Abstract
Background
The combination of bevacizumab and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) could prolong progression-free survival (PFS) in patients with EGFR-mutant advanced non-small-cell lung cancer (NSCLC). Our study investigated the clinical and molecular factors that affect the efficacy of first-generation EGFR-TKI with or without bevacizumab and identify the subset of patients who can benefit from combination therapy.
Methods
Our study included 318 patients with EGFR-mutant locally advanced/advanced NSCLC treated with either first-generation EGFR-TKI combined with bevacizumab (A+T; n = 159) or EGFR-TKI monotherapy (T; n = 159). Two nomogram models to predict PFS and overall survival (OS), respectively, were constructed using two factors that impact EGFR-TKI efficacy: metastatic site and presence of concurrent mutations. The study cohort was stratified into 2 cohorts for training (n = 176) and validation (n = 142) of the nomogram model. Using the median score from the nomogram, the patients were stratified into two groups to analyze their survival outcome.
Results
The A+T group had significantly longer PFS (14.0 vs. 10.5 months; p < 0.001) and OS (37.0 vs. 26.0 months; p = 0.042) than the T group. Among the patients with concurrent mutations in tumor suppressor genes, those in the A+T group had significantly longer PFS and OS than the T group (PFS 14.5 vs. 8.0 months, p < 0.001; OS 39.0 vs. 20.0 months, p = 0.003). The higher scores from the nomograms were associated with the presence of brain/liver/pleural metastasis or concomitant gene mutations, which indicated a higher likelihood of shorter PFS and OS. The validation of the nomogram revealed that patients with lower scores had significantly longer PFS for the T group than those with higher scores (15.0 vs. 9.0 months, p = 0.002), but not for the A+T group (15.9 vs. 13.9 months, p = 0.256).
Conclusions
Using a nomogram, our study demonstrated that the addition of bevacizumab may enhance the therapeutic effectiveness of EGFR-TKI by overcoming the negative impact of certain clinical and molecular factors on the efficacy of EGFR-TKI.
Similar content being viewed by others
Background
Lung cancer remains the leading cause of cancer deaths worldwide [1]. Non–small-cell lung cancer (NSCLC) is the most prevalent histological type, accounting for 85% of all lung cancers [2].
Molecular targeted therapies for specific genetic alterations such as epidermal growth factor receptor (EGFR) have significantly improved the progression-free survival (PFS) and overall survival (OS) and profoundly shifted the treatment landscape in recent decades [3,4,5]. However, most patients who initially respond to EGFR-TKIs will inevitably develop resistance within 1 year [6,7,8]. To prevent or delay the emergence of acquired resistance to EGFR-TKIs, combination therapy with chemotherapy or antiangiogenic agents plus EGFR-TKIs are an emerging trend and have been evaluated in several clinical trials [9,10,11,12]. Bevacizumab is one of the commonly used antiangiogenic monoclonal antibodies that target the vascular endothelial growth factor (VEGF) signaling pathway [13]. Preclinical studies suggested that simultaneous inhibition of the EGFR and VEGF/VEGFR pathways could yield a biologically synergistic effect on antitumor activity and could overcome primary and acquired EGFR-TKI resistance [14, 15]. In the JO25567, NEJ026, and ARTEMIS-CTONG1509 trials, bevacizumab plus erlotinib showed prolonged PFS as compared with erlotinib monotherapy [9,10,11]. Several other studies also showed that bevacizumab plus EGFR-TKIs significantly prolonged PFS with acceptable toxicity profile than EGFR-TKI monotherapy for patients with EGFR-mutant NSCLC [12, 16, 17].
However, data on the effect of concomitant gene mutations at baseline on the clinical efficacy of the EGFR-TKI plus bevacizumab combination therapy are still lacking. Despite treatment with EGFR-TKI, the prognosis remains poor for patients with concomitant gene mutations such as TP53 [18, 19]. The optimal first-line treatment for NSCLC patients with various concomitant mutations remains unclear. Our previous clinical study revealed that EGFR-TKIs plus bevacizumab was associated with a significantly higher systemic and intracranial objective response rate (ORR) and significantly longer systemic and intracranial PFS [20]. On this foundation, we analyzed the therapeutic effect of the addition of bevacizumab to EGFR-TKI among Chinese patients with EGFR-mutant locally advanced/advanced NSCLC. Our main research aim is to evaluate the effect of concurrent gene mutations on patient prognosis. We also constructed two nomogram prediction models for PFS and OS, respectively, based on molecular features such as the presence/absence of concurrent gene mutations, and clinical features such as the specific location of metastasis. We also explored the molecular mechanisms of acquired resistance to the combination therapy.
