Abstract
Background
Hemotropic mycoplasmas (hemoplasmas) are emerging zoonotic pathogens with a worldwide distribution that can cause mild to severe hemolytic anemia, icterus, ill-thrift, infertility, and poor weight gain. However, understanding of the molecular epidemiology of hemoplasmas (Mycoplasma ovis and ‘Candidatus Mycoplasma haemovis’) is limited in sheep and goats, and the hemoplasma strain/species/variant ‘Candidatus M. haemovis’ was poorly studied throughout the world and had never been detected in China until now. Thus, the aim of the present study was to determine the molecular prevalence of hemoplasmas, including M. ovis and ‘Candidatus M. haemovis’ in sheep and goats from seven provinces and one autonomous region of China.
Methods
A total of 1364 blood samples were collected from sheep and goats in seven provinces and one autonomous region of China. All blood samples were tested for hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) by nested PCR amplification based on 16S rRNA gene. Positive specimens underwent nucleotide sequencing and phylogenetic analysis.
Results
Overall, 610 specimens (44.7%, 610/1364) were shown to be hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) -positive by nested PCR amplification based on 16S rRNA gene. The prevalence in goats was 44.1% (379/860), and 45.8% (231/504) in sheep, while that in grazing small ruminants was 54.4% (396/728) and 33.6% (214/636) in house feeding small ruminants. Sequencing of the nearly complete 16S rRNA gene was successful for the 103 randomly selected positive specimens from different farms in different sampling sites of China. Among them, analysis of the 16S rRNA gene sequences identified M. ovis (n = 56) and ‘Candidatus M. haemovis’ (n = 47). Two (KU983740 and KU983746) of the four novel genotypes obtained in this study were closely related to M. ovis, while the other two genotypes (KU983748 and KU983749) had high identity with ‘Candidatus M. haemovis’. Remarkably, the genotype (KU983740) of M. ovis in sheep and goats in this study fell in a clade with two human hemoplasmas from USA (KF313922 and GU230144) and shared 99.8%–99.9% with them.
Conclusions
In this study, ‘Candidatus M. haemovis’ was first detected in Chinese sheep and goats and hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) are highly prevalent, and widely distributed in China.
Similar content being viewed by others
Background
Hemotropic mycoplasmas (hemoplasmas, formerly classified as Haemobartonella and Eperythrozoon spp.) are uncultivated, small, pleomorphic, wall-less bacteria that parasitize on the surface of animal erythrocytes [1,2,3]. Hemoplasmas have now been reclassified into the Mycoplasma genus based on 16S rRNA gene sequence. These pathogens can cause mild to severe hemolytic anemia, icterus, ill-thrift, infertility, and poor weight gain, but death is rare in infected adults [1, 4,5,6]. Worldwide, hemoplasmas have been reported to affect livestock [2, 7,8,9,10], companion animals [11,12,13,14,15,16,17], wildlife [5, 18, 19], and humans [20,21,22,23].
Mycoplasma ovis (formerly Eperythrozoon ovis), which could elicit major health problems and high mortality in lambs, has been identified in sheep and goats [8, 24,25,26]. In 2009, a novel hemoplasma, a 17 bp long deletion and an overall the 16S RNA gene sequence identity of only 97% compared to M. ovis, has been detected in Hungary. Subsequent studies identified this new hemoplasma as ‘Candidatus M. haemovis’ [24]. Subsequently, complete genome sequencing conducted by Deshuillers et al. revealed that M. ovis strain Michigan contains two copies of 16S rRNA gene, one corresponding to M. ovis, and the other to ‘Candidatus M. haemovis’ [27]. Therefore, Deshuillers et al. and other researchers can speculate that M. ovis and ‘Candidatus M. haemovis’ may exist as a single species with two copies of 16S rRNA gene or as two different species [27].
