Abstract
Background
The effect of ginsenosides on the growth and apoptosis of human lens epithelial (HLE) B3 cells exposed to H2O2 was investigated. In addition, the effect of ginsenosides on gene expression in HLE-B3 cells was analyzed using microarray assays to determine its molecular mechanism.
Methods
HLE-B3 cells were treated with 1.75 M H2O2 in the presence or absence of 5, 10 or 20 μM ginsenosides. Cell viability and apoptosis were examined by MTT assays and flow cytometry, respectively, at 24 to 120 h after the treatment. Furthermore, HLE-B3 cells were treated with 20 μM ginsenosides for 8 days and total RNA was isolated and analyzed using the Affymetrix GeneChip Array. Principal component analysis was performed to visualize the microarray data.
Results
Addition of ginsenosides significantly alleviated the growth inhibitory effect of H2O2 on HLE-B3 cells and the percentage of viable cells was increased by more than 3 folds. Flow cytometric analysis showed that 6.16 ± 0.29% of H2O2-treated HLE-B3 cells were early apoptotic cells, and the percentage was reduced to 4.78 ± 0.16% (P < 0.05) in the presence of 20 μM ginsenosides. Principal component analysis revealed that ginsenoside caused extensive changes in gene expression in HLE-B3 cells. A total of 6219 genes showed significant differential expression in HLE-B3 cells treated with ginsenoside; among them, 2552 (41.0%) genes were significantly upregulated, whereas 3667 (59.0%) genes were significantly downregulated. FOXN2, APP and RAD23B were the top three upregulated genes while WSB1, PSME4 and DCAF7 were the top three downregulated genes in HLE-B3 cells treated with ginsenosides.
Conclusion
Ginsenosides induce extensive changes in the expression of genes involved in multiple signaling pathways, including apoptotic signaling pathway and DNA damage response signaling pathway. Ginsenosides alleviate H2O2-induced suppression of the growth of HLB cells and inhibit H2O2-induced apoptosis of HLB cells.
Similar content being viewed by others
Background
It has been estimated that 95 million people worldwide are affected by cataracts [1]. Age-related cataracts remain highly prevalent in the elderly population, and elderly patients with cataracts account for a significant proportion of visually impaired elderly people globally [2]. Surgery has been shown to be effective for cataract correction, but is not without risks and problems [3]. Currently, there is a lack of effective alternative treatment modalities for cataract surgery. Although many risk factors for cataractogenesis have been identified, such as long-term corticosteroid use [4], smoking, excessive UV-B exposure, and diabetes [5], other than a healthy lifestyle such as smoking cessation, there are no effective preventive measures, including pharmacological treatment of cataract formation.
Cataract is a multifactorial eye disease. Although the exact molecular mechanism of cataractogenesis remains elusive, oxidative stress (OS) has been implicated as the main culprit of cataract lens opacity [6]. Hydrogen peroxide (H2O2), a non-free radical member of the active oxygen family, is the major intracellular reactive oxygen species (ROS) in the aqueous humor, which generates hydroxyl radicals that irreversibly damage the lens epithelium. It can activate multiple signaling events such as the activation of apoptosis-associated molecules or pathways, including caspases, the Bcl-2 family, the mitogen-activated protein kinases (MAPKs), and NF-кB pathways, which lead to apoptosis of lens epithelial cells (HLE), ultimately resulting in lens opacification and subsequent cataract development [7, 8]. A variety of antioxidant nutrients, such as flavonoids, phenolic acids, carotenoids, and vitamins, have been tested for their ability to prevent or delay cataract development in animal studies, but their protective effects have not been demonstrated unequivocally [9].