Methods
Patients
All the patients with EGFR sensitizing mutation-positive locally advanced/advanced NSCLC treated with either EGFR-TKI monotherapy or EGFR-TKI plus bevacizumab as first-line therapy were retrospectively enrolled in this study for efficacy evaluation and resistance mechanism investigation. The main inclusion criteria included (1) age of 18–75 years, (2) stage IIIB-IV (according to the 8th American Joint Committee on Cancer Staging System), (3) histologically confirmed lung adenocarcinoma/adenosquamous carcinoma with EGFR activating mutations (exon 19 deletions or L858R), (4) tumor biopsy sample available at baseline and at progression submitted for next-generation sequencing (NGS) detection for panel of 168 cancer-related genes (Additional file 1: Table S1), (5) received at least two cycles of regimen at standard dosing, and (6) having at least one radiological response evaluation according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. The major exclusion criteria included treatment with other previous systemic therapy. The study protocol was approved by the Hunan Provincial Ethics Committee and written informed consent was obtained from each participant for the use of his or her clinical information for research analysis.
Study design and treatments
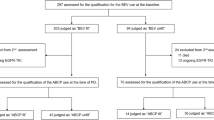
Our study included a total of 318 patients with EGFR-mutant locally advanced/advanced NSCLC who submitted samples for 168-gene panel NGS testing and were treated with either first-generation EGFR-TKI combined with bevacizumab (A+T) or EGFR-TKI monotherapy (single T). For the nomogram model construction, the cohort was further stratified into two groups as training and validation cohorts. The training cohort A was comprised of 176 patients from Hunan Cancer Hospital wherein 88 patients respectively received A+T and T regimens, while the validation cohort B was comprised of 142 patients from Xiangya Hospital wherein 71 patients respectively received A+T and T regimens. To ensure that the two groups had comparable baseline clinical characteristics, the patients included in the T group were selected using propensity score matching with the A+T group using a 1:1 ratio. Retrospective analyses were performed on clinical data, survival outcomes, and mutation profiles at baseline and at progression. Figure 1 illustrates the study design. The data cutoff was August 31, 2020, with median follow-up of 31 months in the training cohort A and 30 months in the validation cohort B. The standard initial dosing in clinical practice is 15 mg/kg, every 21 days for bevacizumab, 150 mg once daily for erlotinib, and 250 mg once daily for gefitinib. Drug dose adjustments were determined by the treating physicians.
Assessment
Clinical information was collected, including but not limited to demographic characteristics, ORR, PFS, adverse events, and somatic mutation profile. Tumor tissue samples were collected at the initial diagnosis (baseline) and at disease progression by resection or needle biopsy and stored as formalin-fixed paraffin-embedded (FFPE) samples. NGS testing was submitted to Burning Rock Biotech.
Tissue DNA isolation, capture-based targeted DNA sequencing, and sequence data analysis
Tissue DNA was isolated from FFPE tumor tissue samples using QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany). The quality and the size of the fragments were assessed using Qubit 2.0 Fluorimeter with the dsDNA high-sensitivity assay kit (Life Technologies, Carlsbad, CA, USA). NGS library was prepared for each sample. Target capture was performed using a 168 gene panel (Lung Plasma, Burning Rock Biotech, Guangzhou, China), which included 68 lung cancer-related genes and 100 other genes related to cancer development, spanning 0.273 megabases (Mb) of the human genome (Table S1). The indexed samples were sequenced on Nextseq500 (Illumina, Inc., CA, USA) with paired-end reads and average sequencing depth of 1000X. DNA sequence data were analyzed using optimized bioinformatics pipelines based on mapping with the reference human genome (hg19) using Burrows-Wheeler Aligner version 0.7.10, local alignment optimization, duplication marking, and variant calling using Genome Analysis Tool Kit version 3.2 and VarScan version 2.4.3. Single nucleotide variations (SNV) and insertion-deletion variations (INDEL) with population frequency over 0.1% in the ExAC, 1000 Genomes, dbSNP, or ESP6500SI-V2 databases were grouped as single nucleotide polymorphisms (SNPs) and excluded from further analysis. Remaining variants were annotated using ANNOVAR (2016-02-01 release) and SnpEff version 3.6. Analysis of structural variations (SVs) was performed using Factera version 1.4.3. Copy number variations (CNV) were analyzed based on the depth of coverage data of capture intervals. The limit of detection for copy number (CN) gain is CN > 2.25 for hotspot genes (such as EGFR, TP53, ERBB2, and MET) and CN > 2.5 for others, while for CN loss is CN < 1.75 for hotspot genes and CN < 1.5 for others.
