Abstract
Background
Herbal medicines are fast gaining popularity. However, their acceptability by modern practitioners is low which is often due to lack of standardization. Several approaches towards standardization of herbals have been employed. The current study attempted to recognize key peaks from 1H NMR spectra which together would comprise of a spectral fingerprint relating to efficacy of Psidium guajava (guava) leaf extract as an antidiarrhoeal when a number of unidentified active principles are involved.
Methods
Ninety samples of guava leaves were collected from three locations over three seasons. Hydroalcoholic (water and ethanol, 50:50) extracts of these samples were prepared and their 1H NMR spectra were acquired. Spectra were also obtained for quercetin, ferulic acid and gallic acid as standards. Eight bioassays reflecting different stages of diarrhoeal pathogenesis were undertaken and based on pre-decided cut-offs, the extracts were classified as ‘good’ or ‘poor’ extracts. The bioactivity data was then correlated with the 1H NMR profiles using Regression or Orthogonal Partial Least Square-Discriminant Analysis (OPLS-DA).
Results
OPLS-DA showed seasonal and regional segregation of extracts. Significant models were established for seven bioassays, namely those for anti-bacterial activity against Shigella flexneri and Vibrio cholerae, adherence of E. coli, invasion of E. coli and S. flexneri and production and binding of toxin produced by V. cholerae. It was observed that none of the extracts were good or bad across all the bioassays. The spectral analysis showed multiple peaks correlating with a particular activity. Based on NMR and LC-MS/MS, it was noted that the extracts contained quercetin, ferulic acid and gallic acid. However, they did not correlate with the peaks that segregated extracts with good and poor activity.
Conclusions
The current study identified key peaks in 1H NMR spectra contributing to the anti-diarrhoeal activity of guava leaf extracts. The approach of using spectral fingerprinting employed in the present study can thus be used as a prototype towards standardization of plant extracts with respect to efficacy.
Similar content being viewed by others
Background
In recent years, phytomedicines have been gaining importance not only in developing countries like India and China but also in the developed countries like USA and Germany. Although their popularity amongst the population is rising, their acceptability by conventional medical practitioners is yet not satisfactory. One of the major reasons for this is the lack of data related to herbal medicine similar to that available for allopathic medicines, especially related to their standardization [1]. An additional factor for the lower acceptance of phytomedicines by the modern practitioners is the restricted acceptance of the holistic concept of herbal medicines which is contrary to the principle of single compound for single therapeutic activity in modern medicine [2].
This holistic approach of herbal medicines is often related to the synergistic action of its constituents. This makes standardization of extracts difficult and is therefore a neglected area leading to lack of uniform efficacy. The identification and quantification of a minimum of four phytoconstituents from a given extract meets the requirement for standardization for phytopharmaceuticals [3]. However, identification of biomarkers in the absence of knowledge of active principles may not be the most suitable approach towards standardization.
Metabolomics is fast becoming the approach of choice across broad fields of sciences as it gives insights into the chemical nature of biological material. In the field of medicinal plant research, the metabolomes of plants are a valuable resource for the evidence-based development of new phytotherapeutics and nutraceuticals. The most commonly used chromatographic techniques for metabolic profiling are Nuclear Magnetic Resonance (NMR) and Mass Spectroscopy (MS). Currently hyphenated techniques like Liquid Chromatography-Mass Spectroscopy (LC-MS) and Gas-Chromatography-Mass Spectroscopy (GC-MS) have also been used for their robustness and higher sensitivity of detection [1]. NMR-based studies due to its reproducibility have been undertaken for plant metabolites grown in varying controlled environments [4] or linkage of one or two activities to the metabolic profile of the extracts [5, 6]. Another application of this has been in quality control [7].
The current study used 1H NMR metabolic profiling and its correlation to anti-diarrhoeal activity of Psidium guajava (guava) leaf extracts, in an attempt to develop a prototype for standardization of extracts. The study was designed to demonstrate the utility of a spectral fingerprint without relying on identification of the active/marker compounds. The choice of guava leaves was on the basis of our previous work which established its anti-diarrheal activity [8]. Leaves of guava have been used as a traditional remedy for gastrointestinal disorders and are rich in phytoconstituents. The presence of several compounds such as sesquiterpene, saponins, sterols, triterpenoid, phenolics, coumarins, alkaloids and carotenoids has been reported in guava leaves [9,10,11]. Phenolics especially flavonoids are abundantly present in the leaves. Common polyphenolics found in guava leaves are protocatechuic acid, caffeic acid, gallic acid, ferulic acid and quercetin [12,13,14]. Though different types of extracts contain diverse types of compounds in varying amounts, Arya et al., reported that ethanolic and hydroalcoholic extracts of guava leaves contain higher quantities of phytochemicals [15]. Quercetin, is the most abundant flavonoid reported to be present in the leaves [16]. Interestingly, quercetin has been documented to be one of the active phytoconstituents of guava leaves and is reported to possess different pharmacological properties including antidiarrhoeal action [17,18,19].
For the present study, guava leaves were collected in different seasons from different locations and their 1H NMR profile were acquired. Eight bioassays representing important stages in diarrhoeal pathogenesis were undertaken with each collection and the results were correlated with the 1H NMR profiles using mathematical models. This work is one of the few studies in metabolomics, which has attempted to correlate the activity of multiple bioassays to 1H NMR signals in plant extracts obtained from an open field environment.
Methods
Plant material, preparation of extract
Mature leaves of the Sardar variety of guava were used for the study. This variety is widely cultivated in the state of Maharashtra, India. Leaves were collected from farms following verbal permission from the owners. Ninety samples of guava leaves were collected from individual trees; 10 from each of the three regions (Shirwal = W, Rahata = R and Dapoli = Da) in over three seasons (B = May 2013; C=October 2013 and D = March 2014). The leaves were authenticated by Dr. P. Tetali, an ethnobotanist. Individual voucher numbers were not obtained since this is a cultivated variety. However, a representative sample from one regional collection was deposited at the herbarium at Naoroji Godrej Centre for Plant Research (NGCPR, Shirwal, Maharashtra) under herbarium number NGCPR 712. All leaf samples were shade dried and powdered. A hydro-alcoholic (water and ethanol, 50:50) extract of each sample was prepared, lyophilized and the dried powdered extracts were then stored at − 80 °C. For all bioassays, extracts were reconstituted in distilled water and for 1H NMR in Deuterated water (D2O) and then used.
Cell culture
HEp-2 (human laryngeal epithelial) cell line was procured from the National Centre for Cell Sciences, Pune, India. The cell line was routinely cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Bio-west), at 37 °C in a 5% CO2 atmosphere and maintained by passaging every 3–4 days.
Bacterial strains used
Five bacterial strains a) enteropathogenic Escherichia coli (EPEC) strain B170, serotype 0111:NH (obtained from Centre for Disease Control, CDC, Atlanta); b) enterotoxigenic E. coli, heat labile toxin producer (ETEC) strain B831–2, serotype unknown (obtained from CDC); c) enteroinvasive E. coli (EIEC) strain EI34, serotype 0136:H- (kindly gifted by Dr. J. Nataro, Veterans Affairs Medical Centre, Maryland, USA); d) Vibrio cholerae Ogawa, serotype 01 (kindly gifted by Dr. S. Calderwood, Massachusetts General Hospital, Boston, USA) and e) Shigella flexneri M9OT, serotype 5 (kindly gifted by Dr. P. Sansonetti, Institut Pasteur, France) were used.
1H nuclear magnetic resonance - based (NMR) spectroscopy
The 1H NMR spectra were acquired at National Facility for High-Field NMR at Tata Institute of Fundamental Research (TIFR), Mumbai. 10 mg of lyophilized hydro-alcoholic extract of guava leaf was dissolved in 600 μl of D2O containing 0.1 mM solution of 3-(Trimethylsilyl)-propionic acid sodium salt (TSP, Sigma Aldrich) as reference. The samples were vortexed for 1 min, centrifuged and then transferred to a 5-mm NMR tube (Norell, NJ, USA). 1H NMR spectra were recorded for all the 90 samples. Spectra were also acquired for three compounds commonly found in guava leaves as standards. These were quercetin (purchased from Sigma), ferulic acid and gallic acid (both kindly gifted by Prof. KS. Laddha, Institute of Chemical Technology, Mumbai). 10 mg of the compound was dissolved in 600 μl D2O containing 0.1 mM TSP for spectral acquisition.