With an increasing number of hemoplasmosis clinical cases, many countries have suffered extensive losses of livestock. However, few epidemiological surveys on hemoplasmosis in sheep and goats have been conducted during the last decade. The prevalence of M. ovis was 6.3% (36/573) in small ruminants in North Africa [28], 20% (4/20) in goats in Switzerland [25], 87% (27/31) in captive cervids in Brazil [26] and 26.3% (5/19) in free-living Japanese serows [18]. Surprisingly, a 49-year-old veterinarian from Texas of USA was found to be co-infected by two M. ovis variants, M. ovis and ‘Candidatus M. haemovis’. In China, M. ovis was found in 41.0% (151/371) and 16.1% (192/1191) in small ruminants from Hubei province and Chongqing city, respectively [29, 30]. However, understanding of the molecular epidemiology of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) is limited in sheep and goats. And ‘Candidatus M. haemovis’ was poorly studied throughout the world and had never been detected in China until now. Thus, the aim of the present study was to determine the molecular prevalence of hemoplasmas, including M. ovis and ‘Candidatus M. haemovis’ in sheep and goats from seven provinces and one autonomous region of China.
Methods
Specimen collection
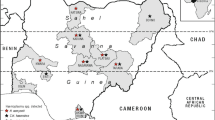
A total of 1364 EDTA-anticoagulated blood samples were collected from jugular vein of sheep and goats in Henan, Guizhou, Shanxi, Shaanxi, Yunnan, Qinghai, Heilongjiang provinces and the Inner Mongolia autonomous region from March 2012 to May 2015 (Fig. 1 and Additional file 1: Figure S1).
For each animal, one blood sample was collected into an EDTA tube marked with breed, gender, age, and feeding habit of the animal. The animals have no obvious clinical symptoms during blood sampling. The specimens were then transported in an ice box to the laboratory for further processing and analysis.
DNA extraction
For each specimen, DNA was extracted from 500 μl of anticoagulated whole blood using a Genome DNA Extraction Kit (LifeFeng, Shanghai, China) according to the manufacturer’s instructions. DNA was eluted from the column with 200 μl Tris-EDTA buffer. After DNA extraction, DNA concentration and quality was measured using absorbance ratio between 260/280 nm (Nanodrop, Thermo Scientific, USA). And all the specimens were subjected to a GAPDH gene conventional PCR assay as described previously to confirm the presence of amplifiable DNA and exclude PCR inhibition [31]. DNA was stored at −20 °C until PCR amplification.
Nested PCR
All samples were tested at least three times for hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) DNA using a nested PCR assay that amplified an approximately 500 bp fragment of the 16S rRNA gene [32]. The base sequences of four primers (A1/A2 and B1/B2) are shown in Table 1 and the binding sites of four primers are shown in Additional file 1: Figure S2. First-round PCR was performed in a total volume of 25 μl, containing 2.5 μl of 10 × LA PCR Buffer II (Mg2+ plus), 4.0 μl of dNTP mixture (2.5 mM each dNTP), 0.5 μl of each A1 and A2 primer (25 μM), 1.25 U of LA Taq polymerase (TaKaRa, Dalian, China), 16.25 μl of double distilled water, and 1.0 μl of DNA template. Cycling conditions were: 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 61 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 7 min. Second-round nested PCR contained 2.5 μl of 10 × PCR Buffer, 2.0 μl of dNTP mixture (2.5 mM each dNTP), 0.5 μl of each B1 and B2 primer (25 μM), 0.75 U of r Taq polymerase (TaKaRa), 18.35 μl of double distilled water, and 1.0 μl of the first-round PCR product (diluted 10-fold). Cycling conditions were: 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 45 s, with a final extension at 72 °C for 7 min. A positive control for M. ovis from a naturally infected Boer goat preserved by our laboratory was used in all reactions, while double distilled water was used as a negative control. Secondary PCR products were examined by 1% agarose gel electrophoresis and visualized after GelRed (Biotium Inc., Hayward, CA) staining.