Statistical analysis
Median survival was calculated by the Kaplan–Meier method. The Cox proportional hazards model was used for multivariable survival analysis. The variables with a p < 0.1 from the univariate analysis were included in the multivariate analysis. Schoenfeld residuals were used to check the proportional hazards assumption. Based on the multivariable analysis, the nomograms were constructed using R version 2.12.1 (The R Foundation for Statistical Computing, Vienna, Austria). The performance of the nomogram was evaluated by calculating the concordance index (C-index). The nomograms were validated using internal and external validation methods. Calibration of the nomogram was performed by comparing the predicted probability and the actual status after bias correction. All p values were two-sided, and p < 0.05 was considered statistically significant, unless stated otherwise. All procedures were performed using either SPSS software (version 24) or R Studio (version 2.12.1).
Results
Patient characteristics
A total of 159 patients, with 88 patients from Hunan Cancer Hospital and 71 patients from Xiangya Hospital with EGFR-mutant locally advanced/advanced NSCLC who were treated with A+T were matched to 159 patients who were treated with single-agent T using propensity score matching with 1:1 ratio (Fig. 1). The two groups and cohorts were comparable for baseline clinical characteristics including age, sex, smoking status, Eastern Cooperative Oncology Group Performance Status (ECOG PS) score, tumor node metastasis (TNM) stage, and tumor histology (Table 1). Cohort A also had comparable baseline somatic mutational characteristics (Additional file 2: Figure S1). In cohort A, baseline brain metastasis was detected in 28 (31.4%) patients in the A+T group and 30 (34.1%) patients in the T group (p = 0.451). No significant difference was observed for the distribution of other metastatic sites in both cohorts (Table 1).
Comparison of PFS and OS in A+T vs. T groups
The A+T group had significantly longer PFS (median PFS: 14.0 vs. 10.5 months; hazard ratio (HR) = 0.55, 95% confidence intervals (CI) = 0.40–0.76; p < 0.001) (Fig. 2A) and improved OS as compared with the T group (median OS 37.0 vs. 26.0 months; HR = 0.65, 95% CI = 0.43–0.98; p = 0.042) (Fig. 2B). Mutational status on certain genes, including TP53 mutation, was associated with significantly different PFS and OS in the T group but not in the A+T group (Additional file 2: Figure S2).
Impact of metastatic sites on therapeutic efficacy
We evaluated the impact of the brain, liver, or pleural metastasis on the therapeutic efficacy of single T and A+T regimens in the training cohort. In the single T group, the presence of brain, liver, or pleural metastasis was associated with significantly shorter PFS and OS (Additional file 2: Figure S3A). However, in the A+T group, the PFS was comparable for patients with brain, liver, or pleural metastasis, but had poor OS outcome for those with brain/liver metastasis (Additional file 2: Figure S3B).
Impact of concomitant mutations on therapeutic efficacy
We evaluated the impact of the concomitant mutations in tumor suppressor or oncogenic driver genes on the therapeutic efficacy of single T and A+T. Patients with only EGFR activating mutations showed comparable PFS with the two treatment regimens (p = 0.568) (Fig. 3A). Patients who concurrently harbor mutations in any tumor suppressor gene achieved longer PFS in the A+T group than in the T group (median PFS 14.5 vs. 8.0 months, p < 0.001) (Fig. 3A). The patients with concurrent oncogenic driver gene mutations had statistically comparable PFS with A+T and T treatments (16.3 vs. 9.0 months, p = 0.665) (Fig. 3A). Similar trends were observed for OS (Fig. 3B).
