Abstract
Background
Body mass index (BMI) has been found to be associated with a decreased risk of non-small cell lung cancer (NSCLC); however, the effect of BMI trajectories and potential interactions with genetic variants on NSCLC risk remain unknown.
Methods
Cox proportional hazards regression model was applied to assess the association between BMI trajectory and NSCLC risk in a cohort of 138,110 participants from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. One-sample Mendelian randomization (MR) analysis was further used to access the causality between BMI trajectories and NSCLC risk. Additionally, polygenic risk score (PRS) and genome-wide interaction analysis (GWIA) were used to evaluate the multiplicative interaction between BMI trajectories and genetic variants in NSCLC risk.
Results
Compared with individuals maintaining a stable normal BMI (n = 47,982, 34.74%), BMI trajectories from normal to overweight (n = 64,498, 46.70%), from normal to obese (n = 21,259, 15.39%), and from overweight to obese (n = 4,371, 3.16%) were associated with a decreased risk of NSCLC (hazard ratio [HR] for trend = 0.78, P < 2×10−16). An MR study using BMI trajectory associated with genetic variants revealed no significant association between BMI trajectories and NSCLC risk. Further analysis of PRS showed that a higher GWAS-identified PRS (PRSGWAS) was associated with an increased risk of NSCLC, while the interaction between BMI trajectories and PRSGWAS with the NSCLC risk was not significant (PsPRS= 0.863 and PwPRS= 0.704). In GWIA analysis, four independent susceptibility loci (P < 1×10−6) were found to be associated with BMI trajectories on NSCLC risk, including rs79297227 (12q14.1, located in SLC16A7, Pinteraction = 1.01×10−7), rs2336652 (3p22.3, near CLASP2, Pinteraction = 3.92×10−7), rs16018 (19p13.2, in CACNA1A, Pinteraction = 3.92×10−7), and rs4726760 (7q34, near BRAF, Pinteraction = 9.19×10−7). Functional annotation demonstrated that these loci may be involved in the development of NSCLC by regulating cell growth, differentiation, and inflammation.
Conclusions
Our study has shown an association between BMI trajectories, genetic factors, and NSCLC risk. Interestingly, four novel genetic loci were identified to interact with BMI trajectories on NSCLC risk, providing more support for the aetiology research of NSCLC.
Trial registration
Similar content being viewed by others
Background
Lung cancer is one of the most common cancers and a leading cause of cancer-related death worldwide [1, 2]. In 2018, there were 2.09 million new cases and 1.76 million deaths of lung cancer worldwide, accounting for 11.6% and 18.4% of all cancer cases and deaths, respectively [3]. In particular, non-small cell lung cancer (NSCLC), the most common type of lung cancer, accounts for approximately 85% of all lung cancer cases [4]. Due to the increasing burden of NSCLC, it is necessary to identify more potential risk factors associated with NSCLC so as to develop individualized prevention strategies.
Obesity, usually defined as body mass index (BMI) ≥ 30 kg/m2, is becoming an increasingly common global health problem [5]. The global prevalence of obesity in adults increased steadily between 1975 and 2016, from 3 to 11% in men and 6 to 15% in women [6]. Several epidemiological studies have demonstrated that a higher BMI is associated with a lower risk of NSCLC in European and Asian populations [2, 7], which was also confirmed by a recent meta-analyses with a sample size of 7,310,130 participants [8]. However, most of these studies only used BMI at a single time point instead of considering the role of longitudinal BMI trajectories across the life course. In addition, a number of studies have shown that BMI trajectory from normal weight to obesity was associated with the risk of multiple cancers, including prostate, colorectal, oesophageal, gastric cardia adenocarcinoma, and even lung cancer [9,10,11].
Although environmental risk factors (e.g. BMI) are the main risk factors for NSCLC [12], genetic susceptibility is also an important contributor [13]. The heritability of lung cancer in European and Asian populations is estimated to be 12–21% [14, 15]. Previous genome-wide association studies (GWAS) identified more than 80 susceptibility variants associated with lung cancer in European and Asian populations, mainly NSCLC, as it is the main type of lung cancer; however, these variants could only explain a small proportion of the overall genetic variance [16, 17]. Interestingly, there is accumulating evidence that gene-environment interactions may be responsible for the missing heritability of cancer and act together with environmental risk factors in the pathogenesis of cancer [18, 19].
However, it remained unclear whether there was evidence to support the joint association between BMI trajectories and genetic variants on NSCLC incidence. In this study, we comprehensively investigated the relationship between BMI trajectories and NSCLC risk in the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. In addition, we applied a genome-wide interaction analysis to further assess the effect of different BMI trajectories in participants stratified according to genetic variants on NSCLC risk, which can provide novel insights into the pathophysiology of NSCLC.
Methods
Study population
The PLCO Cancer Screening Trial is a population-based cohort study aimed to evaluate the accuracy and reliability of screening methods for prostate, lung, colorectal, and ovarian cancer, which randomly recruited 154,897 individuals aged 49–78 years from 10 centres in the USA between 1993 and 2001 [20]. Exclusion criteria included (i) personal history of cancer prior to trial entry (n = 11,803); (ii) individuals with missing BMI at any age (n = 3,504) or BMI < 15 or > 50 kg/m2 (n = 361); (iii) individuals failing to return or complete the baseline questionnaire (n = 669); (iv) individuals at enrolment with age < 50 years (n = 2); and (v) individuals with small cell lung cancer (n = 448). Ultimately, a total of 138,110 participants were retained for analysis. No included individuals had been diagnosed with lung cancer at the time of voluntarily joining the study. The diagnosis of NSCLC was histologically confirmed via medical record reviews, the National Death Index (for completeness), and self-reported annual questionnaires during follow-up [21]. This study was approved by the ethics committees of the PLCO consortium providers (PLCO-424). Additional information for the study subjects is presented in the Additional file 1: Appendix S1 [22].