1D-NMR were recorded at room temperature (300 K) on Bruker Avance 800 MHz. Each spectrum was recorded for 256 scans using the noesygppr1d pulse sequence with an acquisition time 0.82 s, relaxation delay of 3 s and spectral width of 10,000 Hz. The FID (free induction decay) were zero-filled to 32 K data points for processing.
The raw data of the 1H NMR spectra acquired from the Bruker instrument was baseline corrected, phased and calibrated using the TOPSPIN Software (Version 3.6.0). The 1H NMR data post processing was imported into MATLAB software (Version R2014a) and all the spectra were aligned using a recursive segment-wise peak alignment method [20]. Regions showing signals for water were removed prior to multivariate statistical analysis. These spectra were exported from MATLAB in a text file format and bioassay results derived from the same samples were added to the dataset before importing into SIMCA (Version 14.1).
Liquid chromatography–mass spectrometry (LC- MS/MS) analysis
To check the presence of quercetin, ferulic and gallic acid used as standards in 1H NMR, LC–MS/MS analysis was outsourced to Shobhaben Pratapbhai Patel School Of Pharmacy & Technology Management, SVKM’s Narsee Monjee Institute of Management Studies, Mumbai. The analysis was executed in a Shimadzu 8040 (Kyoto, Japan) instrument attached with a UV detector and a binary pump (G4220B). An electrospray ionization was the ionization source. Extract (five microliters, 1000 PPM in methanol) was injected into a Thermofisher C8 column (5 μm × 150 mm × 4.6 mm) with the oven temperature set to 30 °C. The mobile phase comprised of aqueous 0.1% formic acid (solvent A) and acetonitrile (solvent B) run with a gradient program. The standardized gradient method was set as time (min)/% B–0/5; 25/80; 28/80; 40/5; 45/5 with stop time at 45.0 min to ensure elution of all compounds. The flow-rate was set to 1 mL/min. The MS used was Triple Quadrupole in positive and negative ion modes. The optimized MS/MS parameters were: Gas temperature: 250 °C; Gas flow: 3 mL/min; nebulizer: 35 psi; MS range: 100–1000 m/z. LabSolutions® software was used for data acquisition. The name of phytocompound was assigned by comparing their mass and fragmentation patterns.
Bioassays
All methods described below have been well established and validated earlier using suitable positive controls [8]. These included ofloxacin, lactulose and gallic acid for antibacterial activity, effect on bacterial colonization and effect on bacterial toxins respectively.
Determination of concentration of extract used and establishing cut off limits
For each assay, pilot experiments were undertaken to determine an appropriate concentration which could give a wide spectrum of activity. All the extracts were then tested in the presence of the concentration of the guava extract finalized. Cut offs were set empirically to identify extracts with good (higher reduction/inhibition of the parameter) and poor (minimal reduction/inhibition of the parameter) activity. Values that were between the two cut offs were defined as intermediates.
The extract concentration used and the cut off values varied for each parameter and have been stated accordingly.
Antibacterial activity
The antibacterial activity was carried out for S. flexneri and V. cholerae and determined by the agar dilution method [21]. The bacterial strains were plated on Mueller Hinton agar (Himedia laboratories, India) alone (as control) and at concentrations of 150 μg/ml and 600 μg/ml (wt/vol) of the reconstituted extract (as test) for S. flexneri and V. cholerae respectively. Two independent experiments were carried out. In each experiment, triplicate plates were used for control as well as the extract. Data was expressed as percent viability and number of colony forming units (cfu) in the control was taken as 100%.
Effect on bacterial colonization
Effect on adherence
The effect on the adherence of E. coli B170 to HEp-2 cells was assayed as described previously [22]. Briefly, HEp-2 cells cultured on glass coverslips for 48 h were infected with a log phase culture (5 × 107/ml) of the bacteria in DMEM in the absence of the extract (control) and presence of 20 μg/ml the extract (test) and incubated for 3 h. Non-adherent bacteria were washed off, the coverslips fixed and stained with toluidine blue stain (0.1% w/v). HEp-2 cells having typical EPEC microcolonies and/or > 5 adherent bacteria were counted under a light microscope. Two independent experiments were carried out. In each experiment, duplicate coverslips were set up for control and the extract. Data was expressed as percent adherence wherein the HEp-2 cells from the control was taken as 100%.
Effect on invasion
The effect on invasion of E. coli EI34 and S. flexneri to epithelial cells was studied as described elsewhere [23]. Briefly, a 24 h culture of HEp-2 cells grown in a 96-well tissue culture plate was infected with log phase culture (108/ml) of the bacteria in DMEM alone (control) and in the presence of 20 μg/ml of the extract (test) and incubated for 2 h. Following incubation, the extracellular bacteria were washed off and the cells further incubated with DMEM containing gentamicin (100 μg/ml) for 2 h. Thereafter, the medium containing gentamicin was washed off and cells lysed with chilled distilled water. The released intracellular bacteria were enumerated by plating on nutrient agar. Two independent experiments were carried out. In each experiment, triplicate wells were set up for control and the extract. Data was expressed as percent viability and cfu from the control was taken as 100%.
Effect on bacterial enterotoxins
Effect on E. coli heat labile toxin (LT) and cholera toxin (CT)
LT, which is localized in the cell membrane of E. coli B831–2 was obtained by lysing the bacterial cells with polymyxin B sulphate (1 mg/ml) whereas CT, which is released extracellularly, was obtained as a culture supernate of V. cholerae. To determine the effect of the extract on LT production, the E. coli cell lysate after polymyxin treatment following growth in absence of the extract (control) and in presence of 100 μg/ml extract (test) were used for the enzyme linked immunosorbent assay (ELISA). Similarly, for effect on CT production the culture supernatant of V. cholerae following growth in absence of the extract (control) and in presence of 75 μg/ml extract (test) served as the toxin in the ELISA as described below. The concentration of the extract to check the effect of the extract on the binding of the released CT was 50 μg/ml extract.
LT and CT were assayed by the ganglioside monosialic acid enzyme linked immunosorbent assay (GM1-ELISA) [24]. Briefly, the toxins (for production of LT/CT) were added to ELISA plates pre-coated with 1.5 μmol/ml of GM1. Anticholera toxin (1:300) and peroxidase labeled swine anti-rabbit immunoglobulin (1:200) were used as primary and secondary antibodies respectively. Orthophenylene diamine was used as the substrate. The intensity of the color thus developed was read at 492 nm in an ELISA plate reader. The protocol used for studying the effect of the extract on binding of CT was similar, except that the released CT was mixed with PBS (control) or the extract (test) before addition to the GM1 in the ELISA plate.
Two independent experiments were carried out. In each experiment, control and test wells were set up in triplicate. Data was expressed as percent toxin produced/bound and optical density of the control was taken as 100%.
Presentation of data and statistical analysis
Bioassays
The data for each of the assay have been expressed as the Mean ± Standard deviation of the percentage values from two independent experiments. The percentage for each individual experiment was obtained using the formula {(C or T)/C} × 100; C being the mean value of the readings of control group and T being the mean value obtained in presence of the extract. Thus, the value of control is 100% and those of the test are percentages relative to control.
1H NMR spectral data analysis
The spectral data were analysed using unsupervised Principal Component Analysis (PCA) and supervised Orthogonal Partial Least Square-Discriminant Analysis (OPLS-DA) in SIMCA (14.0).
Statistical link between the variation in biologic activity and the biochemical composition of the extracts was established using OPLS-Regression Analysis (OPLS-RA). Additional OPLS-DA analysis was carried out to observe the metabolic differences between the extracts with ‘good’ bioactivity and that with ‘poor’ bioactivity.
