Abstract
Background
The aim of present study was to screen the novel and promising targets of curcumin in hepatocellular carcinoma diagnosis and chemotherapy.
Methods
Potential targets of curcumin were screened from SwissTargetPrediction, ParmMapper and drugbank databases. Potential aberrant genes of hepatocellular carcinoma were screened from Genecards databases. Fifty paired hepatocellular carcinoma patients’ gene expression profiles from the GEO database were used to test potential targets of curcumin. Besides, GO analysis, KEGG pathway enrichment analysis and PPI network construction were used to explore the underlying mechanism of candidate hub genes. ROC analysis and Kaplan-Meier analysis were used to evaluate the diagnostic and prognostic value of candidate hub genes, respectively. Real-time PCR was used to verify the results of bioinformatics analysis.
Results
Bioinformatics analysis results suggested that AURKA, CDK1, CCNB1, TOP2A, CYP2B6, CYP2C9, and CYP3A4 genes served as candidate hub genes. AURKA, CDK1, CCNB1 and TOP2A were significantly upregulated and correlated with poor prognosis in hepatocellular carcinoma, AUC values of which were 95.7, 96.9, 98.1 and 96.1% respectively. There was not significant correlation between the expression of CYP2B6 and prognosis of hepatocellular carcinoma, while CYP2C9 and CYP3A4 genes were significantly downregulated and correlated with poor prognosis in hepatocellular carcinoma. AUC values of CYP2B6, CYP2C9, and CYP3A4 were 96.0, 97.0 and 88.0% respectively. In vitro, we further confirmed that curcumin significantly downregulated the expression of AURKA, CDK1, and TOP2A genes, while significantly upregulated the expression of CYP2B6, CYP2C9, and CYP3A4 genes.
Conclusions
Our results provided a novel panel of AURKA, CDK1, TOP2A, CYP2C9, and CYP3A4 candidate genes for curcumin related chemotherapy of hepatocellular carcinoma.
Similar content being viewed by others
Background
Recently, the incidences of live cancer ranked fifth and ninth in male and female cancer, respectively. Its mortality ranked second and sixth in male and female cancer, respectively [1]. Besides, according to global cancer statistics 2018, China shared 46.7% of global liver cancer cases [2]. Hepatocellular carcinoma serves as the most common type of primary liver cancer, with its high-risk factors at least including metabolic liver disease, hepatitis virus infection, and alcohol abuse [3, 4]. Long-term exposure of hepatitis B/C virus would develop chronic viral hepatitis, then followed by liver cirrhosis and hepatocellular carcinoma [3, 5]. Nonalcoholic fatty liver disease or metabolic associated fatty liver disease, one typical metabolic liver disease, would develop nonalcoholic steatohepatitis, then followed liver cirrhosis and hepatocellular carcinoma [6, 7]. Alcohol abuse would induce chronic liver injury, which developed liver fibrosis and eventually progressed to hepatocellular carcinoma [8].
Earlier-stage clinical symptoms of hepatocellular carcinoma were vague or nonspecific, thus hepatocellular carcinoma patients usually were diagnosis at an intermediate and advanced stage. Surgical resection was the ideal option for earlier- stage hepatocellular carcinoma patients without cirrhosis, while transplantation was the best option for those earlier- stage hepatocellular carcinoma patients with cirrhosis [9, 10]. Systemic therapies at least include chemotherapy, immunotherapy and radiotherapy, which were strongly recommended for hepatocellular carcinoma patients at intermediate and advanced stage. However, up to date, chemoprevention and adjuvant therapy regarded as much less efficient interventions in advanced hepatocellular carcinoma treatments [9, 11].
Natural herb or peptide have exhibited antioxidant, anti-inflammatory, and anti-proliferative effects on disease treatment [12,13,14,15,16]. Accumulating evidence indicated that curcumin was a promising natural compound, which has been extensively investigated and shown multiply therapeutic activities, at least including anticancer, anti-virus, anti-arthritis, anti-amyloid, anti-oxidation, and anti-inflammatory [17]. In the molecular events of liver disease, curcumin inhibits HBV gene expression and replication via down-regulation of PGC-1α [18]. Curcumin had no effect on HCV RNA replication or viral assembly/release, but impaired virus binding and entry into human liver cells [19]. Randomized Controlled Trials showed that curcumin significantly ameliorated nonalcoholic fatty liver disease [20, 21]. Curcumin increased PPARγ to inhibit the expression of SREBP-2 and low-density lipoprotein receptor, which subsequently inactivated hepatic stellate cells, curcumin also increased SREBP-1c to promote lipid storage [22]. Thus, these findings indicated the potential therapeutic value of curcumin in protecting against liver steatosis and fibrosis. Curcumin would decrease the stemness of liver cancer stem cells by attenuating NF-κB/HDAC signaling [23]. Interestingly, curcumin suppressed stromal cell-derived factor-1/CXCR4 signaling to reduce the incidence of circulating gastric cancer cells, and subsequently decreased the risk of secondary liver cancer [24]. Thus, these previous studies highlighted that curcumin might exhibit multifunction in the initiation and progression of liver cancer.
In the present study, we performed bioinformatic analysis to screen the targets of curcumin, which would contribute to initiation and progression of hepatocellular carcinoma. Then we further validated these candidates with gene profiles of hepatocellular carcinoma patients, and tried to answer the underlying mechanism.