BMI and BMI trajectories ascertainment
Height (m) and body weight (kg) at age 20, 50, and enrolment were collected from self-recorded questionnaires completed by the participants in the PLCO study (https://cdas.cancer.gov/datasets/plco/90/). BMI at each age period was calculated using the formula body weight (kg)/height (m2). Individuals were classified according to their BMI in each age period according to the World Health Organization 2000 criteria: underweight (<18.5 kg/m2), normal weight (18.5–24.9 kg/m2), overweight (25.0–29.9 kg/m2), and obesity (>30 kg/m2) [23]. To assess the relationship between pre-diagnostic BMI changes, defined as BMI from the age of 20 or 50 to the entry, and the risk of NSCLC at age 20, 50, and entry, latent class growth model (LCGM) was used to identify longitudinal patterns of BMI change at three-time points during adulthood [24]. Specifically, the LCGM here was fitted using linear and quadratic polynomials with three to five trajectory categories (individuals per trajectory ≥ 1%), and the model with the highest number of fitting categories was selected using the Bayesian Information Criterion (BIC) method and the average posterior probability (AvePP) of each trajectory [25]. Detailed information for the calculation of BMI trajectories is provided in the Additional file 1: Appendix S1 [26, 27].
Genotyping
The PLCO GWAS data were deposited in the database of Genotypes and Phenotypes (dbGaP, phs001286.v1.p1 and phs000336.v1.p1), including a total of 14,497 participants genotyped using Illumina Hap240, Hap300, and Hap550 [28, 29]. The use of the PLCO genetic datasets was approved by both the Internal Review Board of Nanjing Medical University and the dbGaP database administration (#21708 and #21643). Basic information on genotyping and imputation for PLCO GWAS data is shown in the Additional file 1: Appendix S1 [30,31,32]. Additional quality control procedures for individuals and single nucleotide polymorphisms (SNPs) levels are presented in the Additional file 1: Appendix S1. Ultimately, 13,365 individuals remained in the genetic analysis (Additional file 1: Table S1).
Analysis of the interaction between the GWAS-based polygenic risk score (PRS) and BMI trajectories
Based on 81 previously reported GWAS SNPs associated with lung cancer in European and Asian populations [16, 17], and a strict quality control process, including (i) SNPs located within autosomal chromosomes; (ii) minor allele frequency (MAF) ≥ 0.05; (iii) call rate ≥ 95%; (iv) P-value for Hardy-Weinberg Equilibrium (HWE) among non-NSCLC individuals ≥ 1.0×10−6; (v) imputation INFO > 0.3; and (vi) a risk effect consistent with previous results, we identified 19 independent [linkage disequilibrium (LD), r2 < 0.5] GWAS-identified SNPs (Additional file 1: Table S2) to construct the simple-count PRS (sPRS) and weighted PRS (wPRS) [16, 17, 33]. The sPRS is equal to the number of risk alleles, which can be estimated as \(sPRS=\sum \limits_{i=1}^I{G}_i\), where Gi (i.e. 0, 1, or 2) denotes the number of risk alleles of ith SNP. The wPRS was calculated using the formula: \(wPRS=\sum \limits_{i=1}^I{\beta}_i{G}_i\), where βi is the per allele ORs derived from previous studies [16, 17, 33]. Additional information on the analysis of the interaction between the PRSGWAS and BMI trajectory is presented in the Additional file 1: Appendix S1.
Genome-wide interaction analysis (GWIA)
GWIA was performed to test for the gene-environment interactions between genome-wide SNPs and BMI trajectories. The interaction was modelled by determining the multiplicative product of SNP genotype and BMI trajectories in the Cox proportional hazard regression model, adjusting for age, sex, race, family history of lung cancer, education, smoking status, personal history of diabetes, current marital status, study centre, and the first 10 principal components. For GWIA, the P-value of the interaction term < 1.0×10−6 was considered statistically significant [34]. Similar to the construction of PRSGWAS, the GWIA-based sPRS (sPRSGWIA) or wPRS (wPRSGWIA) was also calculated to evaluate the cumulative interaction effects with BMI trajectories, separately.
Functional annotation
Functional annotation was conducted to explore the potential molecular roles of the GWIA-identified loci by (i) pinpointing the most likely candidate genes at the identified loci by identifying cis-expression quantitative trait loci (cis-eQTL) within no more than 1 Mb of each investigated SNP in the Genotype-Tissue Expression project (version 7.0, http://www.gtexportal.org/home/) database from multiple relevant tissues [35, 36] and (ii) using the Encyclopedia of DNA Elements [37], HaploReg (version 4.1) [38], and RegulomeDB (http://www.regulomedb.org/) to further assess the regulatory potential for variants of interest.
Statistical analysis
Cox proportional hazards regression model was used to estimate the hazard ratio (HR) and 95% confidence intervals (CIs) between BMI trajectories and NSCLC risk with adjustments for age, sex, race, family history of lung cancer, education, smoking status, personal history of diabetes, current marital status, and study centre. The proportional hazard assumption was assessed by Schoenfeld residuals [39]. Further, continuous variables were adapted to conduct tests of linear trends. Individual follow-up time was defined as a period from entry until the time of NSCLC occurrence (diagnosis) or censoring defined as the exit of the study due to other causes or death, loss to follow-up, or the end of the study.
