;15:548–56.
Literák I, Vanko R, Dolejska M, Cižek A, Karpýškova R. Antibiotic resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett Appl Microbiol. 2007;45:616–21.
Literák I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, Bzoma S, Cizek A. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea coast of Poland. Appl Environ Microbiol. 2010;276:8126–34.
Jamborova I, Dolejska M, Vojtech J, Guenther S, Uricariu R, Drozdowska J, Papousek I, Pasekova K, Meissner W, Hordowski J, Cizek A, Literak I, Nojiri H. Plasmid-mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Appl Environ Microbiol. 2015;81:648–57.
Silva N, Igrejas G, Rodrigues P, Rodrigues T, Gonçalves A, Felgar AC, Pacheco R, Gonçalves D, Cunha R, Poeta P. Molecular characterization of vancomycin-resistant enterococci and extended spectrum β-lactamase-containing Escherichia coli isolates in wild birds from the Azores archipelago. Avian Pathol. 2011;40:473–9.
Griffin L, Thomas C. The spatial distribution and size of rook (Corvus frugilegus) breeding colonies is affected by both the distribution of foraging habitat and by intercolony competition. Proc R Soc Lond B. 2000;267:1463–7.
Kitowski I, Sándor AD, Czarnecka J, Grzywaczewski G. Diet of rooks Corvus frugilegus and potential seed dispersal in urban and agricultural habitats of Romania and Poland. North-West J Zool. 2017:94–100.
Chmielewski A, Dombrowski A, Jabłoński P, Łukaszewicz M, Nicewicz Ł, Trębicki Ł, Pagórski P, Tabor J. Breeding population of the rook Corvus frugilegus in the Mazovian lowland: current status and changes. Intern Stud Sparrows. 2017;41:4–21.
Bhatt P, Sahni AK, Praharaj AK, Grover N, Kumar M, Chaudhari CN. Khajuria a detection of glycopeptide resistance genes in enterococci by multiplex PCR. Med J Armed Forces India. 2015;71:43–7.
Sivertsen A, Pedersen T, Larssen KW, Bergh K, Rønning TG, Radtke A, Hegstadad K. A silenced vanA gene cluster on a transferable plasmid caused an outbreak of vancomycin-variable enterococci. Antimicrob Agents Chemother. 2016;60:4119–27.
Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, Chen D, Bian H, Li Y, Guangchao YG. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14:45.
Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti-Infect Ther. 2015;12:1221–36.
Depardieu F, Perichon B, Courvalin P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J Clin Microbiol. 2004;42:5857–60.
Harwood VJ, Brownell M, Perusek W, Whitlock JE. Vancomycin-resistant Enterococcus spp. isolated from wastewater and chicken feces in the United States. Appl Environ Microbiol. 2001;67:4930–3.
Heuer OE, Hammerum AM, Collignon P, Wegener HC. Human health hazard from antimicrobial-resistant enterococci in animals and food. Clin Infect Dis. 2006;43:911–6.
Hammerum AH. Enterococci of animal origin and their significance. Clin Microbiol Infect. 2012;18:619–25.
Lozano C, Gonzalez-Barrio D, Camacho MC, Lima-Barbero JF, de la Puente J, Höfle U, Torres C. Characterization of fecal vancomycin-resistant enterococci with acquired and intrinsic resistance mechanisms in wild animals. Spain Microb Ecol. 2016;72:813–20.
Shrestha NK, Tuohy MJ, Hall GS, Isada CM, Procop GW. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J Clin Microbiol. 2002;40:2659–61.
Ramos-Trujillo E, Pérez-Roth E, Méndez-Alvarez S, Claverie-Martín F. Multiplex PCR for simultaneous detection of enterococcal genes vanA and vanB and staphylococcal genes mecA, ileS-2 and femB. Int Microbiol. 2003;6:113–5.
Poulsen AB, Skov R, Pallesen LV. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA detection kit. J Antimicrob Chemoth. 2003;51:419–21.
Vlahović K, Prukner-Radovčić E, Horvatek D, Pavlak M, Gomerčić T, Rumiha Z, Dovč A. Bacterial and fungal flora in faecal samples from rooks (Corvus frugilegus) in the City of Zagreb. Croatia Veterinarski Arhiv. 2010;80:81–92.
Hubálek Z. An annotated checklist of pathogenic microorganisms associated with migratory birds. J Wildl Dis. 2004;40:639–59.
Loncaric I, Stalder GL, Mehinagic K, Rosengarten R, Hoelzl F, Knauer F, Walzer C. Comparison of ESBL- and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS One. 2013;8:e84048. https://doi.org/10.1371/journal.pone.0084048.
Krawiec M, Kuczkowski M, Kruszewicz AG, Wieliczko A. Prevalence and genetic characteristics of Salmonella in free-living birds in Poland. BMC Vet Res. 2015;31:11–5.
Weis AM, Miller WA, Byrne BA, Chouicha N, Walter M, Boyce WM, Townsend AK. Prevalence and pathogenic potential of Campylobacter isolates from free-living, human-commensal american crows. Appl Environ Microbiol. 2014;80:1639–44.
Mirzaei B, Babaei R, Asiabar APD, Bameri Z. Detection of both vanA & vanB genes in vanA phenotypes of Enterococci by Taq man RT-PCR. Braz J Microbiol. 2015;46:161–5.
Papagiannitsis CC, Malli E, Florou Z, Medvecky M, Sarrou S, Hrabak J, Petinaki E. First description in Europe of the emergence of Enterococcus faecium ST117 carrying both vanA and vanB genes, isolated in Greece. J Glob Antimicrob Resist. 2017;11:68–70.
Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–54.
de Niederhäusern S, Bondi M, Messi P, Iseppi R, Sabia C, Manicardi G, Anacarso I. Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr Microbiol. 2011;62:1363–7.
Clinical Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk susceptibility tests; approved standard – 11th ed CLSI document M02-A11. Wayne, Pennsylvania, USA.
Clinical and Laboratory Standards Institute, editor. Performance standards for antimicrobial susceptibility testing; M100S. 28th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 19th informational supplement M100-S19. Wayne, PA: CLSI; 2009.
Acknowledgments
Not applicable.
Funding
The authors received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
AT-S performed molecular biological analyses and participated in writing the paper. MK contributed to the study design and collected the samples. HK collected the samples and analyzed the data. JZ performed the microbiological analyses. JK is the main researcher, who performed the microbiological and molecular biological analyses, and wrote the paper. TU-S was responsible for scientific coordination of the research and data analysis, and participated in writing the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Cloacal swabs were collected from wild-living migratory birds at the time of ringing. The ornithologist ringed the birds with the consent of the General Directorate of Environmental Protection, Poland (nos. 291/2016 and 291/2017) in accordance with decisions DZP-WG.6401.03.98.2016.km, DZP-WG.6401.03.97.2017.jro and Minister of Environment DLP-VIII-6713-21/29762/14/RN. In accordance with the Polish law the Act of January 15th, 2015 on the Protection of Animals Used for Scientific or Educational Purposes (Dz.U.2015.266) fecal and cloacal sampling does not require the consent of the local ethics committee (Article 1.2, point 5).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kutkowska, J., Turska-Szewczuk, A., Kucharczyk, M. et al. Methicillin-resistant Staphylococcus aureus and glycopeptide-resistant enterococci in fecal samples of birds from South-Eastern Poland. BMC Vet Res 15, 472 (2019). https://doi.org/10.1186/s12917-019-2221-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-019-2221-1