Methods
Screening the potential curcumin-related targets for hepatocellular carcinoma therapy
Simplified Molecular Input Line Entry Specification (SMILES) structure of curcumin was obtained from Pubchem website (https://pubchem.ncbi.nlm.nih.gov/). The SMILES structure of curcumin was used to predict the potential Homo Sapiens target from SwissTargetPrediction database and PharmMapper database (version 2017) [25, 26]. We used “curcumin” to screen its verified targets from DrugBank database [27]. Curcumin relative targets were then integrated with the above three databases and removed the repeats. We used “Hepatocellular Carcinoma” and “Hepatocellular Cancer” items to acquire the potential hepatocellular carcinoma relative target from GeneCards database [28], then we transformed their gene ID from Uniprot database (https://www.uniprot.org/) for further analysis. We next merged the curcumin relative target and the hepatocellular carcinoma relative target and picked up the overlapped candidate for further analysis.
Clinical data collection and processing
The hepatocellular carcinoma relative microarray data (GSE14520) was based on GPL3921 platform and downloaded from Gene Expression Omnibus (GEO) database by using GEOquery package (version 2.56.0) [29], the data was firstly normalized by using normalizeBetweenArrays function of limma package (version 3.44.3) [30,31,32]. We then obtained 50 fully paired gene expression matrix of normal adjacent liver tissues and hepatocellular carcinoma tissues from GSE14520. The principal component analysis (PCA) was performed to visually present the data by FactoMineR package (version 2.3) and factoextra package (version 1.0.7) [33]. The gene expression matrix and differential expression analysis were carried out by using pheatmap package (version 1.0.12).
Gene functional annotations
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed by using ClusterProfiler package (version 3.16.1) and visualized by using RColorBrewer package (version1.1-2) [34,35,36]. The Gene Ontology (GO) enrichment analysis, including Biological process (BP) analysis, Cellular component (CC) analysis, and Molecular function (MF) analysis, were performed by using ClusterProfiler package (version 3.16.1) [37], ggplot2 package (version 3.3.3), and stringr package (version 1.4.0). P-value less than 0.05 was regarded as the cutoff value of statistical significance.
Protein-protein interaction (PPI) network construction
The selected candidate gene was uploaded onto Search Tool for the Retrieval of Interacting Genes (STRING, version 11.0) databaseto predict and construct a potential PPI network [38], then network data integration, analysis, and visualization were performed by using Cytoscape software (version 3.7.1).
Expression, prognosis, and diagnosis analysis of selected genes
In order to track the dynamic expression of selected genes in initiation and progression of hepatocellular carcinoma, we downloaded the mRNA expression profile and the corresponding clinical information of hepatocellular carcinoma from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/) by TCGAbiolinks package (version 2.16.3), including normal samples (n = 44), stage I samples (n = 175), stage II samples (n = 86), stage III samples (n = 84), and stage IV samples (n = 5). Expression data of selected genes were processed by GraphPad Prism 9, and analyzed by Student’s t-test. The Kaplan-Meier survival analysis was constructed by Gene Expression Profiling Interactive Analysis (GEPIA) on-line tool [39]. The receiver operating characteristic (ROC) curve was plotted by using dplyr package (version 1.0.3) and pROC package (1.16.2) [40]. P-value less than 0.05 was regarded as the cutoff value of statistical significance.
Cell culture and treatment
HepG2.2.15 cell line was derived from HepG2 cell which was transfected full length DNA of Hepatitis B Virus, purchased from China Center for Type Culture Collection, and which was a gift from Dr. Liufeng Mao. For cell culture, HepG2.2.15 cells were maintained in complete growth Dulbecco’s Modified Eagle Medium (DMEM) containing 4.5 g/L glucose (Life Technologies, Inc., Carlsbad, CA), 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were incubated at 37 °C with 5% CO2. For cell treatment, HepG2.2.15 cells were seeded into 12-well plates at 2 × 105 cells/well overnight, then cells were treated with 1, 4, 10 μM curcumin (CSNpharm, China) for further 24 h, dimethyl sulfoxide (Beyotime, China) was used as the vehicle control.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using the RNAeasy™ Animal RNA Isolation Kit with Spin Column (Beyotime, China) according to manufacturer’s instructions. Total RNA was reversely transcribed amplified using the BeyoFast™ SYBR Green One-Step qRT-PCR Kit (Beyotime, China). All real-time PCR primers were listed as following: AURKA, 5′-CTAACGGCTGAGCTCTTGGA-3′ and 5′-GAACCGACAGGGGACTTGAC-3′. CCNB1, 5′-ACCTTTGCACTTCCTTCGGA-3′ and 5′-TGTTCTTGACAGTCCATTCACCA-3′. CDK1, 5′-GCCCTTTAGCGCGGATCTAC-3′ and 5′-AGGAACCCCTTCCTCTTCACT-3′. TOP2A, 5′-CCGTCACCATGGAAGTGTCA-3′ and 5′-TGTCTGGGCGGAGCAAAATA-3′. CYP2B6, 5′-CCTCAACCTCAACACGCTCT-3′ and 5′-TTTGGCTCGGTCATGAAGCT-3′. CYP2C9, 5′-ACCAGCTGTGCTTCATTCCT-3′ and 5′-GCACAGTGAAACATAGGAAACTCTC-3′. CYP3A4, 5′-GCTTTCCTGCACATTAAGGAGAA AT-3′ and 5′-ATGGGCAAAGTCACAGTGGAT-3′. GAPDH, 5′-AGCCTCAAGATCATCAGC-3′ and 5′-GAGTCCTTCCACGATACC-3′. Relative mRNA expression levels of target genes were normalized by comparing to GAPDH, and calculated using the 2 − ΔΔCt method [41]. Expression data of selected genes were processed by GraphPad Prism 9, and analyzed by Student’s t-test. P-value less than 0.05 was regarded as the cutoff value of statistical significance.