Interaction effects of PRSGWAS, PRSGWIA, or each GWIA-identified SNP with BMI trajectories were further investigated by adding multiplicative interaction terms in the Cox models with adjustment for the first 10 principal components. A cumulative incidence function was estimated using Kaplan-Meier technique to quantify the risk of developing NSCLC over time, stratified by GWIA-identified SNPs, and differences in the full time-to-event distributions between different BMI trajectory groups were compared by a log-rank test [40].
Subgroup analysis was performed to evaluate the heterogeneity of the association between BMI trajectories and NSCLC risk stratified by sex, smoking status, or histological type. Further, several sensitivity analyses were performed to assess the reliability of the primary results. One-sample Mendelian randomization (MR) analysis was also performed to access the causality between BMI trajectories and NSCLC risk, including inverse-variance-weighted (IVW), Mendelian randomization Egger (MR-Egger), and simple median method. P values (two-sided) < 0.05 were deemed significant. All analyses were performed using R 3.5.3 and PLINK 1.90 software. Additional information is presented in the Additional file 1: Appendix S1.
Results
There were 138,110 individuals in the prospective cohort study (Table 1). In total, 2641 NSCLC patients with a mean age of 64.34 years (SD = 5.20) were confirmed, including 2343 (88.72%) whites and 298 (11.28%) non-whites (184 blacks, 32 Hispanics, 63 Asians, and 19 others) populations. Compared with non-NSCLC individuals, NSCLC was more common among participants who were male (HR = 0.61, 95% CI: 0.56 to 0.66, P < 2×10−16), older (HR = 1.06, 95% CI: 1.05 to 1.07, P < 2×10−16), non-Hispanic Blacks (HR = 1.58, 95% CI: 1.36 to 1.84, P = 2.32×10−9), and current (HR = 24.22, 95% CI: 20.93 to 28.03, P < 2×10−16) or ever smoker (HR = 6.94, 95% CI: 6.01 to 8.01, P < 2×10−16); had a family history of lung cancer (HR = 1.83, 95% CI: 1.66 to 2.03, P < 2×10−16); had a low level of education (HR = 0.63, 95% CI: 0.58 to 0.69, P < 2×10−16); had a history of diabetes (HR = 1.25, 95% CI: 1.09 to 1.44, P = 0.001); and were divorced, separated, or widowed (HR = 1.41, 95% CI: 1.30 to 1.54, P = 1.08×10−14).
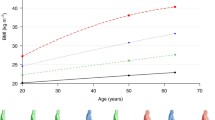
No evidence of departure from the proportional hazard assumption in Cox models for NSCLC (P = 0.166) was found. Cox proportional hazards model showed that a higher BMI at 20 years, 50 years, and the time of enrolment (baseline) were associated with a decreased risk of NSCLC (HR = 0.88, P = 0.001; HR = 0.70, P < 2×10−16; HR = 0.75, P < 2×10−16, respectively), and similar findings were observed in categorical BMI (decreased risk in overweight and obesity, Additional file 1: Table S3). Further, we identified four distinct BMI trajectories by the latent class growth model (Fig. 1). Compared with participants with a stable normal BMI in their adulthood (n = 47,982, 34.74%), the risk of NSCLC decreased in participants who progressed from a normal BMI to an overweight BMI at baseline (n = 64,498, 46.70%, HR = 0.77, 95% CI: 0.70 to 0.84, P = 3.80×10−9), who progressed from a normal BMI to an obese BMI at baseline (n = 21,259, 15.39%, HR = 0.60, 95% CI: 0.53 to 0.69, P = 5.42×10−13), and who were overweight at the onset of adulthood and became obese at baseline (n = 4371, 3.16%, HR = 0.54, 95% CI: 0.40 to 0.74, P = 9.33×10−5). Interestingly, the NSCLC risk decreased gradually across all three BMI trajectories (HR for trend = 0.78, 95%CI: 0.74 to 0.83, P = 2×10−16) compared with subjects who maintained a normal BMI. Sensitivity analyses showed that the primary model retained a stable association between BMI trajectories and NSCLC risk (Additional file 1: Table S4). Furthermore, stratified analyses by sex, smoking status, and histological type showed almost no significant heterogeneity in the effect of age-specific BMI and BMI trajectories on NSCLC risk, although the P value for the heterogeneity test was less than 0.05 among those with BMI < 18.5 at baseline stratified by sex (Additional file 1: Figures S1-S3).
The latent class growth model of BMI trajectories in the PLCO study. A BMI changes for each participant in each trajectory group across three analysed age points (ages of 20 years, 50 years, and baseline). B Each trajectory was calculated at any of the three analysed age points (ages of 20 years, 50 years, and baseline). HR and 95% CI were estimated by Cox proportional hazards regression model with the adjustment for age, sex, race, family history of lung cancer, education, smoking, personal history of diabetes, current marital status, and study centre
Nineteen GWAS-identified SNPs were used to construct the PRS and examine the potential effect of BMI trajectories on NSCLC risk according to the genetic variants. The characteristics of 13,365 individuals from the GWAS are shown in Additional file 1: Appendix S1. Nineteen GWAS-identified SNPs associated with lung cancer were used to construct the sPRS and wPRS (Additional file 1: Table S2). Furthermore, compared with the low tertiles of sPRSGWAS, the middle and high tertiles of sPRSGWAS were associated with a higher probability of NSCLC (HR = 1.13, 95% CI: 1.12 to 1.59, P = 0.001; HR = 1.56, 95% CI: 1.34 to 1.82, P = 1.62×10−8, respectively) (Additional file 1: Table S5). Similar results were obtained for wPRSGWAS, indicating that a higher PRSGWAS was associated with an increased risk of NSCLC. However, there was no significant interaction between BMI trajectories and PRSGWAS with the NSCLC risk (PsPRS= 0.863 and PwPRS= 0.704; Additional file 1: Figure S4). Similar findings were observed for age-specific BMI (Additional file 1: Tables S6-S7).