PCR for 16S rRNA gene and sequencing
One hundred three positive specimens representative of different hosts, farms/flocks and geographic locations were selected and the nearly complete 16S rRNA gene sequences were amplified using the protocol described by Grazziotin et al. [26]. Primers are shown in Table 1. The 25 μl PCR mixture is the same as first-round PCR mixture of the nested PCR. PCR products were purified using Montage PCR filters (Millipore, Bedford, MA) and sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 3730 DNA analyzer (Applied Biosystems). The nucleotide sequences were confirmed by bidirectional sequencing and by sequencing a new PCR product if necessary.
Phylogenetic analysis
Nucleotide sequences of the nearly complete 16S rRNA gene together with reference sequences downloaded from GenBank were aligned using ClustalX 2.0 (http://www.clustal.org/) with further adjustments made manually as necessary. And then Mega 5.05 (http://www.megasoftware.net/) software was applied to conduct phylogenetic and molecular evolutionary analysis. A bootstrap phylogenetic tree demonstrating the relationship of M. ovis and ‘Candidatus M. haemovis’ genotypes to other hemoplasma species was created by the neighbor-joining method using a distance matrix corrected for nucleotide substitutions based on the Kimura 2-parameter model. A bootstrap analysis was used to assess the robustness of the clusters using 1000 replicates.
Statistical analysis
Positive rates were compared using the chi-square test. Differences were considered statistically significant at P < 0.05. Analyses were performed using QuickCalcs software (GraphPad Software Inc., La Jolla, CA). The odds ratios (ORs) of the univariate analysis were calculated using measures of association along with 95% confidence intervals (CIs).
Nucleotide sequence accession numbers
The four novel sequences determined in this study were deposited in GenBank under accession numbers: KU983740, KU983746, KU983748 and KU983749.
Results
Molecular prevalence of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) in sheep and goats
As shown in Fig. 1, of the 1364 blood specimens from sheep and goats, 610 (44.7%) were found to be positive for hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) by PCR amplification of the 16S rRNA gene. And the positive samples distributed in 49 studied farms from seven provinces and one Aut. Reg.. In the 35 sheep and goats from Heilongjiang province, 34 (97.1%) were identified as hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) -positive, while just two (5.0%) of the 40 animals in the Inner Mongolia Aut. Reg. was positive. On the other hand, the animals in three farms, including one in Henan province and two from Yunnan province were 100% positive for hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) (Additional file 1: Table S1).
The infection rates of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) in sheep and goats in Heilongjiang (34/35, 97.1%) and Yunnan (246/269, 91.4%) were significantly higher than that of other provinces/Aut. Reg.. A higher prevalence was observed in grazing sheep and goats (54.4%) than household sheep and goats (33.6%). Male animals had significantly higher hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) infection rates, at 60.0%, than females (41.6%). There were no significant differences between prevalence in different age groups of sheep and goats as well as that between sheep (45.8%) and goats (44.1%) (Table 2). The potential risk factors associated to percentage of infection with hemoplasmas were determined by statistical calculations. Among animal age, gender, species and feeding habits, gender (OR = 2.10, CI = 1.58–2.81) and the use of grazing (OR = 2.35, CI = 1.89–2.93) were identified as potential risk factors for hemoplasmas infection (P < 0.001) (Table 2).
Sequence analysis of 16S rRNA gene
Sequencing of the nearly complete 16S rRNA gene was successful for the 103 randomly selected positive specimens. Among them, analysis of the 16S rRNA gene sequences identified M. ovis (n = 56) and ‘Candidatus M. haemovis’ (n = 47). The origin of the 103 positive specimens was shown in Additional file 1: Table S2. After alignment, the obtained 103 16S rRNA gene sequences formed four sequence clusters KU983740, KU983746, KU983748 and KU983749.