Cell counting kit-8 assay
Cells were seeded into 96-well plates at 10000 cells/well, after culture for 24 h, cells were treated with indicated concentrations of curcumin (CSNpharm, China) for further 48 h, and then the cell vitality was measured by the cell counting kit-8 (Beyotime, China) kit. The cellular viability (%) = OD value (treated cell) / OD value (control cell) × 100%.
Results
Potential targets of curcumin in hepatocellular carcinoma therapy
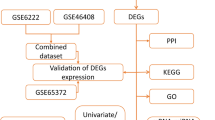
Canonical SMILES of curcumin is COC1 = C(C=CC(=C1)C=CC(=O)CC(=O)C=CC2 = CC (=C(C=C2)O)OC)O (Computed by OEChem 2.1.5, PubChem release). 3D conformer of curcumin was shown in Fig. 1A. Thirteen candidates of curcumin were acquired by Drugbank, 299 candidates of curcumin were acquired by PhamMapper, 100 candidates (Probability> 0) of curcumin were acquired by Swisstarget (Fig. 1B). After deleting the repeats candidates, we obtained 144 candidates of curcumin (Fig. 1B). Five thousand two hundred eighty-five aberrant genes were acquired by GeneCards, when they overlapped with candidates of curcumin, we here obtained 109 potential targets of curcumin in hepatocellular carcinoma therapy (Fig. 1B). KEGG pathway analysis showed that aberrant expression of these 109 potential targets significantly and mainly correlated with viral-induced carcinoma, at least including prostate cancer, hepatocellular carcinoma, pancreatic cancer, and leukemia. They also highly correlated with cytochrome P450-related drug metabolism (Fig. 1C). Biological process (BP) of GO analysis showed that these 109 potential targets mainly regulated xenobiotic-induced protein phosphorylation in long-chain fatty acid metabolic process (Fig. 1D). Cellular component (CC) of GO analysis showed that these 109 potential targets mainly regulated activities of protein kinases in the transcriptional process (Fig. 1D). Molecular function (MF) of GO analysis showed that these 109 potential targets mainly responded to activities of the transmembrane receptor protein kinase and the histone kinase (Fig. 1D).
Screening curcumin-related targets in hepatocellular carcinoma therapy. A 3D conformer of curcumin (acquired by PubChem). B Workflow of Screening curcumin-related targets in hepatocellular carcinoma therapy. C KEGG pathway analysis of 109 selected potential targets. D Biological process (BP), Cellular component (CC), and Molecular function (MF) of GO analysis of 109 selected potential targets
Validating the 109 selected potential targets in GSE14520 dataset
Gene profiles of 50 paired normal adjacent liver tissues and hepatocellular carcinoma tissues were extracted from GSE14520 dataset. Principal component analysis (PCA) showed that there was a clear distinction between normal adjacent liver tissues and hepatocellular carcinoma tissues (Fig. 2A). Correlation coefficient heatmap analysis also showed that there was a significant difference of gene profiles between the two groups. Besides, Genetic variation in normal adjacent liver tissues was smaller than those in hepatocellular carcinoma tissues (Fig. 2B). Among these 109 selected potential targets, 3 selected potential targets (BRAF, TLR9, CDK3) were missing in the GSE14520 dataset. 9 (8.5%) selected potential targets were significantly upregulated in GSE14520 array, 14 (13.2%) selected potential targets were significantly downregulated in GSE14520 dataset, the rest had no significant difference (Fig. 2C).
Identification of the 109 selected potential targets in GSE14520 dataset. A Principal component analysis of the GSE14520 dataset, each turquoise dot represented a normal adjacent liver tissue, and each tangerine triangle represented a hepatocellular carcinoma tissue. B Correlation coefficient heatmap analysis of the GSE14520 dataset. C Expression of 106 selected potential targets in the GSE14520 dataset
PPI network analysis and functional annotations of the selected potential targets
According to PPI network analysis, 105 nodes were found, and the average number of neighbors (degree distribution) was 11.505 (Fig. 3A, Supplemental Table 1). Besides, the top list of hub nodes (Degree distribution>11) were showed in Supplemental Table 2. When we merged these hub genes with differentially expressed genes in GSE14520 dataset and differentially expressed genes of hepatocellular carcinoma in GEPIA database, seven communal genes were found, they were AURKA, CDK1, CCNB1, TOP2A, CYP3A4, CYP2C9, and CYP2B6 (Fig. 3B). We then constructed the core-network of these seven hub genes and their neighbor genes (Fig. 3C and D). Our results further showed that AURKA, CCNB1 and CDK1 involved in Oocyte meiosis and maturation via regulated FoxO signaling and/or p53 signaling. Besides, CCNB1 and CDK1 were also involved in virus-related carcinogenesis. TOP2A was mainly involved in platinum drug resistance (Fig. 3E). CYP3A4, CYP2C9, and CYP2B6 mainly involved in cytochrome P450 related metabolism under physiological or pathological situations (Fig. 3F).
