Abstract
Background
Dental caries remains a serious problem due to its detrimental effects on individual health and quality of life. The bulbs of Myrmecodia pendans (Merr & Perry), native plants of Papua, have been used as natural remedies for tumours, gout, diarrhoea, and fever. In this study, one of the active compounds of M. pendans was isolated, and its biological activity against the formation of Streptococcus mutans ATCC 25175 biofilm was tested.
Methods
M. pendans was extracted with ethyl acetate using a Soxhlet apparatus. The extract was then separated, and chromatographic purification provided the isolated compound. The structure of the active compound was then characterized using UV, IR, NMR, and MS spectrometry. The obtained compound was added to S. mutans biofilms to determine the MBIC and MBEC values.
Results
The compound isolated from M. pendans was determined to be a labdane diterpene derivative with the formula C31H50O3. The MBIC value of the terpenoid towards the S. mutans biofilms was 50 ppm, and the MBEC value for the 1 min induction time was 40%.
Conclusion
The terpenoid extracted from M. pendans has the potential to be developed into an antibacterial agent particularly for preventing the formation of biofilms.
Similar content being viewed by others
Background
Dental caries remains a serious problem because it can trigger further health problems. Dental caries is a multifactorial disease. Interactions between cariogenic agents will impact the health of the tooth due to the presence of cariogenic biofilms on the tooth surface [1]. Streptococcus mutans is considered one of the cariogenic bacteria due to its ability to form biofilms [2, 3].
Currently, caries prevention is aimed at preventing the formation of dental biofilms or reducing the amount of S. mutans in the biofilms [3]. Some chemical compounds are commonly used in many dental products to suppress the growth of the bacteria, but chemical compounds often cause many side effects [4, 5]. Therefore, many studies have been conducted on the use of natural compounds for the prevention of disease.
Myrmecophytes (Myrmecodia pendans, literally “ant-plant”) originates from the Papua Islands, which are located in the eastern part of Indonesia. This plant can also be found in the Malay Peninsula, the Philippines, Cambodia, Sumatera, Java, Cape York, and the Solomon Islands. M. pendans belongs to the Rubiaceae family, which contains five genera, and only two of these genera are associated with ants, Myrmecodia (45 species) and Hydnophytum (26 species). Of all these species, only H. formicarum, M. pendans, and M. bulbosa are often used as natural remedies. M. pendans, known by the Papuans as a medicinal plant, can be used to treat various diseases, such as cancer, tumours, gout, diarrhoea, and fever [6, 7]. Soeksmanto’s research on rats showed that the toxicity of M. pendans extract at a dose of 375 mg/kg bw caused liver degeneration, whereas a dose of 3.750 mg/kg bw caused cell necrosis [8].
Methods
Chemicals and reagents
Silica gel resin 60 Li Chroprep RP-18 (Merck®), Kiesel gel 60 F254 and RP-18 F254S were purchased from Merck® (Darmstadt, Germany). Some of the solvents used in this study were purchased from Merck Co. Ltd., and the rest were purchased from Sigma Aldrich Co. Ltd. (St. Louis, MO, USA).
Plant material collection and determination
Dried bulbs of M. pendans were acquired from Papua and were identified by Mr. Joko (Botanist) at the Laboratory of Plant Taxonomy, Department of Biology, Faculty of Mathematics and Natural Sciences Universitas Padjadjaran, Bandung, Indonesia.
Extraction of the bulbs of M. pendans
The extraction of M. pendans bulbs was performed using a Soxhlet apparatus because the associated procedures are relatively fast and require less solvent than maceration methods [9]. Additionally, based on prior research on the isolation of these compounds, a temperature of 40 °C was used to prevent the decomposition of the compounds in the extract. Heating at 40 °C during the Soxhlet extraction process will not damage the constituents; therefore, the compounds contained in the M. pendans bulb were thermally stable. Ethyl acetate was used in the extraction process because after conducting a qualitative test with a thin-layer chromatography, the target compound was found in the ethyl acetate fraction.