GWIA was subsequently performed to investigate the effect of the genome-wide interaction between each SNP and BMI trajectories on the NSCLC risk. A Manhattan plot was constructed to show the significant SNPs that interacted with BMI trajectories (Additional file 1: Figure S5). Four independent SNPs reached statistically suggestive significance [34] instead of genome-wide significance in GWIA, which were also confirmed in the bootstrap and permutation tests (Additional file 1: Table S8). Among the four SNPs, rs79297227 with the lowest P value (1.01×10−7) located in SLC16A7 (12q14.1) showed a statistically suggestively significant interaction with the BMI trajectories, and the remaining three SNPs, including rs2336652 near CLASP2 (3p22.3, P = 3.92×10−7), rs16018 in CACNA1A (19p13.2, P = 3.92×10−7), and rs4726760 near BRAF (7q34, P = 9.19×10−7) interacted with the BMI trajectories in terms of the NSCLC risk. Similar results were obtained from the analysis stratified by genotype (Table 2). Figure 2B displays the cumulative incidence of NSCLC stratified by GWIA-identified SNPs by the log-rank test. In the sensitivity analysis, a significant interaction was observed between four SNPs and the BMI trajectories by additionally adjusting for occupation and family history of any cancer or performing other sensitivity analyses (almost P < 1.0×10−4, Additional file 1: Table S9). MR sensitivity analyses showed that the correlation direction between BMI trajectories and NSCLC risk was consistent with the above analysis, although no meaningful differences in these results were observed, with no evidence of directional pleiotropy (Additional file 1: Tables S10-S11). For the functional annotation, the search for cis-eQTLs at the four loci detected by GWIA showed that SNP rs4726760 at 7q34 was a strong cis-eQTL for BRAF (P = 0.011, β = 0.073) in the lung tissue. No cis-eQTL was found at the other three loci (rs16018, P = 0.070, β = 0.128; rs2336652, P = 0.854, β = −0.015; rs79297227, P = 0.376, β = −0.042) (Additional file 1: Figure S6A). SNP rs16018 is located on chromosome 19p13.2 in calcium voltage-gated channel subunit alpha1 A (CACNA1A), which is a protein-coding gene involved in calcium channel regulation; SNP rs2336652 at 3p22.3 is located near cytoplasmic linker-associated protein 2 (CLASP2),which is significantly expressed in lung tissue and promotes the stability of microtubules; and SNP rs79297227 at 12q14.1 is located in the solute carrier family 16 member 7 (SLC16A7), which is not only significantly expressed in lung tissues (Additional file 1: Figure S6B) but also expressed in various types of malignant tumours.
Stratifications analysis for the interaction effects between BMI trajectories and GWIA-identified SNPs on NSCLC risk. A The identified four BMI trajectories from the onset of adulthood to the baseline. B Cumulative incidence of NSCLC stratified by GWIA-identified SNPs. P-value was derived from the Log-rank test. C Pathway of the gene (BRAF)-BMI trajectories interaction effect on the risk of NSCLC
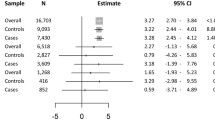
GWIA-based PRS of the four SNPs above was constructed to evaluate the cumulative interaction with BMI trajectories on NSCLC risk (Fig. 3). Although a significant association was identified between BMI trajectories and a higher NSCLC risk among the individuals with high tertiles of wPRSGWIA (HR for trend =1.30, 95% CI = 1.10–1.54), interestingly, BMI trajectories were also associated with a decreased risk of NSCLC among individuals with a low (0.54, 0.47–0.62) or intermediate tertiles of wPRSGWIA (0.85, 0.72–0.99), indicating an obvious interaction between the GWIA-based wPRSGWIA and BMI trajectories. Similar findings were observed for age-specific BMI (Additional file 1: Table S12). The interaction between BMI trajectories and PRSGWIA with the NSCLC risk was significant (PsPRS = 6.61×10−5 and PwPRS = 3.80×10−16; Additional file 1: Figure S4). In addition, individuals with low or intermediate tertiles of wPRSGWIA experienced a gradually decreased cancer risk across the BMI trajectories from normal to normal, normal to overweight, overweight to obese, and normal to obese, while the high tertiles of wPRSGWIA were just the opposite after adjustment for age, sex, race, family history of lung cancer, education, smoking, personal history of diabetes, current marital status, study centre, and first 10 principal components (Fig. 3A, B). Stratification analyses for wPRSGWIA showed that associations between BMI trajectories and NSCLC risk were heterogeneous (I2 = 73.09%, P for heterogeneity < 0.001, Fig. 3B). Similar results were also observed in sPRSGWIA (Additional file 1: Figure S4CD, Table S13).