Apart from using different subsets of data for modeling, OPLS-RA was considered over the other models, since it considers actual values of bioassays. Failure of development of a significant model for certain bioassays using the supervised methods as well as the linear regression analysis led to exploration using Kruskal-Wallis test, a non- parametric model.
Further spectral signals correlating with the bioassay activities, were obtained from the S-plots of the significant models. Those signals that had correlation coefficient values above 0.6 were considered for establishment of the relationship between biological activity and 1H NMR profiles. Since SIMCA identified the different peaks based on the numerical values of the correlation coefficient there was a need to visualize the same in the graphical format. Hence MATLAB was used to overlap the spectra and each of the regions defined by the S-plot in SIMCA were visually checked to determine whether more than 60% of the spectra exhibiting either ‘good’/ ‘poor’ biological activity, used for the development of the model, showed greater height (intensity) with respect to the other spectra.
Results
A representative 1D 1H-NMR acquired for 90 guava extracts is depicted in Fig. 1 and representative 1H NMR spectra for seasonal and regional differentiation have been provided as Additional data (Supplementary Fig. S1 and Fig. S2 respectively). The 1H NMR spectra of quercetin, ferulic acid and gallic acid have also been given as Additional data (Supplementary Fig. S3, S4 and S5 respectively). Table 1 depicts the common ppm for standard compounds as compared to peaks in spectra of the representative guava leaf extracts presented as Additional data (Supplementary Figs. S1, S2, S3, S4, S5). As seen from this table, these three compounds were present. LC-MS/MS data also supported the presence of these three compounds (Table 2). The fragmentation patterns from LC-MS/MS have been provided as Additional data (Supplementary Figs. S6, S7, S8, S9, S10).
Environmental influence
Seasonal differentiation
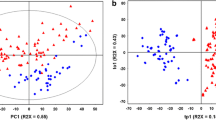
The PCA scores plot (Fig. 2) showed clear clustering based on the three seasons in which the samples were collected. The biggest variation between months May (B) and October (C), and March (D) was observed along the first principal component (PC1), whereas samples from May were separated from the other two seasons along the PC2. The pair-wise comparison were carried out using OPLS-DA analysis and statistically significant models were observed in May vs. October (R2X = 0.10; Q2(cum) = 0.79; p = 1.5 × 10− 013; Fig. 2b), October vs. March (R2X = 0.17; Q2(cum) = 0.748; p = 1.64 × 10− 14; Fig. 2c) and May vs March (R2X = 0.1; Q2(cum) = 0.77; p = 4.21 × 10− 14; Fig. 2d).
Regional differentiation
The PCA scores plot (Fig. 3) showed clear clustering based on the three regions from which the samples were collected. The biggest variation between regions Rahata (R) and Dapoli (Da), and Shirwal (W) was seen along the first principal component (PC1), whereas samples from Dapoli (Da) were separated from other two regions along PC2. The pair-wise comparison were carried out using OPLS-DA analysis and statistically significant models were observed in Shirwal vs Rahata (R2X = 0.09; Q2(cum) = 0.552; p = 8 × 10− 7; Fig. 3b), Rahata vs Dapoli (R2X = 0.13; Q2(cum) = 0.757; p = 3.32 × 10− 13; Fig. 3c) and Shirwal vs Dapoli (R2X = 0.1; Q2(cum) = 0.68; p = 2.75 × 10− 10; Fig. 3d).
The differences observed between the samples from different seasons and regions could be the effect of changes in the environmental conditions such as precipitation, intensity of sunlight and temperature. Similar results have been shown earlier by others [25, 26].
Correlation of 1H NMR spectral data with the biological activity
Correlation of the biological activity results (presented as Additional file Tables S1, S2, S3, S4, S5, S6, S7, S8) with the metabolomic profile of the extracts was undertaken.
The ppm values responsible for the significant differences, as identified from the models established for each bioassay is discussed below. Table 3 consolidates the ppm values of the prominent signals identified for individual bioassays.
Antibacterial activity against S. flexneri
The antibacterial activity of the extract ranged from 0 to 137% as compared to the control. Cell viability ≤40% or ≥ 90% were considered as effective or absence of inhibition of the growth. Based on this criterion, 21 extracts were regarded as good, while 58 extracts were poor and remaining 11 extracts were intermediates (Additional data - Table S1).
The relationship between the metabolites and the antibacterial activity of the extract against S. flexneri was attempted using the regression model. The inclusion of all 90 samples did not develop a significant model. A subsequent OPLS model was established based on the samples exhibiting high and low inhibition. This model including 21 good activity extracts and 58 poor activity extracts showed a significant p-value of 4.73 × 10− 10; R2X = 0.12; Q2 (cum) = 0.561 (Fig. 4). A total of 63 signals had absolute correlation coefficient values greater than 0.6 in the S-plot, as being responsible for the differentiation between the high and low activity samples. Spectra of all the samples used for the development of the model were overlapped. Thirty six of the 63 signals identified by the S-plot, correlated with poor activity samples while 16 signals belonged to samples with good activity (Table 3).
Antibacterial activity against E. coli
E. coli was highly resistant and even at an extract concentration of 1 mg/ml the bacterial growth was only partially inhibited (data not shown). Hence in the absence of marked inhibitory effect, further antibacterial assays and the subsequent metabolomic data analysis with E. coli was not undertaken.
Antibacterial activity against Vibrio cholerae
The antibacterial activity of the extract ranged from 7 to 138% as compared to the control. For this assay, percent viability ≤40% was considered as greater inhibition of growth/ ‘good activity’ and percent values ≥80% was taken as absence of inhibition/ ‘poor activity’. Thus seven extracts were good, 45 extracts were poor and the remaining 38 extracts were intermediates (Additional data - Table S2).
The correlation between antibacterial activity of the extract against V. cholerae and the 1H NMR profiles was established using OPLS regression analysis (p-value of 0.03; R2X = 0.101; Q2 (cum) = 0.151; Fig. 5). The S-plot of this model identified 10 signals that differentiated the extracts based on their activity. Out of the 10 signals identified by the S-plot, in seven the intensity of the signals was higher in spectra of samples with poor activity and remaining 3 signals showed a mix trend (Table 3).
Effect on bacterial adherence
It was noted that the percent bacterial adherence in presence of the extract ranged from 0 to 99% as compared to the control. For this assay, bacterial adherence ≤30% was taken as good inhibitory activity and ≥ 60% was taken as an indicator of poor activity. Hence 25 extracts had good activity, 19 extracts had poor activity and 46 extracts had intermediate activity (Additional data - Table S3).
A significant OPLS-DA model with a p-value of 0; R2X = 0.28; Q2(cum) = 0.564, was successfully developed by considering 21 good activity extracts and 14 poor activity samples after exclusion of confounders (4 good and 5 poor activity extracts, Fig. 6). In the S-plot, the model identified 39 signals that differentiated the samples based on activity; only 12 peaks correlating with poor activity could be identified (Table 3).
Effect on bacterial invasion
EIEC
The percent bacterial invasion in presence of the extract ranged from 7 to 73% as compared to the control. For this assay, percent invasion ≤20% was taken as greater inhibition of bacterial invasion and percent values ≥40% was considered to be absence of inhibition. Hence 22 extracts were considered as good, 13 extracts were poor and 55 extracts had intermediate activity (Additional data - Table S4).
Numerous attempts were made to derive a significant model. Models based on inclusion of all the samples, only the samples showing good and poor activity and even the regression analysis did not show significance. Hence based on the OPLS-DA plot derived from the good versus the poor activity model, certain borderline samples (those in the overlapping region between the good and the poor activity) were omitted (good-7; poor-4). The new set of samples (good-15; poor-9) thus derived were reanalysed and a significant OPLS-DA model with a p-value of 0 was established (Fig. 7). The other parameters of this model were R2X = 0.195 and Q2(cum) = 0.678. In the S plot, the model developed identified 65 signals responsible for differentiation out of which 25 signals showed higher intensities in samples with good activity. The remaining could not be assigned to any group since they were equally present in both (Table 3).