KU983740 (n = 50) and KU983746 (n = 6) genotypes had 99.7% and 99.6% identity, respectively, to M. ovis from sheep in the USA (AF338268), and 99.9% and 99.8% identity, respectively, to a human M. ovis isolate form USA (KF313922). Both genotypes shared a high degree of identity (99.8%) with another human M. ovis isolate (GU230144), while, lower identity of them with sheep ‘Candidatus M. haemovis’ genotypes from Japan (AB617737) (97.3% and 97.4%, respectively) were identified. However, the other two genotypes KU983748 (n = 21) and KU983749 (n = 26) were closely related to ‘Candidatus M. haemovis’ (AB617737), with 99.5% and 99.4% identities, respectively, yet, their identities with M. ovis (AF338268) were 97.4% and 97.5%, respectively. Based on the 16S rRNA gene sequence analysis, the four novel genotypes in this study differed from M. ovis (AF338268) at several nucleotide positions along the full-length of the gene (Table 3).
Phylogenetic analysis
Phylogenetic analysis revealed that the four novel genotypes accompanied by five other ‘Candidatus M. haemovis’ (AB617737, AB617736, EU828580, EU828581 and JF931131), six other M. ovis (GU230144, JF931135, FJ440328, KF313922, AF338268 and EU828582) as well as one Mycoplasma spp. (FJ824847) formed a separate clade. In addition, ‘Candidatus M. haemovis’ and M. ovis genotypes fell into two different groups. The two genotypes KU983740 and KU983746, representing 56 M. ovis sequences from the present study, were grouped in the same clade with two human genotypes from USA (GU230144 and KF313922) (Fig. 2).
Phylogenetic relationships of Mycoplasma ovis and ‘Candidatus M. haemovis’ genotypes identified here and other hemotropic mycoplasmas. Phylogeny was inferred with a neighbor-joining analysis of the nearly complete 16S rRNA gene sequences based on distances calculated with the Kimura 2-parameter model. Mycoplasma pneumoniae was used as an outgroup. Bootstrap values >50% from 1000 replicates are shown on the nodes. The genotypes detected in this study are shown as triangles
Discussion
Globally, hemotropic Mycoplasma spp. are emerging or re-emerging zoonotic pathogens that affect numerous animal species including humans, potentially causing loss of livestock and public health concern. M. ovis is prevalent worldwide, and is causative of hemolytic anemia in sheep and goats [4].
In the present study, the prevalence of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) in sheep and goats was found to be 44.7% (610/1364). A similar prevalence of M. ovis was previously reported in goats in Hubei, China (41.0%, 151/371) [29], while a lower rate was detected in sheep in Tunisia (6.3%, 36/573) [28]. A higher M. ovis prevalence was previously found in sheep in Malaysia (94.0%, 47/50) [33], Argentina (81.8%, 9/11) [9], and Hungary (51.5%, 17/33) [24]. The differences in prevalence between this study and others [9, 24, 28, 29, 33] may be caused by discrepancy in the age groups studied, as well as the number of animals, detection methods, and geographic and ecological environments. In this study, there is a striking difference in prevalence between the north-central sampling sites (Qinghai, 15.0%; Shanxi, 16.7%; Shaanxi, 7.3% and Inner Mongolia, 5.0%) versus the southern and eastern sites (Guizhou, 40.6%; Yunnan, 91.4%; Heilongjiang, 97.1% and Henan, 39.1%). The climate type of north-central sampling sites is monsoon climate of medium latitudes, whereas southern sites are subtropical monsoon climate. Invertebrate vector abundance and vegetation species in north-central sampling sites is relatively lower than southern sites. Therefore, we speculated the different climates, vegetation species and invertebrate vector abundance factors that may contribute to the apparent difference in prevalence in different provinces.
It was reported that the M. ovis prevalence was significantly higher in young sheep than adults (P < 0.001) by Rjeibi et al. [28]. However, in the present study, there was no significant difference in hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) infection rates between different age groups of sheep and goats. Nevertheless, significant differences were detected among the prevalence of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) according to animal gender and feeding habits of the host animals, although no differences were observed between samples obtained from different genders by Song et al. [29]. In this study, potential risk factors were identified as animal gender and feeding habits of the host animals, providing useful information for control of the hemoplasmosis. Grazing flocks are more likely to be exposed to blood-sucking arthropods than household flocks. Household flocks received good feeding and management, which were thought to be important for the prevention from various diseases. Therefore, infections of hemoplasmas may be transmitted by blood-sucking arthropods and reusing needles during herd immunization in China.