Interaction analysis and stratification analysis of BMI trajectories and the PRS constructed by four GWIA-identified SNPs on NSCLC risk. A, BwPRSGWIA were weighted according to the strength of their association with lung cancer. C, DsPRSGWIA were calculated by simple counting. P value for interaction was derived from multivariate-adjusted Cox proportional hazards regression model. PRS, polygenic risk score; GWIA, genome wide interaction analysis; SNP, single nucleotide polymorphism; HR, hazard ratio; CI, confidence interval
Discussion
In this multi-centre study, four distinct trajectories of BMI were identified during adulthood, finding that subjects who progressed from a normal BMI at the onset of adulthood to overweight or obesity at baseline (compared to maintaining a stable BMI) had a lower risk of developing NSCLC in this PLCO cohort (Fig. 2A). In addition, interaction analysis provided evidence that the association between BMI trajectories and NSCLC risk slightly differed according to genetic variation at SNPs rs4726760, rs16018, rs2336652, and rs79297227.
The results of this study suggested that the BMI trajectory from normal weight to overweight or obesity was associated with protective effects against NSCLC development, which was consistent with previous epidemiology studies [1, 2, 41,42,43]. Several hypotheses have been postulated to explain the relationship between leanness and a higher risk of lung cancer. For example, smoking, as a dominant risk factor for lung cancer, usually leads to lower body weight, which may explain the observed inverse BMI-lung cancer association. However, several large prospective studies show a negative association between BMI and lung cancer risk, and this association persists after excluding up to 10 years of follow-up, suggesting that it is not entirely due to smoking [44]. Moreover, never-smokers were more likely to have a stable normal BMI trajectory according to a stratified analysis of smoking status, although never-smokers in each BMI trajectory group accounted for about 50% of our analysis. Likewise, it has been suggested that weight loss represents a preclinical event prior to the clinical manifestation of lung cancer [45]. However, our sensitivity analysis suggested that BMI trajectories resulting in overweight or obesity were associated with a lower risk of lung cancer, even excluding patients who developed the disease during the first, second, or fourth year of follow-up. Interestingly, interaction analysis of PRSGWIA with BMI trajectories on NSCLC risk indicated that BMI progressed from normal to overweight or obesity was associated with higher NSCLC risk among individuals with the high tertiles of wPRSGWIA or sPRSGWIA. Specifically, they experienced a gradually increased NSCLC risk across the BMI trajectories from normal to normal, normal to overweight, overweight to obese, and normal to obese, although the low or intermediate tertiles of wPRSGWIA or sPRSGWIA were just the opposite (Fig. 3). In addition, those identified SNPs were located in or near genes that might be involved in biological pathways leading to lung cancer. The gene BRAF near rs4726760 provides instructions for making a protein that helps transmit chemical signals from outside the cell to the nucleus. This protein is a component of the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway, which regulates several important cell functions including cellular proliferation, differentiation, migration, and apoptosis. Chemical signalling through this pathway is essential for normal development before birth. BRAF also is an oncogene. When mutated, oncogenes have the potential to cause normal cells to become cancerous [46]. BRAF mutations are seen in 3–5% of NSCLC cases [47]. It is generally believed that obese people eat nutrient-rich foods, and studies have found that nutrients (antioxidants) can significantly inhibit the MAPK signalling pathway to reduce the inflammation response related to the risk of cancer [48]. The MAPK pathway plays an important role in the differentiation of adipocytes [49], and ERK is essential for the transcription of gene CCATT/enhancer binding protein α/β/δ and peroxisome proliferator-activated receptor gamma (PPARγ), key factors of adipocyte differentiation. When the ERK signalling pathway is activated, PPARγ is phosphorylated and transcriptional activity is reduced, which inhibits adipocyte differentiation [50]. Decreased adipocyte differentiation reduces the accumulation of adipocytes, thereby reducing the incidence of inflammation that may be related to pathological obesity (Fig. 2C).
The SNP rs16018, a member of the family of voltage-gated calcium channels, is located in the gene CACNA1A which is upregulated in numerous types of cancer including lung cancer [51]. The roles of calcium channels in various cell functions including mitogenesis, cell proliferation, differentiation, inflammation, and metastasis are well recognized [52]. Through calmodulin, intracellular calcium (Ca2+) levels regulate many different kinases, phosphatases, cyclases, esterases, and ion channels. Increased intracellular Ca2+ levels are correlated with cell proliferation, leading to inflammation and promoting carcinogenesis [51]. Subjects with a higher BMI may have sufficient nutritional status, and current studies have demonstrated that people with higher intake of nutrients (e.g. high dietary calcium) can modulate circulating calcitriol, thereby regulating intracellular Ca2+ levels [53], maintaining the balance of intracellular and extracellular Ca2+ concentrations and reducing the risk of lung cancer.
The SNP rs2336652, located near CLASP2, interacts with cytoplasmic linker protein, binds to microtubules, and has microtubule-stabilizing effects [54]. Increasing microtubule instability may cause genetic instability, and altered expression of CLASP2 may induce genetic instability and contribute to the development of lung cancer [55]. The variant rs79297227 is associated with the expression of SLC16A7. The SLC16A family of monocarboxylate transporters is a subfamily of solute carriers that transport monocarboxylate molecules, including L-lactate and pyruvate, across cell membranes [56]. Aberrant expression of SLC16A gene family members occurs in various types of malignant tumours and regulates cell migration, invasion, and proliferation [57,58,59].
MR analysis revealed non-significant associations between genetic polymorphisms affecting BMI and NSCLC. Although MR is considered a powerful tool to infer causality from nature’s randomization, it cannot completely avoid bias and confounders; thus, the results of MR studies warrant a cautious interpretation [60]. For example, BMI is strongly affected by smoking status, age, sex, and ethnicity [61]. However, confounding could not result in the genetic variant, and it is possible that attenuation of a protective effect against NSCLC has been caused by adjustment for mediators actually along the causal pathway or associated with collider bias [62]. In the end, the use of BMI variants in MR as proxies for BMI trajectories had inherent limitations due to the lack of previous GWAS studies on BMI trajectories, and insufficient PLCO genetic data despite the large sample in the PLCO cohort.