PPI network construction and functional annotations of the selected potential targets. A PPI network construction of the 109 selected potential targets, the red square represented the communal gene (Degree distribution≥12), the blue arrow represented the hub gene that degree distribution was greater than or equal 12, the green dot represented the hub gene that degree distribution was less than 12. B Venn diagram showed that seven communal genes were found among these indicated datasets. C Core network construction of AURKA, CCNB1, CDK1 and TOP2A. D Core network construction of CYP2B6, CYP2C9, and CYP3A4. E KEGG pathway analysis of AURKA, CCNB1, CDK1 and TOP2A. F KEGG pathway analysis of CYP2B6, CYP2C9, and CYP3A4
Clinical correlation of selected genes in hepatocellular carcinoma
Transcriptional expression profile of hepatocellular carcinoma patients’ tumor tissues and adjacent normal tissues was extracted from TCGA database. The expression of AURKA, CCNB1, CDK1, or TOP2A in each stage samples was higher than the expression of its normal samples. There was not significantly difference of expression of AURKA, CCNB1, CDK1, or TOP2A between any two different stage samples, except for the expression of CDK1 and TOP2A in stage III samples were slightly and significantly decreased while compared with stage I samples (Fig. 4A-D). The expression of CYP2B6, CYP2C9, or CYP3A4 in each stage samples were less than the expression of its normal samples. There was not significantly difference of expression of CYP2B6, CYP2C9, or CYP3A4 between any two different stage samples (Fig. 4E-G). Besides, AURKA, CCNB1, CDK1 and TOP2A positively and significantly correlated with poor prognosis of hepatocellular carcinoma, while CYP2C9 and CYP3A4 negatively and significantly correlated with poor prognosis of hepatocellular carcinoma, but there was not significant correlation between the expression of CYP2B6 and prognosis of hepatocellular carcinoma (Fig. 4H-N). ROC curves showed that the AUC of AURKA, CCNB1, CDK1, TOP2A, CYP2B6, CYP2C9 and CYP3A4 were 95.7, 98.1, 96.9, 96.1, 96.0, 97.0 and 88.0%, respectively (Fig. 4O).
Expression, prognosis, and diagnosis of selected genes in hepatocellular carcinoma. A-G The mRNA expression of AURKA, CCNB1, CDK1, TOP2A, CYP2B6, CYP2C9 and CYP3A4 in hepatocellular carcinoma patients’ tumor tissues and adjacent normal tissues. H-N Kaplan-Meier analysis of Overall Survival for hepatocellular carcinoma patients based on the expression of AURKA, CCNB1, CDK1, TOP2A, CYP2B6, CYP2C9 and CYP3A4. O ROC curve analysis for AURKA, CCNB1, CDK1, TOP2A, CYP2B6, CYP2C9 and CYP3A4 in hepatocellular carcinoma. ***p < 0.001, tumor tissues of each stage versus adjacent normal tissues; ###p < 0.001 tumor tissues of stage I versus tumor tissues of stage III
The effect of curcumin on regulating the target genes
We next investigated the effect of curcumin on regulating the target genes. Our results showed that curcumin dose-dependently decreased cellular viability in HepG2.2.15 cells, and the cellular viability remained at least 95% when the concentration of curcumin was under or equal to 10 μM (Fig. 5A). The lower dose (from 0 μM to 10 μM) of curcumin was then used for the subsequent experiments. Compared to the vehicle treatment, curcumin significantly decreased mRNA expression of AURKA, CDK1, and TOP2A (Fig. 5B-D). Curcumin had no effect on transcription of CCNB1 (Fig. 5E). Besides, higher concentrations of curcumin significantly increased mRNA expression of CYP2B6, CYP2C9, and CYP3A4 (Fig. 5F-H).
The effect of curcumin on regulating the target genes. A After treating with a series concentration of curcumin for 48 h, the cellular viability of HepG2.2.15 cells were measured by CCK8 assay. B-H After treating HepG2.2.15 cells with a series concentration of curcumin for 24 h, mRNA expression of AURKA, CDK1, TOP2A, CCNB1, CYP2B6, CYP2C9, and CYP3A4 were analyzed by real-time PCR, GAPDH served as housekeeping gene
Discussion
Liver take predominantly advantage in multi-fundamental physiological processes, at least including material metabolism, digestion, detoxification, and coagulation. Carcinoma in liver not only defected its bio-functions, but also attenuated clinical treatments and amplified the toxic and side-effects [9]. As described above, accumulating in vitro and in vivo studies have indicated that curcumin acted as a promising compound to exhibit multifunction in the prevention and treatment of liver cancer. We herein mainly focused and predicted the potential clinical effects of curcumin in hepatocellular carcinoma treatment. In the present study, we then screened and verified several novel and promising curcumin-target genes in hepatocellular carcinoma therapy via bioinformatics analysis approach.