‘Candidatus M. haemovis’ was first demonstrated in a sheep flock with fatal hemolytic anemia in Hungary [24], following studies reported the presence of both M. ovis and ‘Candidatus M. haemovis’ DNAs in sheep in Hungary [24], Japan [8], and the USA [27], in goats in Switzerland [25], and in Capricornis crispus in Japan [18]. However, there was no report documented the existence of M. ovis and ‘Candidatus M. haemovis’ in sheep and goats in China. Therefore, detection of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) by nested PCR and sequence analyses in the present study represents the first molecular evidence of the occurrence of ‘Candidatus M. haemovis’ in sheep and goats in China.
Phylogenetic analysis revealed that ‘Candidatus M. haemovis’ and M. ovis belong to the same clade. 16S rRNA gene homology between M. ovis and ‘Candidatus M. haemovis’ in this study was very high, at 97.3–97.5% (data not shown), revealing the close genetic relationship between them, which suggested that ‘Candidatus M. haemovis’ might not be a new hemoplasma species, but another copy of M. ovis 16S rRNA gene, which was validated by Deshuillers et al.’s report that M. ovis strain Michigan has two different copies of the 16S rRNA gene. The detection of multiple different 16S rRNA gene sequences in a veterinarian in Texas [22], in a severe anemia sheep in Japan [8], in goats in Switzerland [25] and in sheep in Hungary [24], might also be explained by the existence of multiple copies of the 16S rRNA gene within a single strain of M. ovis, each having a variable genetic composition, as reported for other bacteria [34].
In previous studies, only five hemotropic Mycoplasma spp., M. haemofelis-like [21], M. suis-like [35], M. ovis [22], ‘Candidatus Mycoplasma haemohominis’ [36], and ‘Candidatus Mycoplasma haematoparvum’ [37] organisms, were described to be infected by human beings. Sykes et al. reported the first case of human infection with hemotropic mycoplasma (M. ovis and ‘Candidatus M. haemovis’) in a veterinarian in Texas. Additionally, among the 11 hemotropic Mycoplasma spp. identified from 489 patients, M. ovis-like organism was the most prevalent [23], a species which was mainly found in small ruminants, such as deer [26, 38, 39], goats [4, 25, 33], sheep [4, 8, 24, 28, 40], and Japanese serows [18]. It is noteworthy that the nucleotide sequences of M. ovis (KU983740 and KU983746) in the present study had highest homology with M. ovis (GU230144 and KF313922) from humans (99.8% and 99.8–99.9%). The high homology of the M. ovis genotypes obtained in the present study with human genotypes observed here may provide additional evidence for the zoonotic potential of M. ovis, which were raised by Sykes et al. and then verified by Maggi et al.
Conclusions
This study provides further evidence about the prevalence of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’), and presents the first detection of ‘Candidatus M. haemovis’ in sheep and goats in China. Our data showed a high prevalence and widespread distribution of hemoplasmas (M. ovis and ‘Candidatus M. haemovis’) in sheep and goats. Further studies are needed to evaluate the pathogenicity of ‘Candidatus M. haemovis’ in sheep and goats, as well as the zoonotic potential of M. ovis.
Abbreviations
- ‘Candidatus M. haemovis’:
-
‘Candidatus Mycoplasma haemovis’
- 16S rRNA:
-
16S ribosomal RNA
- Aut. Reg.:
-
Autonomous region
- B. henselae :
-
Bartonella henselae
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- HIV:
-
Human immunodeficiency virus
- M. haemofelis :
-
Mycoplasma haemofelis
- M. ovis :
-
Mycoplasma ovis
- M. suis :
-
Mycoplasma suis
- PCR:
-
Polymerase chain reaction
References
Messick JB. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet Clin Pathol. 2004;33:2–13.