Our study had several strengths. First, this study was performed in a multi-centre, large sample size cohort. Second, we not only investigated the association between BMI trajectories and the NSCLC risk but also evaluated the interaction between BMI trajectories and genetic variants in the development of NSCLC. Third, we identified four novel and functionally plausible GWIA-based SNPs, which located near genes that paly critical roles in cell growth, differentiation, and inflammation and were mechanistically linked to BMI and NSCLC genesis. However, limitations of this study have also been identified. Similar to nearly all epidemiologic study on the subject, BMI at age 20 and 50 were obtained from individual’s self-report. However, that information was obtained before the subsequent development of the outcomes of interest, so recall bias could not have been operative. Second, a substantial number of exclusions could limit generalizability, while it constrained our study cohort to those with complete data available that should help mitigate against threats to internal validity. Third, residual for unmeasured confounding cannot be excluded even exhaustive adjustment was performed in the multivariable analyses. And conclusions from further Mendelian randomization, which purportedly provides a methodologic approach for causality inference, should also be treated with caution. Fourth, our findings have not been validated by other larger-sample epidemiological studies, especially the limited sample size of the PLCO GWAS data. Finally, additional functional studies are warranted to elucidate the mechanisms underlying the effects of these loci and BMI trajectories interactions on NSCLC risk.
Conclusions
Our study found that genetic susceptibility may modify the effect of BMI trajectories on the development of NSCLC by regulating cell growth, differentiation and inflammation. Further larger or multi-ethnicity studies should be conducted to validate our findings.
Availability of data and materials
The PLCO phenotypic data and GWAS data analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- DNA:
-
Deoxyribonucleic acid
- eQTL:
-
Expression quantitative trait loci
- GWAS:
-
Genome-wide association study
- GWIA:
-
Genome-wide interaction analysis
- HR:
-
Hazard ratio
- HWE:
-
Hardy-Weinberg equilibrium
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MAPK:
-
Mitogen-activated protein kinase
- MR:
-
Mendelian randomization
- NSCLC:
-
Non-small cell lung cancer
- PLCO:
-
Prostate, lung, colorectal, and ovarian
- PRS:
-
Polygenic risk score
- PRSGWAS :
-
Polygenic risk score constructed by 19 previous GWAS-identified SNPs
- PRSGWIA :
-
Polygenic risk score constructed by 4 GWIA-identified SNPs
- SCLC:
-
Small cell lung cancer
- SD:
-
Standard deviation
- SNP:
-
Single nucleotide polymorphism
- sPRS:
-
Simple-count polygenic risk score
- sPRSGWAS :
-
Simple-count polygenic risk score constructed by 19 previous GWAS-identified SNPs
- sPRSGWIA :
-
Simple-count polygenic risk score constructed by 4 GWIA-identified SNPs
- wPRS:
-
Weighted polygenic risk score
- wPRSGWAS :
-
Weighted polygenic risk score constructed by 19 previous GWAS-identified SNPs
- wPRSGWIA :
-
Weighted polygenic risk score constructed by 4 GWIA-identified SNPs
References
Rauscher GH, Mayne ST, Janerich DT. Relation between body mass index and lung cancer risk in men and women never and former smokers. Am J Epidemiol. 2000;152(6):506–13.
Smith L, Brinton LA, Spitz MR, Lam TK, Park Y, Hollenbeck AR, et al. Body mass index and risk of lung cancer among never, former, and current smokers. J Natl Cancer Inst. 2012;104(10):778–89.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non-small cell lung cancer, Version 6.2015. J Natl Compr Cancer Netw. 2015;13(5):515–24.
Alvarnas A, Alvarnas J. Obesity and cancer risk: a public health crisis. Am J Manag Care. 2019;25(11 Spec No.):Sp332-Spsp3.
Chen C, Ye Y, Zhang Y, Pan XF, Pan A. Weight change across adulthood in relation to all cause and cause specific mortality: prospective cohort study. BMJ. 2019;367:l5584.
Yu D, Zheng W, Johansson M, Lan Q, Park Y, White E, et al. Overall and central obesity and risk of lung cancer: a pooled analysis. J Natl Cancer Inst. 2018;110(8):831–42.
Duan P, Hu C, Quan C, Yi X, Zhou W, Yuan M, et al. Body mass index and risk of lung cancer: Systematic review and dose-response meta-analysis. Sci Rep. 2015;5:16938.
Zheng R, Du M, Zhang B, Xin J, Chu H, Ni M, et al. Body mass index (BMI) trajectories and risk of colorectal cancer in the PLCO cohort. Br J Cancer. 2018;119(1):130–2.
Petrick JL, Kelly SP, Liao LM, Freedman ND, Graubard BI, Cook MB. Body weight trajectories and risk of oesophageal and gastric cardia adenocarcinomas: a pooled analysis of NIH-AARP and PLCO Studies. Br J Cancer. 2017;116(7):951–9.
Abdel-Rahman O. Pre-diagnostic body mass index trajectory in relationship to lung cancer incidence and mortality; findings from the PLCO trial. Expert Rev Respir Med. 2019:1–7.
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85.
Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12(4):399–408.
Dai J, Shen W, Wen W, Chang J, Wang T, Chen H, et al. Estimation of heritability for nine common cancers using data from genome-wide association studies in Chinese population. Int J Cancer. 2017;140(2):329–36.