Previous studies have showed that natural herb, at least including saffron, safranal, salvadora persica, ginger and their metabolites suppressed inflammatory, proliferative, and oxidative pathways, while triggered caspases activities and DNA instability to induced cytotoxicity and apoptosis in the liver cancer cells [42,43,44,45,46]. In the present study, seven common target genes were selected when we overlapped curcumin predicted targets, aberrant genes in initiation and progression of hepatocellular carcinoma, and differentially expressed genes of GSE14520 dataset. Previous studies showed that AURKA promoted proliferation and metastasis of hepatocellular carcinoma cells [47,48,49]. Besides, AURKA also involved in formation of secondary liver cancer [50, 51]. Blocking CDK1/PDK1/β-Catenin signaling would inhibit proliferation and EMT of hepatocellular carcinoma cells [52]. Previous studies showed that CCNB1 was upregulated to promote proliferation of hepatocellular carcinoma via decreased its negative regulators, at least including RNA-binding motif protein 43 (RBM43), miR-199a-3p, or miR-144 [53,54,55]. So far as we know, only one study showed that synthetic resveratrol-curcumin hybrid compound 4c significantly decreased the expression AURKA, AURKB, and CCNB1 in MCF-7 cells, it also significantly inhibited proliferation of MCF-7 cells, A549 cells, and HepG2 cells [56]. Otherwise, accumulating studies have shown that curcumin and its analogs significantly downregulated the expression of CDK1 to induce cell cycle arrest in various human cancers, but not in hepatocellular carcinoma [57,58,59]. In the present study, our results showed that curcumin significantly decreased the transcription of AURKA and CDK1 in HBV-transfected HepG2.2.15 cells, but not CCNB1. Otherwise, previous studies reported the dose of curcumin-induced cytotoxicity in HepG2 cells seemed to be contradictory. Lee and his college reported that no cytotoxicity was observed in HepG2 cells when curcumin was not more than 20 μM for 48 h treatment [60]. But Soni and his college reported that 10 μM curcumin sufficiently and significantly decreased the cell viability of HepG2 cells [61]. In line with Lee’ s result, our results showed that the lower dose (from 0 μM to 10 μM) curcumin has no significant effect on decreasing cellular viability of HepG2.2 cells. Taken these together, our results suggested that the lower dose curcumin treatment might not induced sufficient downregulation of AURKA or CDK1 to inhibit proliferation of hepatocellular carcinoma cells. Alternatively, curcumin-induced downregulation of AURKA and CDK1 might confer the other antitumor effects, but not inhibiting proliferation..
Previous studies showed that inhibition of AURKA would activate NF-κB signaling pathway to confer radio-resistance or chemoresistance in hepatocellular carcinoma and acute myeloid leukemia [62,63,64]. Blocking CDK1/PDK1/β-Catenin signaling would decrease stemness of cancer stem cells of hepatocellular carcinoma cells, and reverse sorafenib chemoresistance [52]. It’s reported that TOP2A was elevated in doxorubicin-resistant hepatocellular carcinoma cells, in line with this finding, the other group showed that TOP2A was elevated in doxorubicin-resistant hepatocellular carcinoma patients, TOP2A inhibitor etoposide would facilitate doxorubicin-induced cytotoxicity in primary cancer cells of hepatocellular carcinoma [65, 66]. TOP2A also conferred platinum resistance in several human cancers [67, 68]. In the present study, our results also showed that curcumin significantly decreased the transcription of TOP2A in HBV-transfected HepG2.2.15 cells. Accumulating studies have shown that curcumin exhibited therapeutic roles by facilitating the cytotoxicity of chemotherapeutic drugs and reversing their chemoresistance [69,70,71,72,73]. Thus, our results indicated that curcumin might decrease the expression of AURKA, CDK1 and TOP2A to reverse chemoresistance in hepatocellular carcinoma treatment.
Cytochrome P450 (CYP) enzymes play important roles in endogenous and xenobiotic metabolism of liver, but their roles in tumorigenesis and progression remain as a complex context [74]. Previous studies have highlighted that most CYP members, such as CYP2C9 and CYP3A4, were defected in hepatocellular carcinoma initiation and progression [75, 76]. CYP2C9 was mainly activated in the metabolism drugs, at least including the activation of cyclophosphamide and tamoxifen, and the clearance of idarubicin [74]. CYP3A4 was activated in the metabolism of procarcinogens, and contributed to the clearance of several chemotherapeutic agents, at least including cisplatin, etoposide or doxorubicin [74, 77]. CYP2B6 also correlated with metabolism of procarcinogens, but it tended to involved in the metabolic activation of anticancer prodrugs [74]. In the present study, our results further showed that curcumin significantly upregulated the expression of CYP2B6, CYP2C9 and CYP3A4. Otherwise, Previous studies have highlighted that curcumin ameliorated side-effects of chemotherapeutic drugs and exhibited hepatoprotective effects [20,21,22, 73]. Thus, the finding of us and the other group together suggested that it should pay more attention to curcumin-induced P450s on regulating the drug activation, inactivation or clearance while consumed with other certain anticancer drugs.
Conclusion
In summary, so far as we known, our results first showed that CDK1, TOP2A, CYP2C9, and CYP3A4 genes correlated to curcumin-related chemotherapy of hepatocellular carcinoma, more than just correlated to diagnosis and prognosis of hepatocellular carcinoma which had highlighted by the previous studies [76, 78]. Our result also showed that AURKA served as a new diagnosis and prognosis of hepatocellular carcinoma, even as also a potential and novel curcumin-related therapeutic target for hepatocellular carcinoma. Taken these together, the present study provided a new insight and proposal for curcumin-related hepatocellular carcinoma therapies.