As much as 1.5 kg of M. pendans bulbs was cut into small pieces to extract the maximum amount of each compound. Bulb pieces were extracted with as much as 3 L of ethyl acetate at 40 °C for 5 × 8 h using a Soxhlet apparatus (CV Ruchi) [9]. The extract was concentrated on a rotatory evaporator (Buchi® brand) at approximately 40 °C until 55.7 g of a concentrated ethyl acetate extract of M. pendans was obtained.
Separation and purification of the ethyl acetate extract of M. pendans bulbs
The concentrated ethyl acetate extract of M. pendans bulbs was separated by liquid column chromatography with stationary G60 silica gel (70–230 mesh) and 10% (v/v) of n-hexane-ethyl acetate as the eluent, and 11 fractions were produced. The contents of each fraction were then analysed using thin-layer chromatography (TLC) [10] with a stationary silica gel phase G60 F254 in determining the suitable solvent composition at subsequent purification process. The TLC results showed that fraction 3 of the first column chromatography step had relatively simple pattern of spots (Rf = 0.87) compared to other fractions. The fraction (5.7 g) was further purified using liquid column chromatography with a stationary G60 (70–230 mesh) silica gel phase and 2.5% (v/v) n-hexane-ethyl acetate as the eluent, and 17 fractions were produced. The fractions of the 2nd column chromatography step were analysed by TLC with a stationary silica gel (G60 F254) phase. The patterns of spots for 3-(7–9) showed a high number of individual components for 3-(7–9) to be considered pure (62.8 mg). The pattern of spots was further analysed by reversed-phase TLC using an octadecyl silane (ODS) stationary phase and 100% methanol as the eluent to determine a suitable solvent for the further purification of these fractions. The results of the analysis of fractions 3-(7–9) showed they had a constituent with the same Rf (0.62) as well as one other constituent. Then, the fraction was purified again by reversed-phase column chromatography using ODS RP-18 as the stationary phase and methanol with a 5% gradient of water (v/v) as the eluent, resulting in 8 fractions. The fractions of the 3rd column chromatography step were analysed using TLC with an ODS stationary phase, and this analysis showed the presence of a single compound, compound 1.
Structural determination
The structure of compound 1 was determined by analysing its 1H NMR, 13C NMR, HMQC, DEPT 135°, 1H-1H COSY and HMBC spectra, which were acquired on a 500 MHz FT-NMR spectrometer (ECA 500 JOEL variant, Japan). We used Delta™ NMR processing and control software, copyright 1990–2004 by JEOL USA, Inc. Version: 4.3.2 [Windows_NT] Network port = 6422. The IR spectrum of the compound was determined on an FT-IR Perkin Elmer Spectrum One spectrometer (Buckinghamshire, UK).
The number of carbon signals in compound 1, hybridization of the carbons (sp3, sp2, and sp), and the electronic environments impacting the chemical shift of each carbon atom in the compound were determined based on its 13C NMR spectrum. Information about the coupling of the signal from each carbon was also obtained from the 13C NMR spectrum using a DEPT parameter at 135°. The signals of the methine and methyl carbon atoms appeared as positive signals, whilst the signals of the methylene carbons appeared as negative signals. Signals from the quaternary carbons do not appear in DEPT spectra. The 2D NMR data were obtained by the DEPT 135° technique and from HMQC measurements. The HMQC spectrum showed the correlation data, or the relationships between protons and carbons one bond removed (1J). Determination of proton-to-proton coupling (1H-1H) across 3 or 4 bonds was performed by analysis of the 1H-1H COSY spectrum. The correlation of protons to protons in the 1H-1H COSY spectrum as indicated by the cross-peaks between the protons that are the result of spin matching.
The proposed structure of compound 1 was confirmed by mass spectrometry (ES-MS Spectrometer, UPLC Type MS/MS TQD, Waters). The mass spectrum was acquired in the negative ion (ES-) mode, which means that the peaks observed will indicate molecular weights slightly lower than the actual molecular weights. Furthermore, the chemical shifts of compound 1 were compared with those of a reference. The diterpenoid structure of compound 1 was confirmed by comparison of the chemical shifts of compound 1 with those of isocupressic acid, and the aliphatic chain was confirmed by the HMBC data of compound 1, and together, these data allowed us to assign all the chemical shifts. The results of the analysis of the two-dimensional NMR data confirmed the fragments and proposed structure of compound 1.