Sampson JN, Wheeler WA, Yeager M, Panagiotou O, Wang Z, Berndt SI, et al. Analysis of heritability and shared heritability based on genome-wide association studies for thirteen cancer types. J Natl Cancer Inst. 2015;107(12):djv279.
Bosse Y, Amos CI. A decade of GWAS results in lung cancer. Cancer Epidemiol Biomark Prev. 2018;27(4):363–79.
Dai J, Lv J, Zhu M, Wang Y, Qin N, Ma H, et al. Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir Med. 2019;7(10):881–91.
Vineis P, Pearce N. Missing heritability in genome-wide association study research. Nat Rev Genet. 2010;11(8):589.
Manuck SB, McCaffery JM. Gene-environment interaction. Annu Rev Psychol. 2014;65:41–70.
Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clinical Pharmacology & Therapeutics. 2005;77(3):145–58.
Oken MM, Hocking WG, Kvale PA, Andriole GL, Buys SS, Church TR, et al. Screening by chest radiograph and lung cancer mortality: the Prostate, Lung, Colorectal, and Ovarian (PLCO) randomized trial. Jama. 2011;306(17):1865–73.
Gohagan JK, Prorok PC, Hayes RB, Kramer BS, Prostate LC. Ovarian Cancer Screening Trial Project T. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin Trials. 2000;21(6 Suppl):251S–72S.
Kelly SP, Graubard BI, Andreotti G, Younes N, Cleary SD, Cook MB. Prediagnostic body mass index trajectories in relation to prostate cancer incidence and mortality in the PLCO Cancer Screening Trial. J Natl Cancer Inst. 2017;109(3).
Armstrong H, Carraro N, Amanda T, Gaudreau P, Louvet B. Latent class growth mdelling: a tutorial; 2009.
Koning M, Hoekstra T, de Jong E, Visscher TL, Seidell JC, Renders CM. Identifying developmental trajectories of body mass index in childhood using latent class growth (mixture) modelling: associations with dietary, sedentary and physical activity behaviors: a longitudinal study. BMC Public Health. 2016;16(1):1128.
Lampousi AM, Moller J, Liang Y, Berglind D, Forsell Y. Latent class growth modelling for the evaluation of intervention outcomes: example from a physical activity intervention. J Behav Med. 2021;44(5):622–9.
Nagin DS, Odgers CL. Group-based trajectory modeling in clinical research. Annu Rev Clin Psychol. 2010;6:109–38.
Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42(Database issue):D975–9.
Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–6.
Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51(1):76–87.
Wang ML, Gu DY, Du ML, Xu Z, Zhang SZ, Zhu LJ, et al. Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat Commun. 2016;7.
Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529.
McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49(7):1126–32.
de Vries PS, Brown MR, Bentley AR, Sung YJ, Winkler TW, Ntalla I, et al. Multiancestry genome-wide association study of lipid levels incorporating gene-alcohol interactions. Am J Epidemiol. 2019;188(6):1033–54.
Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–13.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206.
Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30.
Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81.
Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):515–26.
Hellstrom A, Nilsson AK, Wackernagel D, Pivodic A, Vanpee M, Sjobom U, et al. Effect of enteral lipid supplement on severe retinopathy of prematurity: a randomized clinical trial. JAMA Pediatr. 2021;175(4):359–67.
Lam VK, Bentzen SM, Mohindra P, Nichols EM, Bhooshan N, Vyfhuis M, et al. Obesity is associated with long-term improved survival in definitively treated locally advanced non-small cell lung cancer (NSCLC). Lung Cancer. 2017;104:52–7.
Greenlee H, Unger JM, LeBlanc M, Ramsey S, Hershman DL. Association between body mass index and cancer survival in a pooled analysis of 22 clinical trials. Cancer Epidemiol Biomark Prev. 2017;26(1):21–9.
Shepshelovich D, Xu W, Lu L. Body mass index (BMI), BMI change, and overall survival in patients with SCLC and NSCLC: a pooled analysis of the International Lung Cancer Consortium (Vol 14, pg 1594, 2019). J Thorac Oncol. 2019;14(11):2024.
Wood AM, Jonsson H, Nagel G, Haggstrom C, Manjer J, Ulmer H, et al. The inverse association of body mass index with lung cancer: exploring residual confounding, metabolic aberrations and within-person variability in smoking. Cancer Epidemiol Biomark Prev. 2021;30(8):1489–97.
Ross PJ, Ashley S, Norton A, Priest K, Waters JS, Eisen T, et al. Do patients with weight loss have a worse outcome when undergoing chemotherapy for lung cancers? Br J Cancer. 2004;90(10):1905–11.
Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–21.
Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29(26):3574–9.
Su X, Shen Z, Yang Q, Sui F, Pu J, Ma J, et al. Vitamin C kills thyroid cancer cells through ROS-dependent inhibition of MAPK/ERK and PI3K/AKT pathways via distinct mechanisms. Theranostics. 2019;9(15):4461–73.
Bost F, Aouadi M, Caron L, Binétruy B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie. 2005;87(1):51–6.
Tang QQ, Grønborg M, Huang H, Kim JW, Otto TC, Pandey A, et al. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A. 2005;102(28):9766–71.
Wang CY, Lai MD, Phan NN, Sun Z, Lin YC. Meta-analysis of public microarray datasets reveals voltage-gated calcium gene signatures in clinical cancer patients. PLoS One. 2015;10(7):e0125766.