Availability of data and materials
The datasets analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- AURKA:
-
Serine/Threonine-Protein Kinase Aurora-A
- CDK1:
-
Cyclin-Dependent Kinase 1
- CCNB1:
-
G2/Mitotic-Specific Cyclin-B1
- TOP2A:
-
DNA Topoisomerase 2-Alpha
- CYP2B6:
-
Cytochrome P450 2B6
- CYP2C9:
-
Cytochrome P450 2C9
- CYP3A4:
-
Cytochrome P450 3A4
- AUC:
-
Area Under the Curve
- ROC:
-
Receiver operating characteristic
- HBV:
-
Hepatitis B Virus
- PGC-1α:
-
Peroxisome Proliferative Activated Receptor Gamma Coactivator 1 Alpha
- HCV:
-
Hepatitis C Virus
- PPARγ:
-
Peroxisome Proliferator-Activated Receptor Gamma
- SREBP-2:
-
Sterol Regulatory Element-Binding Protein 2
- NF-κB:
-
Nuclear Factor Kappa-B
- HDAC:
-
Histone Deacetylase
- CXCR4:
-
Chemokine CXC Motif Receptor 4
- SMILES:
-
Simplified Molecular Input Line Entry Specification
- GEO:
-
Gene Expression Omnibus
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene Ontology
- BP:
-
Biological process
- MF:
-
Molecular function
- CC:
-
Cellular component
- STRING:
-
Search Tool for the Retrieval of Interacting Genes
- TCGA:
-
The Cancer Genome Atlas
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- BRAF:
-
B Raf Proto-oncogene Serine/Threonine Kinase
- TLR9:
-
Toll-Like Receptor 9
- CDK3:
-
Cyclin-Dependent Kinase 3
- PDK1:
-
Pyruvate Dehydrogenase Kinase Isoenzyme 1
- RBM43:
-
RNA-Binding Motif Protein 43
- CYP:
-
Cytochrome P450
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Qiu G, Jin Z, Chen X, Huang J. Interpretation of guidelines for the diagnosis and treatment of primary liver cancer (2019 edition) in China. Glob Health Med. 2020;2:306–11.
Blum HE. History and global burden of viral hepatitis. Dig Dis (Basel, Switzerland). 2016;34:293–302.
Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604.
Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68:526–49.
White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol. 2012;10:1342–1359.e1342.
Eslam M, Sanyal AJ, George J. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999–2014.e1991.
Ganne-Carrié N, Chaffaut C, Bourcier V, Archambeaud I, Perarnau JM, Oberti F, et al. Estimate of hepatocellular carcinoma incidence in patients with alcoholic cirrhosis. J Hepatol. 2018;69:1274–83.
Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–62.
Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25:74–85.
Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–53.
Hamza AA, Mohamed MG, Lashin FM, Amin A. Dandelion prevents liver fibrosis, inflammatory response, and oxidative stress in rats. J Basic Appl Zool. 2020;81(1):1–3.
El-Dakhly SM, Salama AAA, Hassanin SOM, Yassen NN, Hamza AA, Amin A. Aescin and diosmin each alone or in low dose- combination ameliorate liver damage induced by carbon tetrachloride in rats. BMC Res Notes. 2020;13(1):259.
Hamza AA, Lashin FM, Gamel M, Hassanin SO, Abdalla Y, Amin A. Hawthorn herbal preparation from Crataegus oxyacantha attenuates in vivo carbon tetrachloride -induced hepatic fibrosis via modulating oxidative stress and inflammation. Antioxidants (Basel, Switzerland). 2020;9(12):1173.
Murali C, Mudgil P, Gan CY, Tarazi H, El-Awady R, Abdalla Y, et al. Camel whey protein hydrolysates induced G2/M cellcycle arrest in human colorectal carcinoma. Sci Rep. 2021;11:7062.
Kamal H, Jafar S, Mudgil P, Murali C, Amin A, Maqsood S. Inhibitory properties of camel whey protein hydrolysates toward liver cancer cells, dipeptidyl peptidase-IV, and inflammation. J Dairy Sci. 2018;101:8711–20.
Zhou H, Beevers CS, Huang S. The targets of curcumin. Curr Drug Targets. 2011;12:332–47.
Rechtman MM, Har-Noy O, Bar-Yishay I, Fishman S, Adamovich Y, Shaul Y, et al. Curcumin inhibits hepatitis B virus via down-regulation of the metabolic coactivator PGC-1alpha. FEBS Lett. 2010;584:2485–90.
Anggakusuma, Colpitts CC, Schang LM, Rachmawati H, Frentzen A, Pfaender S, et al. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2014;63:1137–49.
Panahi Y, Kianpour P, Mohtashami R, Jafari R, Simental-Mendía LE, Sahebkar A. Efficacy and safety of phytosomal curcumin in non-alcoholic fatty liver disease: a randomized controlled trial. Drug Res. 2017;67:244–51.
Rahmani S, Asgary S, Askari G, Keshvari M, Hatamipour M, Feizi A, et al. Treatment of non-alcoholic fatty liver disease with curcumin: a randomized placebo-controlled trial. Phytother Res. 2016;30:1540–8.
Kang Q, Chen A. Curcumin suppresses expression of low-density lipoprotein (LDL) receptor, leading to the inhibition of LDL-induced activation of hepatic stellate cells. Br J Pharmacol. 2009;157:1354–67.
Marquardt JU, Gomez-Quiroz L, Arreguin Camacho LO, Pinna F, Lee YH, Kitade M, et al. Curcumin effectively inhibits oncogenic NF-κB signaling and restrains stemness features in liver cancer. J Hepatol. 2015;63:661–9.