Ginsenosides, also known as ginseng saponins, are isolated from the total saponins of Panax notoginseng and have been tested against various diseases including ischemic stroke [10]. Ginsenosides have antioxidant and antioxidant-related properties in a variety of cell types. Ginsenosides were shown to significantly inhibited UV-B-induced ROS elevation in HaCaT keratinocytes [11]. Ginsenoside Rg1 mediated by ultrasound-targeted microbubble destruction can reduce the level of OS, relieve intraocular pressure and reduce ganglion cell damage in glaucomatous optic nerve of rabbits. Ginsenosides Rb1 and Rd. were also shown to protect the retina from intense light-induced degeneration in BALB/c mice exposed to intense light [12]. However, the possible effect of ginsenosides on cataracts has not been examined. Given that ginsenosides have been shown to protect against UV-B exposure in keratinocytes [13, 14], we hypothesized that ginsenosides may also exert protective effects against OS-induced lens epithelial damage. In the current study, we investigated the effect of ginsenosides on the growth and apoptosis of human lens epithelial (HLE-B3) cells exposed to H2O2. To further determine the molecular mechanism of ginsenosides, we analyzed the effects of ginsenosides on gene expression in HLE-B3 cells using microarray analysis.
Methods
H2O2 treatment
Human lens epithelial (HLE-B3) cells purchased from American Type Culture Collection were cultured in minimal essential medium (MEM) containing 20% of fetal bovine serum in a humidified incubator at 37 °C with 5% CO2. Cells at 50–60% confluency were treated with H2O2 at concentrations from 0.039 to 2.5 μM for 24 h, and the concentration was incremented by a factor of two. Cell viability was examined by the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assays using Cell Proliferation Reagent Kit I (MTT; Roche Applied Science) following the manufacturer’s protocol and as previously described [15]. Each experiment was repeated at least three times independently in quintuplicate. Optical density (OD) was measured at 490 nm using a microplate reader (iMark, USA). A standard curve was drawn to determine IC50 of H2O2. All cells were treated with H2O2 at IC50 for 24 h in all subsequent experiments.
Ginsenoside treatment
HLE-B3 cells (passage 6) were plated at 6 × 103 cells per well in 96-well plates. After the medium was changed, the cells at 30–40% confluency were treated with H2O2 at IC50 in the absence or presence of 5, 10 or 20 μM ginsenoside (Fleton Natural Products, Chengdu, China; HPLC grade pure 99.8%). Ten microliter of MTT (at a final concentration of 5 mg/ml) was added to each well and the cells were incubated for another 4 h, and the supernatant was discarded. DMSO (150 μL) was added to each well to dissolve the precipitate. Cell viability was examined by MTT assays after 24 to 120 h of drug treatment following the manufacturer’s instructions (Sigma, St. Louis, MO, USA). Each experiment was repeated at least three times independently in quintuplicate.
Flow cytometry
Flow cytometry was performed as previously described [16]. Briefly, HLE-B3 cells were seeded in 6-well plates. When the cells were 70% confluent (5 × 105 cells), they were treated with 5, 10 or 20 μM ginsenoside for 48 h and harvested. The treated cells were washed once with phosphate-buffered saline (PBS), trypsinized, and washed again in PBS containing 2% fetal bovine serum and fixed in ice-cold ethanol for at least 1 h at − 20 °C. The cells were washed, and stained with FITC-annexin V (Beyotime Biotechnology Research Institute, China) and propidium iodide (30 μg/mL) and treated with RNase (0.6 mg/mL) in PBS containing 0.5% (v/v) Tween 20 and 2% fetal bovine serum. Fluorescence-activated cell sorting analysis was performed on a FACS Calibur flow cytometer (BD Biosciences) using Cellquest software, and the Mod-Fit program (Verity Software House Inc., Topsham, ME) was used to analyze the percentage of apoptotic cells.
Microarray
HLE-B3 cells were treated with 20 μM ginsenoside for 8 days. Total RNA was extracted using the Recover All™ Total Nucleic Acid Isolation Kit (Ambion, AM1975) following the manufacturer’s protocol. RNA was biotin-labeled using the FlashTag™ Biotin HSR RNA Labeling Kit (Affymetrix). An input of 400 nanograms of total RNA was used for each reaction. Hybridization, washing and staining were performed using the commercially available Affymetrix GeneChip Hybridization, Wash and Stain Kit. All samples were hybridized to the Affymetrix GeneChip Array. Expression data were normalized using the robust multi-array average (just RMA) method where the raw intensity values were background-corrected, log2-transformed and then quartile-normalized. A linear model was fitted to the normalized data to obtain a measure of expression for each probe set on each array.