Lang F, Föller M, Lang KS, Lang PA, Ritter M, Gulbins E, et al. Ion channels in cell proliferation and apoptotic cell death. J Membr Biol. 2005;205(3):147–57.
Zemel MB, Miller SL. Dietary calcium and dairy modulation of adiposity and obesity risk. Nutr Rev. 2004;62(4):125–31.
Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104(6):923–35.
Tai AL, Mak W, Ng PK, Chua DT, Ng MY, Fu L, et al. High-throughput loss-of-heterozygosity study of chromosome 3p in lung cancer using single-nucleotide polymorphism markers. Cancer Res. 2006;66(8):4133–8.
Halestrap AP. The SLC16 gene family – structure, role and regulation in health and disease. Mol Asp Med. 2013;34(2-3):337–49.
Zhang G, Zhang Y, Dong D, Wang F, Ma X, Guan F, et al. MCT1 regulates aggressive and metabolic phenotypes in bladder cancer. J Cancer. 2018;9(14):2492–501.
Wu DH, Liang H, Lu SN, Wang H, Su ZL, Zhang L, et al. miR-124 suppresses pancreatic ductal adenocarcinoma growth by regulating monocarboxylate transporter 1-mediated cancer lactate metabolism. Cell Physiol Biochem. 2018;50(3):924–35.
Sanità P, Capulli M, Teti A, Galatioto GP, Vicentini C, Chiarugi P, et al. Tumor-stroma metabolic relationship based on lactate shuttle can sustain prostate cancer progression. BMC Cancer. 2014;14:154.
Kim EJ, Hoffmann TJ, Nah G, Vittinghoff E, Delling F, Marcus GM. Coffee consumption and incident tachyarrhythmias reported behavior, Mendelian randomization, and their interactions. JAMA Intern Med. 2021;181(9):1185–93.
Hales CM, Fryar CD, Carroll MD, Freedman DS, Aoki Y, Ogden CL. Differences in obesity prevalence by demographic characteristics and urbanization level among adults in the United States, 2013-2016. Jama-Journal of the American Medical Association. 2018;319(23):2419–29.
Cornelis MC, Munafo MR. Mendelian randomization studies of coffee and caffeine consumption. Nutrients. 2018;10(10).
Acknowledgements
We thank all the PLCO study staff for their efforts and dedication to the study and acknowledge all the participants whose contributions made this project possible. We thank Mrs. Yue Wang for language polishing.
Funding
This work is supported by the National Natural Science Foundation of China [Project No. 82173620 to Y.Z., 81530088 to F.C., 81820108028 to H.S., 81922061 to H.M.], and US NIH/NCI [U01CA209414 to D.C.C], Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD).
Author information
Authors and Affiliations
Contributions
D.Y., D.W., Y.W.1 (Yaqian Wu), F.C., and Y.Z. contributed to the study design. D.Y., X.C., and F.S. contributed to the data collection. D.Y., D.W., and Y.W.1 performed the statistical analyses and interpretation and drafted the manuscript. Y.Z., F.C., F.S., Y.W.2 (Yongyue Wei), R.Z., T.L., H.M., H.X., Z.H., D.C.C., and H.S. revised the manuscript. All authors contributed to the critical revision of the manuscript and approved its final version. Financial support and study supervision were provided by Y.Z. and F.C.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethics committees of the PLCO consortium providers (PLCO-424) and approved by the ethics committees of Nanjing Medical University (Jiangsu, China). All patients provided written informed consent to participate in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
David C. Christiani is the senior author who supervised the work.
Supplementary Information
Additional file 1: Appendix S1.
Supplementary methods. Table S1. Characteristics of the study subjects for GWAS. Table S2. Summary of the 19 GWAS-identified SNPs associated with lung cancer. Table S3. Association between age-specific BMI and NSCLC risk. Table S4. Sensitivity analysis for the association between BMI trajectory and NSCLC risk. Table S5. Association of sPRSGWAS and wPRSGWAS with NSCLC risk. Table S6. Interaction analysis between age-specific BMI and the sPRSGWAS. Table S7. Interaction analysis between age-specific BMI and the wPRSGWAS. Table S8. Summary of four independent SNPs identified by GWIA. Table S9. Sensitivity analyses for the interaction between BMI trajectory and rs79297227, rs2336652, rs16018 and rs79297227. Table S10. Single-nucleotide polymorphisms used as instrumental variables in the multivariable Mendelian randomization analyses of BMI trajectory. Table S11. Sensitivity analysis of the relationship between BMI trajectory and lung cancer using one-sample Mendelian randomization. Table S12. Interaction analysis between age-specific BMI/BMI trajectories and the sPRSGWIA. Table S13. Interaction analysis between age-specific BMI/BMI trajectories and the wPRSGWIA. Figure S1. Stratification analysis for age-specific BMI and BMI trajectory on NSCLC risk by sex. Figure S2. Stratification analysis for age-specific BMI and BMI trajectory on NSCLC risk by smoking status. Figure S3. Stratification analysis for age-specific BMI and BMI trajectory on NSCLC risk by histological type. Figure S4. Association of multivariate-adjusted NSCLC risk with BMI trajectories according to PRSGWIA categories. Figure S5. Circle Manhattan Plot for interaction analysis between SNPs and BMI trajectory in regard to NSCLC risk. Figure S6. Analysis of the four loci and related gene expression in lung tissue.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
You, D., Wang, D., Wu, Y. et al. Associations of genetic risk, BMI trajectories, and the risk of non-small cell lung cancer: a population-based cohort study. BMC Med 20, 203 (2022). https://doi.org/10.1186/s12916-022-02400-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-022-02400-6