Progression-free survival (PFS) (A) and overall survival (OS) (B) of patients with EGFR-mutant advanced NSCLC without concomitant mutations (EGFR_only) and those with concomitant gene mutations in either tumor suppressor genes (EGFR_Suppressor) or oncogenic driver genes (EGFR_multiDriver) treated with either EGFR-TKI with bevacizumab (A+T) or EGFR-TKI monotherapy (T-single)
As the most common genetic alteration observed in our cohort, the presence of TP53 mutation had a negative impact on the efficacy of single T treatment. Patients who harbored TP53 mutations had inferior PFS in comparison to those who had wild-type TP53; however, it did not impact the efficacy of A+T treatment (Additional file 2: Figure S4A). Expectedly, as compared with single T, A+T significantly prolonged the PFS of TP53-mutant patients (median PFS 15.0 vs. 8.0 months, p < 0.001), while no difference was observed among TP53 wild-type patients (Additional file 2: Figure S4A). These observations were also consistent for OS (Figure S4B). As compared to single T, significantly longer PFS and OS were observed with A+T treatment of patients with TP53 mutations found in exons 5-8 (Additional file 2: Figure S5A-B), but not TP53 mutations resulting in loss-of-function (Additional file 2: Figure S5C-D) or those with concomitant mutations in TP53 and RB1 (Additional file 2: Figure S5E-H).
Furthermore, two patients with stage IV lung adenocarcinoma detected with EGFR exon 19 deletion and either concurrent KRAS K117N (n = 1) or concurrent de novo EGFR T790M (n = 1) achieved partial response with A+T regimen, with PFS of > 16.0 months and 10.3 months, respectively. Another patient was detected with concurrent EGFR exon 21 L858R concurrent with de novo EGFR T790M and had best response of progression disease and PFS of 1 month with single-agent gefitinib.
Nomograms
In training cohort A, through statistical analysis and in consideration of clinical significance, the presence/absence of concurrent genetic mutation (i.e., exclusive EGFR activating mutations, concurrent oncogenic driver gene mutation, or concurrent tumor suppressor gene mutation) and the location of metastasis (i.e., locally pleural, brain, or liver metastasis) were selected as the molecular and clinical factors to generate the nomograms for estimating the 12-month and 18-month PFS rates of T treatment (Fig. 4A). As shown in the nomogram, a lower total score/point was associated with having only EGFR mutations and/or having only intrathoracic metastasis and indicated a lower risk of disease progression with T treatment, whereas a higher score indicated a higher risk of disease progression due to the presence of concurrent gene mutations and/or presence of brain or liver metastasis. To validate the prognostic utility of the nomogram model, we used the nomogram to score the patients in either the T or the A+T group in the training cohort A or validation cohort B and used the median score as the cutoff to divide the patients as low-risk (< median, low score) and high-risk (≥ median, high score). As expected, patients with lower scores presented with significantly longer PFS than those with higher scores (15.0 vs. 9.0 months, p = 0.002) (Fig. 4B). In contrast, the scores from the nomogram were not predictive of PFS with A+T treatment as shown by the comparable PFS between the subgroups (15.9 vs. 13.9 months, p = 0.2556), suggesting that the factors included in the nomogram did not impact the efficacy of A+T (Fig. 4C). Consistently, patients with higher scores in the T group of the validation cohort B had significantly shorter PFS than those with lower scores (14.5 vs. 8.0 months, p < 0.001) (Fig. 4D), while the patients in the A+T group also had comparable PFS for those with high and low scores (16.8 vs. 12.3 months, p = 0.9042), suggesting that the factors included in the nomogram did not impact the efficacy of A+T (Fig. 4E).
Nomograms were constructed for predicting the risk of 12-month and 18-month progression-free survival (PFS) (A). Survival analyses of the training cohort A (B, C) and validation cohort B (D, E) were performed using Kaplan-Meier curves to compare the PFS of patients in the T group (B, D) or A+T (C, E) group, which were further subgrouped according to the median score into low-risk (< median score) and high-risk (≥ median score)
Likewise, we constructed a model for estimating the 12-month and 24-month OS of T treatment, using the same molecular and clinical factors included in the nomogram model for PFS (Additional file 2: Figure S6A). Similarly, lower scores predicted significantly prolonged OS in the T group (36.0 vs. 21.0 months, p = 0.0051) (Additional file 2: Figure S6B); however, the scores were not predictive of the OS for the patients in the A+T group (38.7 vs. 28.0 months, p = 0.1221) (Additional file 2: Figure S6C).
The fact that this model was not applicable for the A+T-treated patients suggested that these molecular and clinical factors negatively impacted the efficacy of T treatment but did not affect A+T treatment. Hence, we further evaluated each variable included in the nomograms on their impact on A+T efficacy using Cox multivariable analysis. We found that none of these factors impacted the PFS for A+T treatment (Additional file 2: Figure S7A), while the presence of brain metastasis (HR = 2.96, 95% CI = 1.51–5.83, p = 0.002) and liver metastasis (HR = 2.31, 95% CI = 1.02–5.22, p = 0.044) significantly deteriorated the OS of A+T-treated patients (Additional file 2: Figure S7B).