Hoelzle L. Haemotrophic mycoplasmas: recent advances in Mycoplasma suis. Vet Microbiol. 2008;130:215–26.
Biondo AW, Dos Santos AP, Guimarães AM, Vieira RF, Vidotto O, Macieira Dde B, et al. A review of the occurrence of hemoplasmas (hemotrophic mycoplasmas) in Brazil. Rev Bras Parasitol Vet. 2009;18:1–7.
Neimark H, Hoff B, Ganter M. Mycoplasma ovis comb. nov. (formerly Eperythrozoon ovis), an epierythrocytic agent of haemolytic anaemia in sheep and goats. Int J Syst Evol Microbiol. 2004;54:365–71.
Mascarelli PE, Keel MK, Yabsley M, Last LA, Breitschwerdt EB, Maggi RG. Hemotropic mycoplasmas in little brown bats (Myotis lucifugus). Parasit Vectors. 2014;7:117.
Willi B, Meli ML, Luthy R, Honegger H, Wengi N, Hoelzle LE, et al. Development and application of a universal Hemoplasma screening assay based on the SYBR green PCR principle. J Clin Microbiol. 2009;47:4049–54.
Dieckmann SM, Winkler M, Groebel K, Dieckmann MP, Hofmann-Lehmann R, Hoelzle K, et al. Haemotrophic Mycoplasma infection in horses. Vet Microbiol. 2010;145:351–3.
Tagawa M, Takeuchi T, Fujisawa T, Konno Y, Yamamoto S, Matsumoto K, et al. A clinical case of severe anemia in a sheep coinfected with Mycoplasma ovis and Candidatus Mycoplasma haemovis' in Hokkaido, Japan. J Vet Med Sci. 2012;74:99–102.
Aguirre D, Thompson C, Neumann R, Salatin A, Gaido A, de Echaide S. Clinical mycoplasmosis outbreak due to Mycoplasma ovis in sheep from Shalta, Argentina. Clinical, microbiological and molecular diagnosis. Rev Argent Microbiol. 2008;41:212–4.
Fujihara Y, Sasaoka F, Suzuki J, Watanabe Y, Fujihara M, Ooshita K, et al. Prevalence of hemoplasma infection among cattle in the western part of Japan. J Vet Med Sci. 2011;73:1653–5.
Torkan S, Aldavood SJ, Rafie SM, Hejazi H, Shirani D, Momtaz H. Prevalence and risk factor analysis of Haemobartonella felis in cats using direct blood smear and PCR assay. Comp Clin Pathol. 2013;22:1103–9.
Sykes JE. Feline hemotropic mycoplasmas. Vet Clin North Am Small Anim Pract. 2010;20:62–9.
Compton S, Maggi RG, Breitschwerdt EB. Candidatus Mycoplasma haematoparvum and Mycoplasma haemocanis infections in dogs from the United States. Comp Immunol Microbiol Infect Dis. 2012;35:557–62.
Willi B, Novacco M, Meli M, Wolf-Jäckel G, Boretti F, Wengi N, et al. Haemotropic mycoplasmas of cats and dogs: transmission, diagnosis, prevalence and importance in Europe. Schweiz Arch Tierheilkd. 2010;152:237.
Roura X, Peters IR, Altet L, Tabar M-D, Barker EN, Planellas M, et al. Prevalence of hemotropic mycoplasmas in healthy and unhealthy cats and dogs in Spain. J Vet Diagn Investig. 2010;22:270–4.
Sykes JE, Ball LM, Bailiff NL, Fry MM. ‘Candidatus Mycoplasma haematoparvum’, a novel small haemotropic mycoplasma from a dog. Int J Syst Evol Microbiol. 2005;55:27–30.