S. flexneri
The percent bacterial invasion in presence of the extract ranged from 2 to 140% as compared to the control. For this assay, percent invasion ≤20% was taken as greater inhibition of bacterial invasion and percent values ≥40% was considered as absence of inhibition. Thus, 28 extracts had good activity, 19 extracts had poor activity and remaining 43 were intermediates (Additional data - Table S5).
A similar approach as that applied to analyse the data for invasion by E. coli into HEp-2 cell lines was applied to this activity data set. The number of omitted samples were 12 (good-7; poor-5) and a significant OPLS-DA model was generated with 35 samples (good-21; poor-14). A p-value of 0, R2X = 0.137 and Q2 (cum) = 0.507, was established (Fig. 8). The model identified 75 signals as those responsible for the differentiation out of which in 14 signals, samples with good activity showed higher intensities (Table 3). None of the signals correlated with samples with poor activity. The remaining peaks comprised of a mixture of good and poor activity samples.
1
Effect on CT production
The toxin production in presence of the extract, ranged from 30 to 83% as compared to the control. For this assay, values ≤40% was considered as greater inhibition in toxin production and values ≥60% was taken as absence of inhibition. Thus 35 extracts were good, 22 extracts were poor and 33 had intermediate activity (Additional data - Table S6).
A significant regression model with a p-value of 0, R2X = 0.139 and Q2 (cum) = 0.183, was developed using all 90 samples (Fig. 9). Out of the 52 signals identified by the S-plot as those responsible for differentiation, only 23 signals could be assigned to extracts with good activity. The remaining peaks comprised of extracts with both good and poor activity (Table 3).
Effect on CT binding
The toxin binding in presence of the extract, ranged from 27 to 72% as compared to the control. For this assay, values ≤40% was taken as indicator of greater reduction in toxin binding and values ≥50% was considered as absence of activity. Thus 42 extracts had good activity, 27 extracts had poor activity and 21 were in the intermediate range (Additional data - Table S7).
An OPLS-DA model with a significant p-value of 1.15 × 10− 6, R2X = 0.07 and Q2(cum) = 0.53, including 42 extracts with good activity and 27 with poor activity was developed (Fig. 10). Only two signals got identified as those responsible for the differentiation and in both the extracts with good activity showed a higher intensity (Table 3).
Effect on LT production
The toxin production in presence of 100 μg/ml extract, ranged from 27 to 102% as compared to the control (toxin production in media alone was considered as 100%). Values ≤40% indicate greater reduction in toxin production and values ≥60% were considered to have no effect on toxin production. Thus 22 extracts had good activity, 35 extracts had poor activity and 33 extracts were in the intermediate range (Additional data - Table S8).
All methods listed earlier were tried but a successful model could not be developed.
Discussion
The aim of this study was to develop a prototype for standardization of crude extracts using the anti-diarrhoeal activity of guava leaves as an example. Diarrhoea was selected due to the high mortality rates, increased incidence of antibiotic resistance and its widespread prevalence in developing countries. Diarrhoea is the eighth leading cause of mortality accounting for over 1.6 million deaths [27]. Increased antibiotic resistance in diarrhoeal pathogens has been reported [28,29,30,31,32] necessitating the development of alternatives. It is envisaged that medicinal plants can fulfil this niche.
Plant extracts are complex mixtures and contain a number of compounds. Some of these may be present in small amounts but nevertheless contribute to efficacy which often may be through some synergistic action with other constituents [33]. Hence as a holistic approach, the total profiling of an extract in the form of a representative fingerprint is required. Fingerprint analysis or characteristic profiling has been accepted by World Health Organization (WHO) and other organizations (Food and Drug Administration, FDA; State Food and Drug Administration of China, SFDA and European Medicines Agency, EMA) as a methodology for the quality control of herbal samples [34,35,36,37]. Fingerprinting is considered distinctive and forms a benchmark for a particular extract especially when the identity of the active principle(s) is unknown [38]. Hence it is in tune with the concept that the entire plant could represent a drug itself and not just a single compound [39]. In high resolution spectra such as 1H NMR used in the present study, fingerprinting ignores the assignment problem arising due to the multitude of signals and on the contrary, through multivariate analysis compares set of spectra and thus the samples from which the spectra were obtained [40]. The current study aimed at the spectral fingerprint profile of guava leaf extract for characterization of extracts showing antidiarrhoeal activity.
Guava leaves have been used globally in the treatment of gastrointestinal disorders [41, 42]. Our work which included a clinical trial has confirmed that guava is a promising anti-diarrhoeal plant exhibiting a wide spectrum of activity [8, 43,44,45,46]. However, its functional components largely remained unknown, mainly due to chemical complexity and possible synergism.
The hydroalcoholic extract of guava leaves have been reported to have several phytoconstituents that include flavonoids, tannins, triterpenoids, saponins, sterols, alkaloids and carbohydrates [15, 47]. All these constituents have varied pharmacological actions. Phenolic acids such as gallic and ferulic acid are reported to possess antimicrobial, anti-carcinogenic, anti-inflammatory, hepatoprotective activities [48]. Similarly, anti-inflammatory and anti-oxidant activities are also associated with ferulic acid, catechins and quercetin [49, 50]. Flavonoids play a significant role in scavenging different reactive oxygen species [51]. Ellagic acid also has several pharmacological activities such as antioxidant, antimicrobial, anti-inflammatory and antiestrogenic [52]. Due to the presence of these compounds, guava leaves have been attributed with several biological activities.
Besides the above pharmacological properties, the anti-diarrhoeal activity of the compounds has also been documented. Chen et al., [53] have reported that gallic acid can prevent binding of E. coli toxin to ganglioside receptor. Presence of gallic acid has also been associated with antidiarrhoeal action due to its anti-secretory properties [54]. Quercetin is known to have antispasmolytic activity [17]. Protective effect of ellagic acid in castor oil and magnesium sulfate induced diarrhoea in mice has also been studied [52, 55]. The antispasmodic activity of saponins using guinea pig ileum has been demonstrated [56]. Other flavonoids such as rutin, kaempferol and morin have been reported to reduce the intestinal transit time in the mouse model [57]. Rutin is also said to have antidiarrhoeal properties as it prevents entry of rotavirus and/or viral interactions with target cells in the gut [58]. Studies by Yi et al., [59] on senna induced diarrhoea in mice showed that presence of flavonoids like rutin, isoquercitrin, quercitrin, myricetin exhibit anti-diarrhoeal properties by controlling the abnormal increase in vascular permeability and alleviating the inflammatory response. Epicatechin has been shown to prevent cholera toxin induced diarrhoea [60].
It is possible that different constituents of the guava leaf extract would be responsible for individual activities. Hence multiple compounds in the whole extract could act against different parameters resulting in an additive effect which would lead to a wider spectrum of antidiarrhoeal activity. Alternatively, some compounds my act as inhibitors and mask the activity of some other compounds.
Besides this additive feature, synergism could also play a role. In studies related to infectious diarrhoea caused by S. flexneri, it has been shown that quercetin alone is not responsible for the action; rather the efficacy of the whole extract is due to the synergistic effect of two or more phytoconstituents [8, 61]. This has also been exemplified by the observation that bio-guided fractionation often results in loss of activity [62]. In the present study, the 1H NMR spectra also highlighted this, for example 14 peaks were identified to be responsible for enhanced inhibition of invasion by S. flexneri into HEp-2 cells (Table 3).
Standardization of herbal extracts remains a challenge and appropriate methods are required. These methods would then ensure proper quality control, reproducibility and accountability of the materials along with the establishment of efficacy and safety profiles [1].
Profiling of the extracts using different analytical techniques, so as to decipher the components is a multistep process due to the large number of metabolites present and varied chemical nature of the individual components [1]. Identification of metabolites is not only challenging but also time consuming [63] necessitating development of high through put methods. This has led to the development of metabolomics towards a detailed, holistic and systemic analysis of all metabolites present [64]. Various techniques such as NMR, FTIR, MS, LC-MS, GC-MS have been used. As the amount of data obtained through these platforms is often large, it demands the use of multivariate statistics [64]. This combination has emerged as a promising tool towards detailed evaluation of metabolite data especially in case of complex/multicomponent mixtures. In targeted metabolomics, specific metabolites are profiled with known marker compounds not necessarily related to bioactivity [65]. The data set obtained using the target approach is simple and permits quantification of the desired metabolite(s) [66]. On the other hand, untargeted metabolomics profiles all possible metabolites present and thus provides a fingerprint rather than identifying any particular metabolite(s) [67]. Thus, the untargeted metabolomics is not limited by the need to know which chemical components are indicators of efficacy.