Gu X, Zhang Q, Zhang W, Zhu L. Curcumin inhibits liver metastasis of gastric cancer through reducing circulating tumor cells. Aging. 2019;11:1501–9.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–w364.
Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–w360.
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–d1082.
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.31–31.30.33.
Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics (Oxford, England). 2007;23:1846–7.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10:946–63.
Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25(1):1–8.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–51.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–d551.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–7.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8.
Ashktorab H, Soleimani A, Singh G, Amin A, Tabtabaei S, Latella G, et al. Saffron: the golden spice with therapeutic properties on digestive diseases. Nutrients. 2019;11(5):943.
Al-Hrout AA, Chaiboonchoe A, Khraiwesh B, Murali C, Amin A. Safranal induces DNA double-strand breakage and ER-stress-mediated cell death in hepatocellular carcinoma cells. Sci Rep. 2018;8(1):1–5.
Al-Dabbagh B, Elhaty IA, Murali C, Madhoon A, Amin A. Salvadora persica (Miswak): antioxidant and promising antiangiogenic insights. Am J Plant Sci. 2018;09:1228–44.
Hamza AA, Heeba GH, Hamza S, Abdalla A, Amin A. Standardized extract of ginger ameliorates liver cancer by reducing proliferation and inducing apoptosis through inhibition oxidative stress/ inflammation pathway. Biomed Pharmacother. 2021;134:111102.
Amin A, Farrukh A, Murali C, Soleimani A, Praz F, Graziani G, et al. Saffron and its major ingredients’ effect on colon cancer cells with mismatch repair deficiency and microsatellite instability. Molecules. 2021;26(13):3855.
Li X, Xu W, Kang W, Wong SH, Wang M, Zhou Y, et al. Genomic analysis of liver cancer unveils novel driver genes and distinct prognostic features. Theranostics. 2018;8:1740–51.
Wang LL, Jin XH, Cai MY, Li HG, Chen JW, Wang FW, et al. AGBL2 promotes cancer cell growth through IRGM-regulated autophagy and enhanced Aurora A activity in hepatocellular carcinoma. Cancer Lett. 2018;414:71–80.
Chen C, Song G, Xiang J, Zhang H, Zhao S, Zhan Y. AURKA promotes cancer metastasis by regulating epithelial-mesenchymal transition and cancer stem cell properties in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;486:514–20.
Goos JA, Coupe VM, Diosdado B, Delis-Van Diemen PM, Karga C, Beliën JA, et al. Aurora kinase A (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. Br J Cancer. 2013;109:2445–52.
Nasri Nasrabadi P, Nayeri Z, Gharib E, Salmanipour R, Masoomi F, Mahjoubi F, et al. Establishment of a CALU, AURKA, and MCM2 gene panel for discrimination of metastasis from primary colon and lung cancers. PLoS One. 2020;15:e0233717.
Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY, Chan ACY, et al. Blocking CDK1/PDK1/β-catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics. 2018;8:3737–50.
Feng H, Liu J, Qiu Y, Liu Y, Saiyin H, Liang X, et al. RNA-binding motif protein 43 (RBM43) suppresses hepatocellular carcinoma progression through modulation of cyclin B1 expression. Oncogene. 2020;39:5495–506.
Jin J, Xu H, Li W, Xu X, Liu H, Wei F. LINC00346 acts as a competing endogenous RNA regulating development of hepatocellular carcinoma via modulating CDK1/CCNB1 axis. Front Bioeng Biotechnol. 2020;8:54.
Gu J, Liu X, Li J, He Y. MicroRNA-144 inhibits cell proliferation, migration and invasion in human hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int. 2019;19:15.
de Freitas Silva M, Coelho LF, Guirelli IM, Pereira RM, Ferreira-Silva G, Graravelli GY, et al. Synthetic resveratrol-curcumin hybrid derivative inhibits mitosis progression in estrogen positive MCF-7 breast cancer cells. Toxicol In Vitro. 2018;50:75–85.
Rutz J, Maxeiner S, Juengel E, Bernd A, Kippenberger S, Zoller N, et al. Growth and proliferation of renal cell carcinoma cells is blocked by low curcumin concentrations combined with visible light irradiation. Int J Mol Sci. 2019;20(6):1464.
Su CC, Lin JG, Chen GW, Lin WC, Chung JG. Down-regulation of Cdc25c, CDK1 and cyclin B1 and up-regulation of Wee1 by curcumin promotes human colon cancer Colo 205 cell entry into G2/M-phase of cell cycle. Cancer Genomics Proteomics. 2006;3:55–61.
Xia YQ, Wei XY, Li WL, Kanchana K, Xu CC, Chen DH, et al. Curcumin analogue A501 induces G2/M arrest and apoptosis in non-small cell lung cancer cells. Asian Pac J Cancer Prev. 2014;15:6893–8.
Lee SC, Jee SC, Kim M, Kim S, Shin MK, Kim Y, et al. Curcumin suppresses the lipid accumulation and oxidative stress induced by benzo[a]pyrene toxicity in HepG2 cells. Antioxidants (Basel, Switzerland). 2021;10(8):1314.
Soni VK, Mehta A, Ratre YK, Chandra V, Shukla D, Kumar A, et al. Counteracting action of curcumin on high glucose-induced chemoresistance in hepatic carcinoma cells. Front Oncol. 2021;11:738961.