Hornok S, Meli M, Gönczi E, Ignits É, Willi B, Lutz H, et al. First molecular identification of ‘Candidatus mycoplasma haemominutum’ from a cat with fatal haemolytic anaemia in Hungary. Acta Vet Hung. 2008;56:441–50.
Ohtake Y, Nishizawa I, Sato M, Watanabe Y, Nishimura T, Matsubara K, et al. Mycoplasma ovis detected in free-living Japanese serows, Capricornis crispus. J Vet Med Sci. 2011;73:371–3.
Maggi RG, Mascarelli PE, Balakrishnan N, Rohde CM, Kelly CM, Ramaiah L, et al. “Candidatus Mycoplasma haemomacaque” and Bartonella quintana Bacteremia in Cynomolgus Monkeys. J Clin Microbiol. 2013;51:1408–11.
Hu Z, Yin J, Shen K, Kang W, Chen Q. Outbreaks of hemotrophic mycoplasma infections in China. Emerg Infect Dis. 2009;15:1139.
dos Santos AP, dos Santos RP, Biondo AW, Dora JM, Goldani LZ, de Oliveira ST, et al. Hemoplasma infection in HIV-positive patient, Brazil. Emerg Infect Dis. 2008;14:1922–4.
Sykes JE, Lindsay LL, Maggi RG, Breitschwerdt EB. Human coinfection with Bartonella henselae and two hemotropic mycoplasma variants resembling Mycoplasma ovis. J Clin Microbiol. 2010;48:3782–5.
Maggi RG, Compton SM, Trull CL, Mascarelli PE, Mozayeni BR, Breitschwerdt EB. Infection with hemotropic Mycoplasma species in patients with or without extensive arthropod or animal contact. J Clin Microbiol. 2013;51:3237–41.
Hornok S, Meli ML, Erdos A, Hajtos I, Lutz H, Hofmann-Lehmann R. Molecular characterization of two different strains of haemotropic mycoplasmas from a sheep flock with fatal haemolytic anaemia and concomitant Anaplasma ovis infection. Vet Microbiol. 2009;136:372–7.
Hornok S, Hajtos I, Meli ML, Farkas I, Gonczi E, Meili T, et al. First molecular identification of Mycoplasma ovis and 'Candidatus M. haemoovis' from goat, with lack of haemoplasma PCR-positivity in lice. Acta Vet Hung. 2012;60:355–60.
Grazziotin AL, Santos AP, Guimaraes AMS, Mohamed A, Cubas ZS, De Oliveira MJ, et al. Mycoplasma ovis in captive cervids: prevalence, molecular characterization and phylogeny. Vet Microbiol. 2011;152:415–9.
Deshuillers PL, Santos AP, do Nascimento NC, Hampel JA, Bergin IL, Dyson MC, et al. Complete genome sequence of Mycoplasma ovis strain Michigan, a hemoplasma of sheep with two distinct 16S rRNA genes. Genome Announc. 2014;2:e01235–13.
Rjeibi MR, Darghouth MA, Omri H, Souidi K, Rekik M, Gharbi M. First molecular isolation of Mycoplasma ovis from small ruminants in North Africa. Onderstepoort J Vet Res. 2015;82:912.
Song W, Song Q, He L, Zhou Y, Zhao J. The establishment and application of a semi-nested PCR assay for the detection of Mycoplasma ovis. Small Rumin Res. 2014;119:176–81.
ZuoYong Z, Kui N, Shi Jun H, Hong Lin L, Ming T, You Lan H, et al. Infection rate and risk factors analysis of Haemotrophic Mycoplasma (formerly Eperythrozoon ovis) in Chongqing area. Chinese Journal of Preventive Veterinary Medicine. 2010;32:563–6.
Frota IM, Leitão CC, Costa JJ, Brito IR, Van dHR SJR. Stability of housekeeping genes and expression of locally produced growth factors and hormone receptors in goat preantral follicles. Zygote. 2011;19:71–83.