Bacterial strain and inocula preparation
The bacteria used was Streptococcus mutans ATCC 25175, and the bacteria were streaked on Muller Hinton agar (MHA) and incubated at 37 °C for 48 h under facultative anaerobic conditions (5% CO2). For the inoculum preparation, one inoculating loop of bacteria was grown in liquid MHA medium overnight at 37 °C and adjusted to the appropriate optical density (at 595 nm) using a UV-VIS (Shimazu® brand) spectrophotometer. The bacterial suspension was then diluted until it reached the McFarland standard of 0.5 or it contained approximately 108 CFU/mL [11].
Determination of the minimum inhibitory concentration (MIC)
The MIC of compound 1 from the bulbs of M. pendans was determined using a series of broth microdilutions according to the procedure described by Eloff [12] with a slight modification. Compound 1 was serially diluted in a 96-well microplate with MH broth as the solvent until a 1–100 ppm concentration was obtained. One hundred microliters of inoculum (bacterial suspension) was added to the well, and then the plate was sealed with parafilm and incubated for 24 h at 37 °C. The MIC values were examined after the addition of 50 μL of crystal violet and incubation for an additional 30 min at 37 °C. The presence of bacterial growth was determined based on the colour change of the suspension in the well, and a reduction in the intensity of the violet colour indicated the activity of compound 1 (terpenoid) [13, 14].
Analysis of the minimum biofilm inhibitory concentration (MBIC)
The effect of this terpenoid from M. pendans bulbs on S. mutans biofilm formation was measured using a modified Perumal method [15]. Each well of a 96-well microplate was filled with brain heart infusion (BHI) and 1% sucrose medium between 20 μL and 160 μL of bacterial culture with a cell density of 108 CFU/mL. To each well, 20 μL of terpenoid compound was added to give final concentrations of the compound from 1 to 100 ppm. Blank bacterial culture was used as a negative control, and the addition of 0.2% chlorhexidine was used as a positive control. After incubation for 24 h, the culture and the test compounds were removed, and each well was twice washed with 200 μL of phosphate-buffered saline (PBS). Afterwards, staining was performed by the addition of up to 200 μL of crystal violet stain and incubation for 30 min. The excess crystal violet stain was removed by rinsing three times with as much as 200 μL of PBS. Finally, up to 200 μL of 10% glacial acetic acid was added to each well to dissolve the crystal violet attached to the biofilm. Then, the absorbance was measured at a wavelength of 595 nm using a microplate reader.
Minimum biofilm eradication concentration (MBEC)
The ability of the terpenoid from M. pendans bulbs to eradicate S. mutans biofilm was analysed using microdilution with a modified version of the method described by LaPlante [16]. The process of S. mutans biofilm production was as described for MBIC analysis. After biofilm production, compound 1 (a terpenoid) was added, and the samples were left to stand for between 1 and 30 min. Wells containing bacteria were used as the negative control, and chlorhexidine was used as the positive control. The MBEC value was the lowest terpenoid concentration that was able to eradicate the biofilm, and the value was determined from the change in the colour of the well from the crystal violet and the MIC value.
Results
Thin-layer chromatography analysis
The pattern of spots from the TLC of fractions 3-(7–9)- (7&8) from the third column chromatography purification with 100% methanol as the solvent is shown in Fig. 1. The fraction was found to have a component with an Rf = 0.6. This compound was subsequently named compound 1, and 33.2 mg of the compound was isolated.
NMR analysis
Compound 1 was a yellow, methanol-soluble oil that fluoresced under UV light (254 nm and 365 nm). These characteristics showed that there were n → π* and π → π * transitions available in compound 1. The transition of n → π* indicated that compound 1 had a carbonyl group, and the transition of π → π* indicated that compound 1 had a double bond. The results of the analysis of the 1H NMR, 13C NMR, HMQC, DEPT 135°, 1H-1H COSY, and HMBC spectra are shown in Table 1.