Shen ZT, Chen Y, Huang GC, Zhu XX, Wang R, Chen LB. Aurora-a confers radioresistance in human hepatocellular carcinoma by activating NF-κB signaling pathway. BMC Cancer. 2019;19:1075.
Zhang K, Chen J, Chen D, Huang J, Feng B, Han S, et al. Aurora-A promotes chemoresistance in hepatocelluar carcinoma by targeting NF-kappaB/microRNA-21/PTEN signaling pathway. Oncotarget. 2014;5:12916–35.
Wei TW, Wu PY, Wu TJ, Hou HA, Chou WC, Teng CJ, et al. Aurora a and NF-κB survival pathway drive chemoresistance in acute myeloid leukemia via the TRAF-interacting protein TIFA. Cancer Res. 2017;77:494–508.
Pang E, Hu Y, Chan KY, Lai PB, Squire JA, Macgregor PF, et al. Karyotypic imbalances and differential gene expressions in the acquired doxorubicin resistance of hepatocellular carcinoma cells. Lab Invest. 2005;85:664–74.
Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–52.
Ferrandina G, Petrillo M, Carbone A, Zannoni G, Martinelli E, Prisco M, et al. Prognostic role of topoisomerase-IIalpha in advanced ovarian cancer patients. Br J Cancer. 2008;98:1910–5.
Ito F, Furukawa N, Nakai T. Evaluation of TOP2A as a predictive marker for endometrial cancer with taxane-containing adjuvant chemotherapy. Int J Gynecol Cancer. 2016;26:325–30.
Najafi M, Mortezaee K, Rahimifard M, Farhood B, Haghi-Aminjan H. The role of curcumin/curcuminoids during gastric cancer chemotherapy: a systematic review of non-clinical study. Life Sci. 2020;257:118051.
Soni VK, Shukla D, Kumar A, Vishvakarma NK. Curcumin circumvent lactate-induced chemoresistance in hepatic cancer cells through modulation of hydroxycarboxylic acid receptor-1. Int J Biochem Cell Biol. 2020;123:105752.
Ashrafizadeh M, Zarrabi A, Hashemi F, Moghadam ER, Hashemi F, Entezari M, et al. Curcumin in cancer therapy: a novel adjunct for combination chemotherapy with paclitaxel and alleviation of its adverse effects. Life Sci. 2020;256:117984.
Khatoon E, Banik K, Harsha C, Sailo BL, Thakur KK, Khwairakpam AD, et al. Phytochemicals in cancer cell chemosensitization: current knowledge and future perspectives. Semin Cancer Biol. 2020;S1044-579X(20)30150–4. https://doi.org/10.1016/j.semcancer.2020.06.014.
Gupta N, Verma K, Nalla S, Kulshreshtha A, Lall R, Prasad S. Free radicals as a double-edged sword: the cancer preventive and therapeutic roles of curcumin. Molecules (Basel, Switzerland). 2020;25(22):5390.
Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25:1679–91.
Yan T, Lu L, Xie C, Chen J, Peng X, Zhu L, et al. Severely impaired and dysregulated cytochrome P450 expression and activities in hepatocellular carcinoma: implications for personalized treatment in patients. Mol Cancer Ther. 2015;14:2874–86.
Braeuning A, Schwarz M. Regulation of expression of drug-metabolizing enzymes by oncogenic signaling pathways in liver tumors: a review. Acta Pharm Sin B. 2020;10:113–22.
Goldstein I, Rivlin N, Shoshana OY, Ezra O, Madar S, Goldfinger N, et al. Chemotherapeutic agents induce the expression and activity of their clearing enzyme CYP3A4 by activating p53. Carcinogenesis. 2013;34:190–8.
Gao X, Wang X, Zhang S. Bioinformatics identification of crucial genes and pathways associated with hepatocellular carcinoma. Biosci Rep. 2018;38(6):BSR20181441.
Acknowledgments
Not applicable.
Funding
This work was supported by National Key Clinical Specialty Construction Project (Clinical Pharmacy) and High Level Clinical Key Specialty (Clinical Pharmacy) in Guangdong Province (YWYH-2021-206), Natural Science Foundation of Guangdong Province (2018A030313639), Guangdong Basic and Applied Basic Research Foundation (2020A1515110615), Guangdong Medical Research Foundation (B2020043), Doctoral Scientific Research Funding of The First Affiliated Hospital of Guangdong Pharmaceutical University (KYQDJF201801, KYQDJF201807), Scientific Developing Research Funding of Meizhou People’s Hospital (PY-C2019018). All funders had no role in the design of the study and collection, analysis, interpretation of data and in writing the manuscript, the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Naihua Liu conceived and designed the study. Tingting Yang performed the experiments, Tingting Yang, Yibiao Chen, Jinyuan Li, Hong Liu performed bioinformatics analysis. Hong Liu prepared and Naihua Liu submitted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was carried out in accordance with the Helsinki Declaration. The study had not involved in any human or animal study.
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest in this work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplemental Table 1.
Simple parameters of PPI network analysis. Supplemental Table 2. The top list of hub nodes in PPI network (Degree distribution≥12).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, T., Chen, Y., Xu, J. et al. Bioinformatics screening the novel and promising targets of curcumin in hepatocellular carcinoma chemotherapy and prognosis. BMC Complement Med Ther 22, 21 (2022). https://doi.org/10.1186/s12906-021-03487-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-021-03487-9