Resistance mechanism
Resistance mechanisms after disease progression from single T and A+T regimens were only evaluable for 79 and 63 patients included in the training cohort A, respectively.
At progression, EGFR T790M was the major resistance mechanism in both groups and was detected from 41% of the T group and 34% of the A+T group. The distribution of acquired mutations in the T group included exclusively EGFR T790M (30%), EGFR T790M plus cell cycle gene mutations (6%), EGFR T790M plus ERBB2 amplification (2%), EGFR T790M plus MET amplification plus cell cycle gene mutation (2%), and EGFR T790M plus TP53 mutation plus RB1 mutation (1%) (Fig. 5A). The distribution of acquired mutations in the A+T group included exclusively EGFR T790M (29%), EGFR T790M plus cell cycle gene mutation (3%), and EGFR T790M plus ERBB2 amplification (2%) (Fig. 5B). As compared to the A+T group, the T group frequently presented with multiple resistant mutations.
Among the 63 patients who progressed from A+T regimen, three patients were detected with RB1 mutation wherein two patients (67%) were confirmed with histological transformation to small-cell lung cancer (SCLC). Among the 79 patients who progressed from single T regimen, six patients were detected with RB1 mutations, of which four patients (67%) were confirmed to have SCLC transformation.
Discussion
Prior clinical trials have demonstrated the improvement of PFS with the addition of bevacizumab to the EGFR-TKI treatment [9,10,11,12, 21]. However, a major challenge remains in the selection of patients who would derive more benefit from the addition of bevacizumab. In addition, the mechanism underlying this improvement is still largely unknown. In this exploratory study, we identified the landscape of the concurrent genomic alterations in EGFR-mutant NSCLCs and explored the subset of patients who can benefit from the addition of bevacizumab into first-line EGFR-TKI regimen. Our study demonstrated that the addition of bevacizumab may overcome the clinical and molecular factors that negatively impact EGFR-TKI efficacy and thus enhance its therapeutic effectiveness.
In our study, the median PFS was 14.0 months in the A+T group and 10.5 months in the T group, which were shorter than the reported outcomes of the NEJ026 study (median PFS 16.9 months in erlotinib plus bevacizumab group and 13.3 months in erlotinib alone group) and the ARTEMIS-CTONG1509 (median PFS 17.9 months in erlotinib plus bevacizumab group and 11.2 months in erlotinib alone group [10, 11]. We consider our observation to be acceptable due to the complicated situations in the real-world clinical practice. Interestingly, our study suggested a significantly longer OS in the A+T group than in the T group, which is in contrast to the lack of OS difference observed in the JO25567, NEJ026, and ARTEMIS-CTONG1509 studies [9,10,11]. We recommend further studies with a larger sample size and longer follow-up to clarify this inconsistency.
The concomitant genetic alterations in either tumor suppressor or oncogenic driver genes, or EGFR amplification, were frequently detected in our cohort. Among these genetic alterations, we observed that patients having concomitant mutations in the tumor suppressor genes had longer PFS in the A+T group than in the T group, indicating the favorable role of the combination treatment strategy in patients with concurrent tumor suppressor gene mutations. Previous study proved that harboring concurrent tumor suppressor gene mutation was a negative prognostic factor for EGFR-TKI treatment [22]. Our study further demonstrated that these patients could benefit from the addition of bevacizumab, which to some extent can improve not only the PFS, but also the OS of this subset of patients.
Of the tumor suppressor gene alterations, we observed that TP53 mutation, which frequently occurs in many human malignancies and is associated with cancer development and progression, was also the most common concurrent genetic alteration in our cohort (41%). This may contribute to the clinical significance of the addition of bevacizumab to EGFR-TKI regimen in our population, particularly among the patients with concomitant mutations in TP53 or other tumor suppressor genes. Retrospective studies have shown that the presence of mutant TP53 is associated with poorer PFS with EGFR-TKI therapy [18, 19, 23,24,25]. Consistent with these studies, our study suggested that concomitant TP53 mutation negatively impact the PFS for EGFR-TKI monotherapy; however, this was not observed in the A+T group. Besides, compared with EGFR-TKI monotherapy, A+T prolonged the PFS in TP53-mutant patients, which was not observed among TP53 wild-type patients. We speculated that the addition of bevacizumab counteracted the negative effect of TP53 mutation on EGFR-TKI efficacy and TP53 mutation may serve as a biomarker for predicting the subset of patients who may benefit from the bevacizumab combined with EGFR-TKI. This conclusion was to some extent supported by some prior studies evaluating the impact of TP53 mutation on the efficacy of bevacizumab combined with chemotherapy in other solid tumor types including metastatic colorectal cancer and advanced endometrial cancer [26, 27]. Studies have also reported that TP53 mutation was positively correlated with VEGF-A expression [28].