Gene analysis
Principal component analysis was carried out to visualize the microarray data. All genes were plotted on the first and second principal components. The first principal component (PC1) measured the grand mean expression and the second (PC2) measured the ginsenosides-induced expression changes. In addition, a scatter plot of principal component analysis of differential gene expression patterns in ginsenosides-treated cells and control cells was drawn. The Pearson correlation of gene expression patterns in HLE-B3 cells treated with ginsenoside was calculated between the vectors pointing from the overall mean of the entire dataset and the respective group mean. For hierarchical clustering, genes in the dataset were subjected to complete-linkage hierarchical clustering using a Euclidean distance metric. The pathway and function analyses were performed using KEGG and Gene Ontology (GO).
Results
Ginsenoside reverses H2O2-induced growth inhibition of HLE-B3 cells
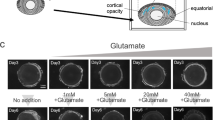
MTT assays showed that H2O2 exhibited a dose-dependent inhibitory effect on the viability of HLE-B3 cells. The mean inhibition rate of H2O2 steadily increased from 6.6% at a concentration of 0.039 μM to 15.3% at a concentration of 1.25 μM and rapidly reached to 85.4% at the final concentration of 2.5 μM (Fig. 1). The IC50 of H2O2 was 1.75 μM (ranging from 1.59 to 1.927 μM). Addition of ginsenosides significantly alleviated the growth inhibitory effect of H2O2 (1.75 μM) on HLE-B3 cells and the percentage of viable cells was increased by more than three folds (Fig. 1). There was no statistical difference in the percentage of viable HLE-B3 cells treated with low (5 μM), mid (10 μM) and high (20 μM) dose of ginsenosides (P > 0.05).
Ginsenoside reverses H2O2-induced growth inhibition of HLE-B3 cells. HLE-B3 cells were treated with 1.75 μM H2O2 and low (5 μM), mid (10 μM) and high dose (20 μM) of ginsenoside. Viabilities of HLE-B3 cells were examined by MTT assays as detailed in Methods. Ginsenoside significantly alleviated the growth inhibitory effect of H2O2 on HLE-B3 cells with more than 3-fold increase in the percentage of viable HLE-B3 cells. No statistical difference was observed in the percentage of viable HLE-B3 cells treated with low (5 μM), mid (10 μM) and high dose (20 μM) of ginsenoside (P > 0.05)
Ginsenosides induce extensive changes in gene expression
Principal component analysis revealed that ginsenosides caused extensive changes in gene expression in HLE-B3 cells (Fig. 2). The differential gene expression patterns in ginsenoside-treated and control cells were further shown in a principal component analysis scatter plot (Fig. 3), indicating a linear relationship in gene expression patterns. The Pearson correlation matrix of gene expression patterns in HLE-B3 cells treated with ginsenosides is shown in Fig. 4. In addition, the volcano plot showed that more genes were downregulated than upregulated in ginsenosides-treated HLE-B3 cells (Fig. 5). Hierarchical clustering of gene expression patterns in HLE-B3 cells treated with ginsenosides further detailed the upregulated and downregulated genes (Fig. 6).