Over the years, metabolomics is being increasingly employed in herbal medicine for applications such as assessing quality, detecting adulterants, evaluating biological efficacy and determining optimum conditions for cultivation [68, 69]. A review by Ning et al., [70] comprehensively describes the various applications of metabolomics citing the examples from traditional Chinese Medicine.
However, there is a dearth of reports on the use of non-targeted metabolomic fingerprinting as a standardization technique. Standardization of Ayurvedic polyherbal formulations Triphala and Trikatu used targeted metabolite profiling [71, 72]. Another polyherbal formulation by trade name ‘Linkus’ containing Adhatoda vasica as an ingredient was standardized by a combination of untargeted and targeted metabolomics using chemometric as well as regression analysis of A. vasica collected from different locations [73].
The present work differs from the existing studies in the field of metabolomics. It used untargeted metabolomics using 1H NMR along with multivariate analysis for guava leaf extracts collected over different seasons and different locations. The aim was to correlate the spectral data with anti-diarrhoeal activity of guava leaves using representative bioassays. It also employed multiple assays to arrive at the identification of fingerprints representing efficacious extracts and aid differentiation of good vs poor batch of leaves. The study did not primarily intend to recognize any singular biomarker(s) correlating with the anti-diarrhoeal activity of guava leaves, rather it aimed at identifying a spectral fingerprint comprising of the desired peaks. The importance of fingerprinting has also been stressed by other workers involved in the study of traditional Chinese medicine [74, 75].
Thus NMR was the method of choice since the results are easily reproducible and sample preparation minimal thus it is more frequently used. Although 1H NMR profiling has lower sensitivity and resolution power it is preferred over MS due to its other advantages. 1H NMR profiling with its minimal and non-destructive sample preparation has higher reproducibility with quantitation being possible without the availability of a standard. It can be used for all types of compounds including non-ionizable compounds which cannot be detected by MS [1]. An added advantage of 1H NMR spectroscopy is that it helps in identifying both the primary and secondary metabolites simultaneously without the need for any kind of fractionation/separation. The primary metabolites usually are represented by signals in the aliphatic region (0–4.6 ppm) while the secondary metabolites are found in the aromatic region (5-11 ppm) [76]. Thus, 1H NMR profiles of the extracts indicate all the possible signals due to presence of 1H in the metabolites irrespective of the matrix in which they are present. Despite a single peak not necessarily representing a single compound and often multiple peaks being the signature of a single compound which is also reflected in the 1H NMR spectra acquired for the standard compounds (Supplementary Figs. S3, S4 and S5), 1H NMR spectroscopy was chosen for the current study because of its advantages.
Statistical analysis comprising of supervised and unsupervised methods (PCA and OPLS-DA) were used to establish the relationship between the different signals in the 1H NMR spectra of the extracts and the biological activity associated with that extract. Since the samples were collected from open field conditions, there was a high degree of variation. Hence statistical models could not sometimes be successfully developed considering the data from all the extracts and a subset was used to develop a significant relationship.
Firstly, the analysis tried to correlate the results of a single bioassay with the signals from the spectra. The results showed that multiple signals were identified correlating with individual activity. Some of these signals were predominantly seen in good activity extracts while some signals correlated with samples having poor activity. However, some signals did not show predominance in either of the good or poor activity extracts despite contributing to the differentiation in the model developed. This was assumed to be due to the limitations associated with the statistical models used and also the large variation in the metabolite profile of the extracts due to the open environment the guava trees were exposed to.
The 1H NMR spectral data for the guava extracts showed signals in the aliphatic and aromatic regions, suggesting that both primary and secondary metabolites were present in these hydroalcoholic extracts. Though The OPLS-DA analysis could clearly distinguish between the different regions and seasons as also reported earlier by others [5,6,7], the results showed that no extract showed either good or poor activity across all seven bioassays undertaken. Even with respect to a single assay, multiple peaks were identified correlating with the activity (Table 3). Quercetin, ferulic acid and gallic acid were present in the extracts based on NMR (Table 1) and LC-MS/MS (Table 2). However, they did not correlate with the peaks that segregated extracts with good and poor activity and thus they could be only used as markers.
Moreover due to the large number of primary and secondary metabolites present in the extract there are overlapping signals which may not clearly allow identification of the individual components involved. Additionally, it was observed that some peaks correlated strongly with poor activity implying that they may be inhibitory constituents. Thus, it was not possible to identify compounds that were responsible for the anti-diarrhoeal activity as a whole or for the individual bioassays. Previous studies attempting to identify the compound(s) responsible for activity through fractionation found that the activity is often significantly reduced in the isolated fraction [62]. Another approach wherein presence of markers has been used for the standardization of extracts has also run into difficulty since while the marker compounds could be present, the active component(s) may or may not be present in a significant amount to show the desired efficacy. As seen from Table 3 a number of peaks were identified for a single bioactivity which suggests an interplay of compounds in achieving the final outcome. This justifies our approach of relying on a fingerprint consisting of the desired peaks and not identifying compound(s) thus bypassing the above limitations. The choice of NMR was therefore crucial due to its reproducibility and minimal sample preparation. Thus, the current study has demonstrated that it is possible to standardize extracts with respect to activity based on a fingerprint rather than relying on markers.
Conclusion
With increase in use of traditional medicine, innovative methods and approaches towards their standardization are needed in phytomedicine research. The current study attempted to recognize key peaks from 1H NMR spectra which together would comprise of a spectral fingerprint relating to efficacy of guava leaf extract when a number of unidentified active principles are involved. This was met through the identification of key signals responsible for the differentiation of extracts with good and poor activity, establishing it as a prototype for standardization of a plant extract.
Availability of data and materials
All the data has been either included in the manuscript or given as Additional file.
Abbreviations
- D2O:
-
Deuterated water
- EPEC:
-
Enteropathogenic Escherichia coli
- EIEC:
-
Enteroinvasive Escherichia coli
- ETEC:
-
Enterotoxigenic Escherichia coli
- LT:
-
Labile toxin
- CT:
-
Cholera toxin
- NMR:
-
Nuclear Magnetic Resonance
- LC-MS:
-
Liquid Chromatography- Mass Spectroscopy
- PCA:
-
Principal Component Analysis
- OPLS-DA/RA:
-
Orthogonal Partial Least Square-Discriminant Analysis/Regression Analysis
- GM1-ELISA:
-
Ganglioside monosialic acid enzyme linked immunosorbent assay
References
Lajis N, Maulidiani M, Abas F, Ismail IS. Metabolomics approach in pharmacognosy A2 – Badal, Simone. In: Delgoda R, editor. Pharmacognosy. Boston, MA: Academic Press; 2017. p. 597–616.
Chugh NA, Bali S, Koul A. Integration of botanicals in contemporary medicine: road blocks, checkpoints and go-ahead signals. Integrative Med Res. 2018;7(2):109–25. https://doi.org/10.1016/j.imr.2018.03.005.
Ministry of Health and Family Welfare Gazette Notification, Government of India, New Delhi, 2015. https://cdsco.gov.in/opencms/opencms/system/modules/CDSCO.WEB/elements/download_file_division.jsp?num_id=MTA3MA== Accessed 2 June 2018.
Kim SH, Cho SK, Hyun SH, Park HE, Kim YS, Choi HK. Metabolic profiling and predicting the free radical scavenging activity of guava (Psidium guajava L.) leaves according to harvest time by 1H-nuclear magnetic resonance spectroscopy. Biosci Biotech Biochem. 2011;75(6):1090–7. https://doi.org/10.1271/bbb.100908.