We constructed nomograms for predicting the risk of progression (PFS) and the risk of death (OS) of the T group based on two significant factors: the presence/absence of concurrent genetic mutations in tumor suppressor genes and oncogenic driver genes and the location of metastasis (locally pleural, brain or liver metastases). Since the models did not impact the PFS or OS for the A+T group, further analyses revealed that the factors included in the nomogram had no effect on PFS of A+T group, while the presence of brain or liver metastasis shortened their OS. This suggested that the factors that negatively impact the efficacy of EGFR-TKI treatment became insignificant when bevacizumab was added to the regimen. Hence, we concluded that the addition of bevacizumab may improve the efficacy of EGFR-TKI through overcoming the negative impact of the clinical and molecular factors which originally affect its efficacy. In the era of precision oncology, other clinical and molecular factors should be taken into account when deciding the treatment strategy. According to the nomogram model, patients with lower scores, or those without concurrent mutation or only have local/pleural metastasis could benefit from single-agent EGFR-TKI regimen. Meanwhile, patients with higher scores, or those with concurrent mutations or have distant metastasis could benefit from A+T combination therapy.
Limitations should be noted in our study. Although the propensity score matching system was used to identify the patients with similar baseline clinical characteristics between the two groups, selection bias was inevitable due to the retrospective nature of our study. In addition, the genomic mutational status was exploratory and had not been verified by another technology. To verify our results, further prospective multi-center studies are recommended.
Conclusion
In conclusion, we identified that the subset of patients with EGFR-mutant advanced NSCLC with concomitant mutations in tumor suppressor genes could derive more survival benefit from the addition of bevacizumab to their EGFR-TKI regimen. Our results also suggest that the addition of bevacizumab may improve the efficacy of EGFR-TKI by overcoming the negative impact of clinical and molecular factors that affect EGFR-TKI efficacy. These observations suggest that the presence of concomitant mutation in tumor suppressor genes could potentially serve as a predictive biomarker for bevacizumab combination therapy. Our study highlighted the importance of interpreting the molecular landscape of the tumor for personalized therapy.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- A+T:
-
Bevacizumab and EGFR-TKI combination regimen
- CI:
-
Confidence intervals
- ECOG PS:
-
Eastern Cooperative Oncology Group Performance Status
- EGFR:
-
Epidermal growth factor
- HR:
-
Hazard ratio
- NGS:
-
Next-generation sequencing
- NSCLC:
-
Non-small-cell lung cancer
- ORR:
-
Objective response rate
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- T alone:
-
EGFR-TKI monotherapy regimen
- TKI:
-
Tyrosine kinase inhibitor
- TNM:
-
Tumor node metastasis status
- VEGF:
-
Vascular endothelial growth factor
- VEGFR:
-
VEGF receptor
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Noone AM, Cronin KA, Altekruse SF, Howlader N, Lewis DR, Petkov VI, et al. Cancer incidence and survival trends by subtype using data from the surveillance epidemiology and end results program, 1992-2013. Cancer Epidemiol Biomarkers Prev. 2017;26(4):632–41.
Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–42.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46.
Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–8.
Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447–e59.
Redmond KL, Papafili A, Lawler M, Van Schaeybroeck S. Overcoming resistance to targeted therapies in cancer. Semin Oncol. 2015;42(6):896–908.
Janne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–99.
Seto T, Kato T, Nishio M, Goto K, Atagi S, Hosomi Y, et al. Erlotinib alone or with bevacizumab as first-line therapy in patients with advanced non-squamous non-small-cell lung cancer harbouring EGFR mutations (JO25567): an open-label, randomised, multicentre, phase 2 study. Lancet Oncol. 2014;15(11):1236–44.
Saito H, Fukuhara T, Furuya N, Watanabe K, Sugawara S, Iwasawa S, et al. Erlotinib plus bevacizumab versus erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomised, multicentre, phase 3 trial. Lancet Oncol. 2019;20(5):625–35.