Principal component analysis for visualization of microarray data. All genes are plotted on to the first and second principal components. The first principal component (PC1) measures total average expression, and the second (PC2) measures changes in expression induced by ginsenoside. The ellipse at the center contains 95% of the genes. Pink color represents genes from ginsenoside-treated cells and green color indicates genes from control cells
A principal component analysis scatter plot of differential gene expression patterns in ginsenoside-treated cells and control cells. Genes with equal expression values line up on the diagonal identity line and higher expression values are further away from the origin. Points below the diagonal represent genes with higher expression in ginsenoside-treated cells plotted on the x-axis. Points above the diagonal represent genes with higher expression values in control cells plotted on the y-axis. The further away a point-of-interest is from the diagonal line the larger is the difference in expression in ginsenoside-treated cells compared with the control cells
Correlation matrix of gene expression patterns in HLE-B3 cells treated with ginsenoside. The Pearson correlation was calculated between the vectors pointing from the overall mean of the entire dataset to the respective group mean. The values at each position in the matrix characterize the expression level of a particular gene under a particular experimental condition. Each box is a log ratio of gene expression of HLE-B3 cells treated with ginsenoside/the control cells. Rows represent genes and columns represent measurements from individual arrays
The volcano plot shows ginsenoside-induced changes of gene expression in HLE-B3 cells. The x axis shows fold changes in gene expression while the y axis shows statistical significance (−log10 of P values). The horizontal green line shows where P = 0.05 with points above the line having P < 0.05 and points below the line having P > 0.05. The volcano plot is color coded; those points having a fold-change less than 2 (log2 = 1) are shown in gray. Red indicates points-of-interest that display statistically significant differences. Points-of-interest left to the vertical green line represent downregulated genes and those that lie right to the vertical green line are upregulated genes
Hierarchical clustering. Genes in the data set were subjected to complete-linkage hierarchical clustering using a Euclidean distance metric. Genes that are upregulated appear in red, and those that are downregulated appear in green, with the relative log2 (ratio) reflected by the intensity of the color
A total of 6219 genes showed significant differential expression in ginsenosides-treated and control HLE-B3 cells. Among them, 2552 (41.0%) genes were significantly upregulated while 3667 (59.0%) genes were significantly downregulated.
The 10 most upregulated and downregulated genes are shown in Tables 1 and 2, respectively. FOXN2, APP and RAD23B are the top three upregulated genes while WSB1, PSME4 and DCAF7 are the top three downregulated genes in HLE-B3 cells treated with ginsenosides. Ginsenoside caused one to two-fold increase in the ten most upregulated genes and one to two-fold decrease in the ten most downregulated genes in HLE-B3 cells.
Ginsenosides upregulate the expression of genes involved in apoptosis and DNA damage response
Gene ontology analysis showed that three of the top ten upregulated genes were related to apoptosis, including APP, LMNB1 and MAPK8 (Table 3). Besides, three genes were involved in DNA damage response, including RAD23B, MAPK8 and TLK2. Specifically, MAPK8 is involved in the cellular response to hydrogen peroxide and APP to OS. Furthermore, PSME4, which is one of the top ten downregulated genes, is involved in the negative regulation of apoptosis (Table 4).
Ginsenosides reduce H2O2-induced apoptosis of HLE-B3 cells
Since gene ontology analysis revealed that ginsenosides modulated the expression of apoptosis-related genes, such as LMNB1 and PSME4, we examined the effect of ginsenosides on H2O2-induced apoptosis of HLE-B3 cells. Flow cytometry analysis showed that 6.16 ± 0.29% of H2O2-treated HLE-B3 cells were early apoptotic cells (Fig. 7a), and the percentage was significantly reduced to 5.22 ± 0.59%, 4.98 ± 0.29% and 4.78 ± 0.16% by the presence of low (5 μM), mid (10 μM) and high (20 μM) dose of ginsenosides (P < 0.05), respectively (Fig. 7b to e), suggesting that ginsenosides reduce H2O2-induced apoptosis in HLE-B3 cells.
Ginsenoside reduces H2O2-induced apoptosis of HLE-B3 cells. HLE-B3 cells were treated with 1.75 μM H2O2 (a) and low (5 μM) (b), mid (10 μM) (c) and high dose (20 μM) (d) of ginsenoside. The percentage of apoptosis cells was examined by flow cytometry. e Data are expressed as mean ± SD of at least three independent experiments. P < 0.05 of the treatment groups versus 1.75 μM H2O2. Representative histograms are shown in (a) to (d)
Discussion
In this study, we presented the first experimental evidence that ginsenosides could protect against H2O2-induced growth inhibition and apoptosis in HLE-B3 cells. Furthermore, ginsenosides caused widespread changes in gene expression, including changes in genes involved in DNA damage response and apoptosis, suggesting that ginsenosides act through multiple molecular mechanisms.