Amirova KM, Dimitrova P, Marchev AS, Aneva IY, Georgiev MI. Clinopodium vulgare L. (wild basil) extract and its active constituents modulate cyclooxygenase-2 expression in neutrophils. Food Chem Toxicol. 2019;124:1–9. https://doi.org/10.1016/j.fct.2018.11.054.
Nazari P, Dowlatabadi-Bazaz R, Mofid MR, et al. The antimicrobial effects and metabolomic footprinting of carboxyl-capped bismuth nanoparticles against Helicobacter pylori. Appl Biochem Biotechnol. 2014;172:570–9. https://doi.org/10.1007/s12010-013-0571-x.
Spraul M, Schütz B, Humpfer E, Mörtter M, Schäfer H, Koswig S, Rinke P. Mixture analysis by NMR as applied to fruit juice quality control. Magn Reson Chem. 2009;47:S1307. https://doi.org/10.1002/mrc.2528.
Birdi T, Daswani P, Brijesh S, Tetali P, Natu A, Antia N. Newer insights into the mechanism of action of Psidium guajava L. leaves in infectious diarrhoea. BMC Complement Altern Med. 2010;10(1):33.
Ngbolua KN, Lufuluabo LG, Moke LE, Bango GN, Liyongo CI, Ashande CM, et al. A review on the phytochemistry and pharmacology of Psidium guajava L. (Myrtaceae) and future direction. Discov Phytomed. 2018;5:7–13. https://doi.org/10.15562/phytomedicine.2018.58.
Shao M, Wang Y, Huang XJ, Fan CL, Zhang QW, Zhang XQ, et al. Four new triterpenoids from the leaves of Psidium guajava. J Asian Nat Prod Res. 2012;14(4):348–54. https://doi.org/10.1080/10286020.2011.653964.
Liang Q, Qian H, Yao W. Identification of flavonoids and their glycosides by high-performance liquid chromatography with electrospray ionization mass spectrometry and with diode array ultraviolet detection. Eur J Mass Spectrom (Chichester). 2005;11(1):93–101. https://doi.org/10.1255/ejms.710.
Koriem KMM, Arbid MS, Saleh HN. Antidiarrheal and protein conservative activities of Psidium guajava in diarrheal rats. J Integr Med. 2019;17(1):57–65. https://doi.org/10.1016/j.joim.2018.12.001.
Hsieh C, Lin Y, Yen G, Chen H. Preventive effects of guava (Psidium guajava L.) leaves and its active compounds against a-dicarbonyl compounds-induced blood coagulation. Food Chem. 2007;103:528–35. https://doi.org/10.1016/j.foodchem.2006.08.022.
Chen H-Y, Yen G-C. Antioxidant activity and free radical-scavenging capacity of extracts from guava (Psidium guajava L.) leaves. Food Chem. 2007;101(2):686–94. https://doi.org/10.1016/j.foodchem.2006.02.047.
Arya V, Thakur VM, Kashyap C. Preliminary phytochemical analysis of the extracts of Psidium leaves. J Pharmacogn Phytochem. 2012;1:1–5.
Gutiérrez RM, Mitchell S, Solis RV. Psidium guajava: a review of its traditional uses, phytochemistry and pharmacology. J Ethnopharmacol. 2008;117(1):1–27. https://doi.org/10.1016/j.jep.2008.01.025.
Díaz-de-Cerio E, Verardo V, Gómez-Caravaca AM, Fernández-Gutiérrez A, Segura-Carretero A. Health effects of Psidium guajava L. leaves: An overview of the last decade. Int J Mol Sci. 2017;18(4):897. https://doi.org/10.3390/ijms18040897.
Zhang WJ, Chen BT, Wang CY, Zhu QH, Mo ZX. Mechanism of quercetin as an antidiarrheal agent. Di Yi Jun Yi Da Xue Xue Bao. 2003;23(10):1029–31.
Lutterodt GD. Inhibition of gastrointestinal release of acetylcholine by quercetin as a possible mode of action of Psidium guajava leaf extracts in the treatment of acute diarrhoeal disease. J Ethnopharmacol. 1989;25(3):235–47. https://doi.org/10.1016/0378-8741(89)90030-5.
Veselkov KA, Lindon JC, Ebbels TM, Crockford D, Volynkin VV, Holmes E, Davies DB, Nicholson JK. Recursive segment-wise peak alignment of biological (1) H NMR spectra for improved metabolic biomarker recovery. Anal Chem. 2009;81(1):56–66. https://doi.org/10.1021/ac8011544.
Cruickshank R, Duguid JP, Marmion BP, Swain RHA, editors. Medical microbiology. Great Britain: Longman Group Ltd; 1975.
Cravioto A, Grass RJ, Scotland SM, Gran RJ, Scotland SM, Rowe B. An adhesive factor found in strains of E. coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979(3):95–9. https://doi.org/10.1007/BF02602439.
Vesikari T, Bromisrska J, Maki M. Enhancement of invasiveness of Yersinia enterocolitica and Escherichia coli to HEp-2 cells by centrifugation. Infect Immun. 1982;36:834–6.
Svennerholm A-M, Wilkund G. Rapid GM1-enzyme-linked immunosorbent assay with visual reading for identification of Escherichia coli heat-labile enterotoxins. J Clin Microbiol. 1983;17:596–600.
Sampaio B, Edrada-Ebel R, Da Costa F. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: a model for environmental metabolomics of plants. Sci Rep. 2016;6:29265. https://doi.org/10.1038/srep29265.
Zhao J, Khan IA, Combrinck S, Sandasi M, Chen W, Viljoen AM. (1) H-NMR and UPLC-MS metabolomics: functional tools for exploring chemotypic variation in Sceletium tortuosum from two provinces in South Africa. Phytochem. 2018;152:191–203. https://doi.org/10.1016/j.phytochem.2018.03.013.
Gill CJ, Thea DM, Hibberd P. Diarrhoeal disease trends in the GBD 2015 study: optimism tempered by scepticism. Lancet. 2017;17:884–5. https://doi.org/10.1016/S1473-3099(17)30336-5.
Chellapandi K, Dutta TK, Sharma I, et al. Prevalence of multi drug resistant enteropathogenic and enteroinvasive Escherichia coli isolated from children with and without diarrhea in Northeast Indian population. Ann Clin Microbiol Antimicrob. 2017;16(1):49. https://doi.org/10.1186/s12941-017-0225-x.
Rathaur VK, Pathania M, Jayara A, Yadav N. Clinical study of acute childhood diarrhoea caused by bacterial enteropathogens. J Clin Diagn res. 2014;8:PC01–5. J Clin Diagn Res 2014;8(5): PC01–05. doi: https://doi.org/10.7860/JCDR/2014/6677.4319.
Aggarwal P, Uppal B, Ghosh R, et al. Highly-resistant E. coli as a common cause of paediatric diarrhoea in India. J Health Popul Nutr. 2013;31(3):409–12. https://doi.org/10.3329/jhpn.v31i3.16835.
Brander RL, Walson JL, John-Stewart GC, Naulikha JM, Ndonye J, Kipkemoi N, et al. Correlates of multi-drug non-susceptibility in enteric bacteria isolated from Kenyan children with acute diarrhea. PLoS Neglect Trop Dis. 2017;11:e0005974. https://doi.org/10.1371/journal.pntd.0005974.
Yismaw G, Negeri C, Kassu A. A five-year antimicrobial resistance pattern of Shigella isolated from stools in the Gondar University hospital, northwest Ethiopia. Trop Doc. 2008;38(1):43–5. https://doi.org/10.1258/td.2007.060215.
Goodarzi M, Russell PJ, Vander HY. Similarity analyses of chromatographic herbal fingerprints: a review. Anal Chim Acta. 2013;804:16. https://doi.org/10.1016/j.aca.2013.09.017.
World Health Organization. Guidelines for for the assessment of herbal medicines. Geneva: WHO; 1991. http://apps.who.int/iris/bitstream/10665/58865/1/WHO_TRM_91.4.pdf?ua=1. Accessed 26 Dec 2020
Food and Drug Administration. Guidance for Industry: Botanical Drug Products. www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm458484.pdf. Accessed 29 Dec 2020.