Zhou Q, Xu CR, Cheng Y, Liu YP, Chen GY, Cui JW, et al. Bevacizumab plus erlotinib in Chinese patients with untreated, EGFR-mutated, advanced NSCLC (ARTEMIS-CTONG1509): a multicenter phase 3 study. Cancer Cell. 2021;S1535-6108(21)00382–2.
Rosell R, Dafni U, Felip E, Curioni-Fontecedro A, Gautschi O, Peters S, et al. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicentre, single-arm, phase 2 trial. Lancet Respir Med. 2017;5(5):435–44.
Ninomiya T, Takigawa N, Ichihara E, Ochi N, Murakami T, Honda Y, et al. Afatinib prolongs survival compared with gefitinib in an epidermal growth factor receptor-driven lung cancer model. Mol Cancer Ther. 2013;12(5):589–97.
Schicher N, Paulitschke V, Swoboda A, Kunstfeld R, Loewe R, Pilarski P, et al. Erlotinib and bevacizumab have synergistic activity against melanoma. Clin Cancer Res. 2009;15(10):3495–502.
Naumov GN, Nilsson MB, Cascone T, Briggs A, Straume O, Akslen LA, et al. Combined vascular endothelial growth factor receptor and epidermal growth factor receptor (EGFR) blockade inhibits tumor growth in xenograft models of EGFR inhibitor resistance. Clin Cancer Res. 2009;15(10):3484–94.
Gridelli C, Rossi A, Ciardiello F, De Marinis F, Crino L, Morabito A, et al. BEVERLY: rationale and design of a randomized open-label phase III trial comparing bevacizumab plus erlotinib versus erlotinib alone as first-line treatment of patients with EGFR-mutated advanced nonsquamous non-small-cell lung cancer. Clin Lung Cancer. 2016;17(5):461–5.
Nakagawa K, Garon EB, Seto T, Nishio M, Ponce Aix S, Paz-Ares L, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(12):1655–69.
Labbe C, Cabanero M, Korpanty GJ, Tomasini P, Doherty MK, Mascaux C, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer. 2017;111:23–9.
Hou H, Qin K, Liang Y, Zhang C, Liu D, Jiang H, et al. Concurrent TP53 mutations predict poor outcomes of EGFR-TKI treatments in Chinese patients with advanced NSCLC. Cancer Manag Res. 2019;11:5665–75.
Jiang T, Zhang Y, Li X, Zhao C, Chen X, Su C, et al. EGFR-TKIs plus bevacizumab demonstrated survival benefit than EGFR-TKIs alone in patients with EGFR-mutant NSCLC and multiple brain metastases. Eur J Cancer. 2019;121:98–108.
Pander J, Gelderblom H, Guchelaar HJ. Pharmacogenetics of EGFR and VEGF inhibition. Drug Discov Today. 2007;12(23-24):1054–60.
Wang Z, Cheng Y, An T, Gao H, Wang K, Zhou Q, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. 2018;6(9):681–90.
Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49(12):1693–704.
Yu HA, Suzawa K, Jordan E, Zehir A, Ni A, Kim R, et al. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res. 2018;24(13):3108–18.
Cheng Y, Ma L, Liu Y, Zhu J, Xin Y, Liu X, et al. Comprehensive characterization and clinical impact of concomitant genomic alterations in EGFR-mutant NSCLCs treated with EGFR kinase inhibitors. Lung Cancer. 2020;145:63–70.
Hsu HC, You JF, Chen SJ, Chen HC, Yeh CY, Tsai WS, et al. TP53 DNA binding domain mutations predict progression-free survival of bevacizumab therapy in metastatic colorectal cancer. Cancers (Basel). 2019;11(8):1079.
Leslie KK, Filiaci VL, Mallen AR, Thiel KW, Devor EJ, Moxley K, et al. Mutated p53 portends improvement in outcomes when bevacizumab is combined with chemotherapy in advanced/recurrent endometrial cancer: An NRG Oncology study. Gynecol Oncol. 2021;161(1):113–21.
Schwaederle M, Lazar V, Validire P, Hansson J, Lacroix L, Soria JC, et al. VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: implications for antiangiogenesis therapy. Cancer Res. 2015;75(7):1187–90.
Acknowledgements
The authors would like to thank all the patients and their families for their cooperation and participation. We would also like to thank the other clinical practitioners, nurses, and other hospital staff of all the participating institutions for their invaluable support to our patients.