Microarray data showed that FOXN2, APP and RAD23B were the top three upregulated genes while WSB1, PSME4 and DCAF7 were the top three downregulated genes by ginsenosides in HLE-B3 cells. FOXN2 is a member of the Forkhead box transcription factors. Its role in cataractogenesis has not been confirmed. A recent study showed that FOXN2 could suppress the proliferation of lung cancer cells [17]. RAD23B, MAPK8 and TLK2 have been shown to be involved in DNA damage response [18]. Effect of ginsenosides on MAPK has been well documented [19,20,21]. A Chinese herbal medicine containing ginsenosides was found to attenuate H2O2-induced injury in PC12 cells by inhibiting Akt and MAPK signaling pathways [22]. H2O2 in the aqueous humor can activate MAPK signaling in HLE cells [7, 23], but the exact effect of ginsenosides on MAPK in HLB cells remains to be elucidated.
We also showed that ginsenosides reduced the percentage of early apoptotic HLB cells. This is consistent with our finding that ginsenosides modulate the expression of apoptosis-related genes such as LMNB1 and PSME4. In fact, PSME4 is among the top ten downregulated genes and is involved in negative regulation of apoptosis. In addition, three of the top ten upregulated genes are also related to apoptosis, including APP, LMNB1 and MAPK8. Apoptosis of HLB cells is increased during cataractogenesis [24], while ginsenosides may attenuate cataractogenesis by inhibiting H2O2–induced expression of apoptosis-related genes in HLB cells.
Conclusion
Ginsenosides can induce widespread changes in the expression of genes involved in multiple signaling pathways, including apoptotic signaling and DNA damage response signaling. Ginsenosides can alleviate H2O2-induced growth inhibition and inhibit H2O2-induced apoptosis in HLB cells.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Abbreviations
- H2O2 :
-
Hydrogen peroxide
- HLE:
-
Human lens epithelial
- MAPKs:
-
Mitogen-activated protein kinases
- OS:
-
Oxidative stress
- PBS:
-
Phosphate-buffered saline
- ROS:
-
Reactive oxygen species
References
Liu YC, et al. Cataracts. Lancet. 2017;390(10094):600–12.
Lee CM, Afshari NA. The global state of cataract blindness. Curr Opin Ophthalmol. 2017;28(1):98–103.
Lansingh VC, Eckert KA, Strauss G. Benefits and risks of immediately sequential bilateral cataract surgery: a literature review. Clin Exp Ophthalmol. 2015;43(7):666–72.
Prokofyeva E, Wegener A, Zrenner E. Cataract prevalence and prevention in Europe: a literature review. Acta Ophthalmol. 2013;91(5):395–405.
Robman L, Taylor H. External factors in the development of cataract. Eye (Lond). 2005;19(10):1074–82.
Kubota M, et al. Mitochondrial oxygen metabolism in primary human lens epithelial cells: association with age, diabetes and glaucoma. Free Radic Biol Med. 2016;97:513–9.
Tang X, et al. Honokiol inhibits H(2)O(2)-induced apoptosis in human lens epithelial cells via inhibition of the mitogen-activated protein kinase and Akt pathways. Eur J Pharmacol. 2011;650(1):72–8.
Truscott RJ. Age-related nuclear cataract-oxidation is the key. Exp Eye Res. 2005;80(5):709–25.
Sunkireddy P, et al. Natural antioxidant biomolecules promises future nanomedicine based therapy for cataract. Colloids Surf B Biointerfaces. 2013;112:554–62.