State Drug Administration of China. Technical requirements for chromatographic fingerprints of traditional Chinese medicinal injection. Chin Tradit Pat Med. 2000;22:671–8.
European Medicines Agency. Guidance on Quality of Herbal Medicinal Products / Traditional Herbal Medicinal Products. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2011/09/WC500113209.pdf. Accessed 17 Dec 2020.
Rajani M, Niranjan S, Kanaki NS. Phytochemical standardization of herbal drugs and poly herbal formulations. In: Ramawat KG, Merillon JM, editors. Bioactive molecules and medicinal plants. India: Springer; 2008. p. 349–69.
Upton R, David B, Gafner S, Glasl S. Botanical ingredient identification and quality assessment: strengths and limitations of analytical techniques. Phytochem Rev. 2019;19:1–21. https://doi.org/10.1007/s11101-019-09625-z.
Krishnan P, Kruger NJ, Ratcliffe RG. Metabolite fingerprinting and profiling in plants using NMR. J Exp Bot. 2005;56:255–65. https://doi.org/10.1093/jxb/eri010.
Daswani PG, Gholkar MS, Birdi TJ. Psidium guajava: a single plant for multiple health problems of rural Indian population. Pharmacogn Rev. 2017;11(22):167–74. https://doi.org/10.4103/phrev.phrev_17_1.
Kafle A, Mohapatra SS, Reddy I, Chapagain M. A review on medicinal properties on Psidium guajava. J Med Plants Stud. 2018;6:44–7. https://doi.org/10.1186/1472-6882-10-33.
Birdi TJ, Daswani PG, Brijesh S, Tetali P. In vitro antigiardial and antirotaviral activity of Psidium guajava L. leaves. Indian J Pharmacol. 2011;43:616–7. https://doi.org/10.4103/0253-7613.84990.
Brijesh S, Tetali P, Birdi TJ. Study on effect of anti-diarrheal medicinal plants on enteropathogenic Escherichia coli induced interleukin-8 secretion by intestinal epithelial cells. Altern Med Stud. 2011;1:e16. https://doi.org/10.4081/ams.2011.e16.
Gupta P, Birdi T. Psidium guajava leaf extract prevents intestinal colonization of Citrobacter rodentium in the mouse mode. J Ayurveda Integr Med. 2015;61:50–2. https://doi.org/10.4103/0975-9476.146557.
Birdi T, Krishnan GG, Kataria S, Gholkar M, Daswani P. A randomized open label efficacy clinical trial of oral guava leaf decoction in patients with acute infectious diarrhoea. J Ayurveda Integr Med. 2020;11:163–72. https://doi.org/10.1016/j.jaim.2020.04.001.
Dakappa SS, Adhikari R, Timilsina SS, Sunita S. A review on the medicinal plant Psidium guajava Linn. (Myrtaceae). J Drug Deliv Ther. 2013;3:162–8.
Borges A, Ferreira C, Saavedra MJ, Simoes M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb Drug Resist. 2013;19(4):256–65. https://doi.org/10.1089/mdr.2012.0244.
Zhang L, Dong M, Xu G, Tian Y, Tang H, Wang Y. Metabolomics reveals that dietary ferulic acid and quercetin modulate metabolic homeostasis in rats. J Agric Food Chem. 2018;66(7):1723–31. https://doi.org/10.1021/acs.jafc.8b00054.
Bernatoniene J, Kopustinskiene DM. The role of catechins in cellular responses to oxidative stress. Molecules. 2018;23(4):965. https://doi.org/10.3390/molecules23040965.
Islam MK, Mahmud I, Saha S, Sarker AB, Mondal H, Monjur-Al-Hossain AS, et al. Preliminary pharmacological evaluation of Alocasia indica Schott tuber. J Integr Med. 2013;11(5):343–51. https://doi.org/10.3736/jintegrmed2013045.
Zhao SS, Ma DX, Zhu Y, Zhao JH, Zhang Y, Chen JQ, et al. Antidiarrheal effect of bioactivity-guided fractions and bioactive components of pomegranate (Punica granatum L.) peels. Neurogastroenterol Motil. 2018;30(7):e13364. https://doi.org/10.1111/nmo.13364.
Chen JC, Ho TY, Chang YS, Wu SL, Hsiang CY. Anti-diarrheal effect of Galla chinensis on the Escherichia coli heat-labile enterotoxin and ganglioside interaction. J Ethnopharmacol. 2006;103(3):385–91. https://doi.org/10.1016/j.jep.2005.08.036.
Pandey G, Gupta SS, Bhatia A, Sidhu OP, Rawat AK, Rao CV. Grilling enhances antidiarrheal activity of Terminalia bellerica Roxb. Fruits. J Ethnopharmacol. 2017;202:63–6. https://doi.org/10.1016/j.jep.2016.12.003.
Xiao HT, Tsang SW, Qin HY, Choi FF, Yang ZJ, Han QB, Chen HB, Xu HX, Shen H, Lu AP, Bian ZX. A bioactivity-guided study on the anti-diarrheal activity of Polygonum chinense Linn. J Ethnopharmacol. 2013;149(2):499–505. https://doi.org/10.1016/j.jep.2013.07.007.
Cimanga RK, Mukenyi PN, Kambu OK, Tona GL, Apers S, Totté J, Pieters L, et al. The spasmolytic activity of extracts and some isolated compounds from the leaves of Morinda morindoides (baker) Milne-Redh. (Rubiaceae). J Ethnopharmacol. 2010;127(2):215–20. https://doi.org/10.1016/j.jep.2009.11.018.
Di Carlo G, Autore G, Izzo AA, Maiolino P, Mascolo N, Viola P, et al. Inhibition of intestinal motility and secretion by flavonoids in mice and rats: structure-activity relationships. J Pharm Pharmacol. 1993;45(12):1054–9. https://doi.org/10.1111/j.2042-7158.1993.tb07180.x.
Kawahara T, Tomono T, Hamauzu Y, Tanaka K, Yasui H. Inhibitory effect of a hot-water extract of leaves of Japanese big-leaf Magnolia (Magnolia obovata) on rotavirus-induced diarrhea in mouse pups. Evid Based Complement Alternat Med. 2014;2014:365831. https://doi.org/10.1155/2014/365831.
Yi R, Tian Y, Tan F, Li W, Mu J, Long X, et al. Intervention effect of Malus pumila leaf flavonoids on senna-induced acute diarrhea in BALB/c mice. Food Sci Nutr. 2020;8(5):2535–42. https://doi.org/10.1002/fsn3.1549.
Calzada F, Correa-Basurto J, Barbosa E, Mendez-Luna D, Yepez-Mulia L. Antiprotozoal constituents from Annona cherimola Miller, a plant used in Mexican traditional medicine for the treatment of diarrhea and dysentery. Pharmacogn Mag. 2017;(49):13, 148–152. https://doi.org/10.4103/0973-1296.197636.
Hirudkar JR, Parmar KM, Prasad RS, Sinha SK, Jogi MS, Itankar PR, et al. Quercetin a major biomarker of Psidium guajava L inhibits SepA protease activity of Shigella flexneri in treatment of infectious diarrhoea. Microb Pathog. 2020;138:103807. https://doi.org/10.1016/j.micpath.2019.103807.
Aquil L, Ahmad I, Owais M. Evaluation of anti-methicillin-resistant Staphylococcus aureus (MRSA) activity and synergy of some bioactive plant extracts. Biotechnol J. 2006;1(10):1093–102. https://doi.org/10.1002/biot.200600130.
Guo JM, Lu YW, Shang EX, Li T, Liu Y, Duan JA, Qian DW, Tang YP. Metabolite identification strategy of non-targeted metabolomics and its application for the identification of components in Chinese multicomponent medicine Abelmoschus manihot L. Phytomed. 2015;22(5):579–87. https://doi.org/10.1016/j.phymed.2015.02.002.
Quansah E, Karikari TK. Potential role of metabolomics in the improvement of research on traditional African medicine. Phytochem Lett. 2016;17:270–7. https://doi.org/10.1016/j.phytol.2016.08.004.