Funding
This work was funded by the Natural Science Foundation of China (grant number: 82003206), Natural Science Foundation of Hunan Province (grant numbers: 2018RS3106, 2018SK50901, kq1801102, 2019-TJ-N04, 2019JJ50357, 2019SK4010, 2020JJ5340, and 2020JJ3025), and Guangdong Association of Clinical Trials (GACT)/Chinese Thoracic Oncology Group (CTONG) and Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (grant number: 2017B030314120). The funding agencies had no role in the study design, data collection, analysis, interpretation, manuscript writing, and decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
YZ and NY: conceptualization, organization, data collection, auditing, supervision, project administration, funding acquisition, writing—reviewing, and editing. LZ, YL, LL, and XZ: data curation, methodology, formal analysis, writing—original draft preparation, writing—reviewing, and editing. QX, HY, TH, and LQ: software, validation, writing—reviewing, and editing. JL and WJ: formal analysis and visualization, writing—reviewing, and editing. AL: critical comments and suggestions, writing—reviewing and editing. LP: formal analysis, writing—reviewing, and editing. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Approval was obtained from the Hunan Cancer Hospital Institutional Review Board Committee (2017YYQ-SSB-159). All patients provided written informed consent for the use of their clinical and molecular data for research purposes.
Consent for publication
Not applicable
Competing interests
AL, LQ, and TH are employed by Burning Rock Biotech. All the other authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
The 168 panel genes list.
Additional file 2: Figure S1.
Baseline mutational profile of Training cohort A. The patients were grouped according to the treatment received; A+T, EGFR-TKI with bevacizumab; T, single-agent EGFR-TKI. Figure S2. Heat map illustrating the association between gene mutation or pathway (y-axis) and clinical characteristics, progression-free survival (PFS), and overall survival (OS) (x-axis) for single-agent EGFR-TKI group (A) and EGFR-TKI plus bevacizumab group (B). Blue indicates no statistical difference. The intensity of red color indicates the level of statistical significance with corresponding p-values indicated. Figure S3. Patients in cohort A with brain, liver, or pleural metastasis had significantly shorter progression-free survival (PFS) and overall survival (OS) with single-agent EGFR-TKI (A) but not on EGFR-TKI plus bevacizumab combination (B). Figure S4. Patients with concomitant TP53 mutation had significantly longer progression-free survival (PFS) and overall survival (OS) with EGFR-TKI plus bevacizumab combination. Kaplan-Meier curves for PFS (A) and OS (B) of patients with EGFR-mutant advanced NSCLC with concurrent TP53 mutations (TP53+, red color) or wild-type TP53 (TP53-, blue color) treated with either EGFR-TKI with bevacizumab (A+T, solid lines) or EGFR-TKI monotherapy (T-single, dashed lines). The table below summarizes the p-values. Figure S5. Patients with concomitant TP53 mutations located in exons 5-8 had significantly longer progression-free survival (PFS) and overall survival (OS) with EGFR-TKI plus bevacizumab combination. Kaplan-Meier curves for PFS (A, C) and OS (B, D) of patients with EGFR-mutant advanced NSCLC with or without concurrent TP53 mutations located between exon 5-8 (TP53_hot; A-B) or mutations that result in loss-of-function (TP53_LOF; C-D) and were treated with either EGFR-TKI with bevacizumab (A+T) or EGFR-TKI monotherapy (T-single). Kaplan-Meier curves for PFS (E, G) and OS (F, H) of patients with concomitant TP53 and RB1 mutations who were treated with either T-only (E, F) or A+T (G, H). Figure S6. Nomograms were constructed for predicting the risk of 12-month and 24-month overall survival (OS) (A). Survival analysis of the training cohort A was performed using Kaplan-Meier curves to compare the OS of patients in the T group (B) or A+T (C) group, which were further subgrouped according to the median score into low-risk (< median score) and high-risk (≥ median score). Figure S7. Tabulated summary of the hazard ratios for each molecular and clinical features calculated for the patients treated with EGFR-TKI with bevacizumab (A+T) using Cox multivariable analysis for progression-free survival (A) and overall survival (B).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, Y., Zeng, L., Zhang, X. et al. Clinical and molecular feature-based nomogram model for predicting benefit from bevacizumab combined with first-generation EGFR-tyrosine kinase inhibitor (TKI) in EGFR-mutant advanced NSCLC. BMC Med 19, 245 (2021). https://doi.org/10.1186/s12916-021-02118-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-021-02118-x