Liu X, et al. Efficacy and safety of ginsenoside-Rd for acute ischaemic stroke: a randomized, double-blind, placebo-controlled, phase II multicenter trial. Eur J Neurol. 2009;16(5):569–75.
Shin D, et al. Defensive properties of Ginsenoside re against UV-B-induced oxidative stress through up-regulating glutathione and superoxide dismutase in HaCaT keratinocytes. Iran J Pharm Res. 2018;17(1):249–60.
Bian M, et al. Combination of ginsenoside Rb1 and Rd protects the retina against bright light-induced degeneration. Sci Rep. 2017;7(1):6015.
Cai BX, et al. Ginsenoside Rb1 suppresses ultraviolet radiation-induced apoptosis by inducing DNA repair. Biol Pharm Bull. 2009;32(5):837–41.
Oh Y, et al. Ginsenoside Rc protects against UVBinduced photooxidative damage in epidermal keratinocytes. Mol Med Rep. 2017;16(3):2907–14.
Sahin E, et al. Cytotoxic, apoptotic and cell migration inhibitory effects of atranorin on SPC212 mesothelioma cells. Asian Pac J Tropl Biomed. 2019;9(7):299–306.
Ittiudomrak T, et al. α-Mangostin and apigenin induced the necrotic death of BT474 breast cancer cells with autophagy and inflammation. Asian Pac J Trop Biomed. 2018;8(11):519–26.
Ma J, et al. beta-Trcp ubiquitin ligase and RSK2 kinase-mediated degradation of FOXN2 promotes tumorigenesis and radioresistance in lung cancer. Cell Death Differ. 2018;25(8):1473–85.
Starostenko LV, et al. Interaction of nucleotide excision repair protein XPC-RAD23B with DNA containing Benzo[a]pyrene-derived adduct and Apurinic/Apyrimidinic site within a cluster. Biochemistry (Mosc). 2016;81(3):233–41.
Chen F, et al. Octyl Ester of Ginsenoside Rh2 induces apoptosis and G1 cell cycle arrest in human HepG2 cells by activating the extrinsic apoptotic pathway and modulating the Akt/p38 MAPK signaling pathway. J Agric Food Chem. 2016;64(40):7520–9.
Lv S, et al. Ginsenoside Rh2-B1 stimulates cell proliferation and IFN-gamma production by activating the p38 MAPK and ERK-dependent signaling pathways in CTLL-2 cells. Immunopharmacol Immunotoxicol. 2014;36(1):43–51.
Siddiqi MH, et al. Inhibition of osteoclast differentiation by Ginsenoside Rg3 in RAW264.7 cells via RANKL, JNK and p38 MAPK pathways through a modulation of Cathepsin K: an in Silico and in vitro study. Phytother Res. 2015;29(9):1286–94.
Cao GS, et al. A combination of four effective components derived from sheng-mai san attenuates hydrogen peroxide-induced injury in PC12 cells through inhibiting Akt and MAPK signaling pathways. Chin J Nat Med. 2016;14(7):508–17.
Peng J, et al. p-Coumaric Acid Protects Human Lens Epithelial Cells against Oxidative Stress-Induced Apoptosis by MAPK Signaling. Oxidative Med Cell Longev. 2018;2018:8549052.
Lu B, et al. miR-211 promotes lens epithelial cells apoptosis by targeting silent mating-type information regulation 2 homolog 1 in age-related cataracts. Int J Ophthalmol. 2018;11(2):201–7.
Acknowledgements
None declared.
Funding
None.
Author information
Authors and Affiliations
Contributions
JC designed the study, ZW prepared the manuscript, SZ and XH collected the data and performed data analysis, all authors gave the final approval.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study did not involve patients or animals, so ethical approval and consent to participate were not required.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, Z., Zhou, S., Hu, X. et al. Ginsenosides induce extensive changes in gene expression and inhibit oxidative stress-induced apoptosis in human lens epithelial cells. BMC Complement Med Ther 20, 44 (2020). https://doi.org/10.1186/s12906-020-2826-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-020-2826-8