Commisso M, Strazzer P, Toffali K, Stocchero M, Guzzo F. Untargeted metabolomics: an emerging approach to determine the composition of herbal products. Comput Struct Biotechnol J. 2013;4:e201301007. https://doi.org/10.5936/csbj.201301007.
Yi L, Dong N, Yun Y, Deng B, Ren D, Liu S, Liang Y. Chemometric methods in data processing of mass spectrometry-based metabolomics: A review. Anal Chim Acta. 2016;914:17–34. https://doi.org/10.1016/j.aca.2016.02.001.
Moco S, Vervoort J, Bino RJ, De Vos RC, Bino R. Metabolomics technologies and metabolite identification. Trends Anal Chem. 2007;26(9):855–66. https://doi.org/10.1016/j.trac.2007.08.003.
Hall RD. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol. 2006;169(3):453–68. https://doi.org/10.1111/j.1469-8137.2005.01632.x.
Wolfender JL, Rudaz S, Hae Choi Y, Kyong KH. Plant metabolomics: from holistic data to relevant biomarkers. Curr Med Chem. 2013;20(8):1056–90. https://doi.org/10.2174/092986713805288932.
Ning Z, Lu C, Zhang Y, Zhao S, Liu B, Xu X, Liu Y. Application of plant metabonomics in quality assessment for large-scale production of traditional Chinese medicine. Planta Med. 2013;79(11):897–908. https://doi.org/10.1055/s-0032-1328656.
Ponnusankar S, Pandit S, Babu R, Bandyopadhyay A, Mukherjee PK. Cytochrome P450 inhibitory potential of Triphala – a rasayana from Ayurveda. J Ethnopharmacol. 2011;133:120–5. https://doi.org/10.1016/j.jep.2010.09.022.
Harwansh RK, Mukherjee K, Bhadra S, Kar A, Bahadur S, Mitra A, et al. Cytochrome P450 inhibitory potential and RP-HPLC standardization of trikatu- A Rasayana from Indian Ayurveda. J Ethnopharmacol. 2014;153:674–81. https://doi.org/10.1016/j.jep.2014.03.023.
Rahman S, Haq FU, Ali A, Khan MN, Shah SM, Adhikhari A, et al. Combining untargeted and targeted metabolomics approaches for the standardization of polyherbal formulations through UPLC–MS/MS. Metabolomics. 2019;15(9):116. https://doi.org/10.1007/s11306-019-1582-6.
Yu Y, Gong D, Zhu Y, Wei W, Sun G. Quality consistency evaluation of Isatidis folium combined with equal weight quantified ratio fingerprint method and determination of antioxidant activity. J Chromatogr B Anal Technol Biomed Life Sci. 2018;1095:149–56. https://doi.org/10.1016/j.jchromb.2018.07.031.
Yang L, Sun G, Guo Y, Hou Z, Chen S. Holistic evaluation of quality consistency of Ixeris sonchifolia (Bunge) Hance injectables by quantitative fingerprinting in combination with antioxidant activity and chemometric methods. PLoS One. 2016;11:e0148878. https://doi.org/10.1371/journal.pone.0148878.
Jayasundar R, Ghatak S, Makhdoomi MA, Luthra K, Singh A, Velpandian T. Challenges in integrating component level technology and system level information from Ayurveda: Insights from NMR phytometabolomics and anti-HIV potential of select Ayurvedic medicinal plants. J Ayurveda Integra Med. 2019;10(2):94–101. https://doi.org/10.1016/j.jaim.2017.06.002.
Acknowledgments
We thank Mrs. Mamata Joshi, National Facility for High Field NMR, Tata Institute of Fundamental Research (TIFR), Mumbai for extending help in recording the NMR fingerprints of the extracts and the standard compounds. The financial support from Tata Education and Development Trust (TEDT) and Godrej Foundation towards training of MSG at Imperial College, London is also acknowledged. We also thank Prof. KS. Laddha, Institute of Chemical Technology, Mumbai for generously providing ferulic acid and gallic acid used as standard compounds for 1H NMR spectra. We are also grateful to Dr. Amisha K. Vora, Assistant Professor, Department of Pharmaceutical Chemistry, Shobhaben Pratapbhai Patel School Of Pharmacy & Technology Management, SVKM’s Narsee Monjee Institute of Management Studies, Mumbai (Deemed to be University) for undertaking the LC-MS-MS analysis of the representative guava extracts.
Funding
This work was financially supported by Zoetis Pharmaceuticals Research Private Limited, India.
Author information
Authors and Affiliations
Contributions
MSG, PGD performed the bioassays. MSG was responsible for NMR studies; JVL trained MSG & contributed to drafting the manuscript; MSG, JVL, PGD, TJB drafted the manuscript; PT provided the guava leaves; TJB was responsible for the overall study. All authors have read and given their final approval for the version submitted for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
Representative 1H NMR plots for seasonal differentiation. Individual plots are representative 1H NMR spectrum of the guava hydroalcoholic extract prepared from leaves collected from Shirwal (W region) in. a) season B (May 2013); b) season C (October 2013); c) season D (March 2014).
Additional file 2: Fig. S2.
Representative 1H NMR plots for regional differentiation. Individual plots are representative 1H NMR spectrum of the guava hydroalcoholic extract prepared from leaves collected in season D (March 2014) from. a) Shirwal (W region); b) Rahata (R region); c) Dapoli (Da region).
Additional file 3: Fig. S3.
1H NMR plot acquired for quercetin.
Additional file 4: Fig. S4.
1H NMR plot acquired for ferulic acid.
Additional file 5: Fig. S5.
1H NMR plot acquired for gallic acid.
Additional file 6: Fig. S6.
Fragmentation patterns for compounds identified by LC-MS/MS in extract WB. W: Leaves collected from Shirwal region; B: May 2013 collection.
Additional file 7: Fig. S7.
Fragmentation patterns for compounds identified by LC-MS/MS in extract WC. W: Leaves collected from Shirwal region; C: October 2013 collection.
Additional file 8: Fig. S8.
Fragmentation patterns for compounds identified by LC-MS/MS in extract WD W: Leaves collected from Shirwal region; D: March 2014 collection.
Additional file 9: Fig. S9.
Fragmentation patterns for compounds identified by LC-MS/MS in extract RD. R: Leaves collected from Rahata region; D: March 2014 collection.
Additional file 10: Fig. S10.
Fragmentation patterns for compounds identified by LC-MS/MS in extract DaD. Da: Leaves collected from Dapoli region; D: March 2014 collection.
Additional file 11: Table S1.
Values represent Mean ± SD of % bacterial (Shigella flexneri) viability from two independent experiments done in triplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Additional file 12: Table S2.
Values represent Mean ± SD of % bacterial (Vibrio cholerae) viability from two independent experiments done in triplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Additional file 13: Table S3.
Values represent Mean ± SD of % bacterial (EPEC) adherence from two independent experiments done in duplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Additional file 14: Table S4.
Values represent Mean ± SD of % bacterial (E. coli) invasion from two independent experiments done in duplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Additional file 15: Table S5.
Values represent Mean ± SD of % bacterial (S. flexneri) invasion from two independent experiments done in duplicates. W: Shirwal; R: Rahata; Da: Dapol
Additional file 16: Table S6.
Values represent Mean ± SD of % cholera toxin (CT) production from two independent experiments done in triplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Additional file 17: Table S7.
Values represent Mean ± SD of % CT binding from two independent experiments done in triplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Additional file 18: Table S8.
Values represent Mean ± SD of % labile toxin production from two independent experiments done in triplicates. B: May 2013 collection; C: October 2013 collection; D: March 2014 collection. W: Shirwal; R: Rahata; Da: Dapoli
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gholkar, M.S., Li, J.V., Daswani, P.G. et al. 1H nuclear magnetic resonance-based metabolite profiling of guava leaf extract: an attempt to develop a prototype for standardization of plant extracts. BMC Complement Med Ther 21, 95 (2021). https://doi.org/10.1186/s12906-021-03221-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-021-03221-5