Xiaoxing WANG, Yanyan CUI, Fuchun JIAN, Zhongyi ZHANG, Jianfeng NING, Longxian ZHANG, et al. Establishment and application of a nested-PCR method assay for detection of Mycoplasma ovis. J Henan Agricultural University. 2016;50:506–11.
Jesse FFA, Jazid NHBA, d Mohamme K, Tijjani A, ELT C, Abba Y, et al. Hemotropic Mycoplasma ovis infection in goats with concurrent gastrointestinal parasitism in Malaysia. J Adv Vet Anim Res. 2015;2:464–8.
Michon A-L, Aujoulat F, Roudiere L, Soulier O, Zorgniotti I, Jumas-Bilak E, et al. Intragenomic and intraspecific heterogeneity in rrs may surpass interspecific variability in a natural population of Veillonella. Microbiology. 2010;156:2080–91.
Yuan CL, Liang AB, Yao CB, Yang ZB, Zhu JG, Cui L, et al. Prevalence of Mycoplasma suis (Eperythrozoon suis) infection in swine and swine-farm workers in Shanghai, China. Am J Vet Res. 2009;70:890–4.
Steer JA, Tasker S, Barker EN, Jensen J, Mitchell J, Stocki T, et al. A novel hemotropic Mycoplasma (hemoplasma) in a patient with hemolytic anemia and pyrexia. Clin Infect Dis. 2011;53:e147–e51.
Maggi RG, Mascarelli PE, Havenga LN, Naidoo V, Breitschwerdt EB. Co-infection with Anaplasma platys, Bartonella henselae and Candidatus Mycoplasma haematoparvum in a veterinarian. Parasit Vectors. 2013;6:103.
Maggi RG, Chitwood MC, Kennedy-Stoskopf S, DePerno CS. Novel hemotropic Mycoplasma species in white-tailed deer (Odocoileus virginianus). Comp Immunol Microbiol Infect Dis. 2013;36:607–11.
Boes KM, Goncarovs KO, Thompson CA, Halik LA, Santos AP, Guimaraes A, et al. Identification of a Mycoplasma ovis-like organism in a herd of farmed white-tailed deer (Odocoileus virginianus) in rural Indiana. Vet Clin Pathol. 2012;41:77–83.
Suzuki J, Sasaoka F, Fujihara M, Watanabe Y, Tasaki T, Oda S, et al. Molecular identification of 'Candidatus Mycoplasma haemovis' in sheep with hemolytic anemia. J Vet Med Sci. 2011;73:1113–5.
Acknowledgements
Veterinary officers and farmers are appreciated for helping with sample collection in China. We are grateful to Dr. Junqiang Li and Fuchang Yu for their constructive comments during initiation of the study.
Funding
This work was supported by the Earmarked Fund for China Modern Agro-Industry Technology Research System (nycytx-39) and the Collaborative Innovation Center of Modern Animal Husbandry, Henan Province, China.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Authors’ contributions
CSN conceived and designed the experiments; XXW, YYC, YZ and KS performed the experiments; XXW, YZ, KS and YQY analyzed the data; XXW, FCJ, LXZ, RJW, and CSN wrote the manuscript. All the authors have read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
This research was conducted according to the Chinese Laboratory Animal Administration Act (1988) after review and approval of its protocol by the Research Ethics Committee of Henan Agricultural University. Appropriate permission was obtained from the farm owners before collection of blood samples from their sheep and goats.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
Prevalence of hemoplasmas (Mycoplasma ovis and ‘Candidatus Mycoplasma haemovis’) of sheep and goats in different farms. Table S2. The origins of the 103 selected positive specimens. Figure S1. Collection sites (indicated by black circles) in Henan province. Figure S2. The binding sites of nested PCR primers. (DOCX 97 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, X., Cui, Y., Zhang, Y. et al. Molecular characterization of hemotropic mycoplasmas (Mycoplasma ovis and ‘Candidatus Mycoplasma haemovis’) in sheep and goats in China. BMC Vet Res 13, 142 (2017). https://doi.org/10.1186/s12917-017-1062-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-017-1062-z