The infrared spectrum (IR) of compound 1 from a KBr pellet showed the presence of an absorption band at 3427 cm− 1, which is indicative of a hydroxyl group [17, 18]. The presence of the absorption at 2935 cm− 1 indicated the presence of an sp3 C-H bond. Additionally, there was a band at 1715 cm− 1 characteristic of a C=O (carbonyl) moiety [18]. There were also absorptions indicative of the gem dimethyl fragment at 1384 cm− 1 and 1450 cm− 1. The presence of the C-O bond in compound 1 was indicated by an absorption band at 1172 cm− 1.
The 13C NMR spectrum of compound 1 showed 31 carbon signals consisting of 22 sp3 carbon signals from δC 12.9–78.9 ppm, 8 sp2 carbon signals, and 1 carbonyl moiety (C=O) at δC 177.1 ppm. The peak with a shift of 78.9 ppm was thought to be from an oxygenated carbon [19].
The DEPT-135° spectrum of compound 1 (Fig. 4.7) showed that compound 1 had 7 methyl (CH3) signals at δC 12.9, 14.6, 17.1, 18.0, 19.9, 22.7, and 26.0 ppm; 10 methylene (CH2) signals composed of 8 sp3 methylene carbons (δC 22.2, 27.0, 29.0, 30.2, 34.1, 36.4, 39.4, and 40.5 ppm) and 2 sp2 methylene carbons (δC 110.5 and 114.5 ppm); and 7 methine (CH) signals comprising 4 sp3 methine carbons (δC 35.4, 42.7, 45.8 and 49.7 ppm), 1 oxygenated methyl carbon at δC 78.9 ppm and 2 sp2 methyl carbons at δC 118.4 and 123.6 ppm. There were also 7 quaternary carbons (Cq) comprising two sp3 quaternary carbon at δC 30.8 and 44.5 ppm, 4 sp2 quaternary carbons (δC 104.6, 133.0, 136.5, and 146.8 ppm), and 1 carbonyl quaternary carbon at δC 177.1 ppm.
The HMQC spectrum confirmed the numbers carbons and hydrogens determined from the 13C NMR and 1H NMR spectra as well as the environmental information determined from the proton signals. There was a one-bond correlation between H-22 (3H; 1.48 ppm) and C-22 (18.0 ppm) in compound 1, and these signals can be attributed to a methyl group. This correlation was supported by the DEPT-135° data, which showed a positive signal for C-22. Additionally, the binding of 3 protons confirmed that C-22 was a methyl group and not a methine group. A one-bond correlation was also observed from H-2 (2H, 1.29 ppm) to C-2 (27.0 ppm) and from H-6 (2H, 1.29 ppm) to C-6 (30.2 ppm), which were methylene groups. The one-bond correlations of H-21 (2H, 4.49 ppm) with C-21 (114.56 ppm) and H-18 (1H, 4.59 ppm; 1H, 4.37 ppm) with C-18 (110.52 ppm) indicated that compound 1 contained an sp2 methylene carbon.
The 1H NMR spectrum showed the number, type, and environment of each of the protons in the compound. The protons in compound 1 consisted of 42 protons in the region characteristic of protons attached to sp3 carbons, 1 proton with a chemical shift suggesting it was part of a hydroxy moiety, 1 proton in the chemical shifting region characteristic of protons attached to oxygenated sp3 carbons, and 6 protons in the region characteristic of protons attached to sp2 hybridized carbons. Protons in the sp2 proton chemical shift region were olefinic protons at δH 4.54 ppm. Many overlapping signals were observed for the protons in the sp3 proton chemical shift region, such as from δH 1.23–1.29 and δH 1.44–1.56 ppm. This overlap was caused by the similar chemical shifts in the sp3 region; thus, the 1H NMR spectrum in this region was difficult to interpret without any supporting data from other experiments such as two-dimensional NMR. The overlapping signals in the δH 1.23–1.56 ppm region were attributed to protons attached to the terpenoid carbon chains [20, 21].
The three-bond correlation between the proton at C-25 and the proton at C-26 confirmed that the protons were in the same spin system, and the correlation between the proton at C-28 and the proton at C-29 proved they were in the same system as well. These correlations showed that these protons were adjacent. The HMBC correlations were used to determine the positions of the functional groups and the partial structure of compound 1, and the HMBC spectrum showed 2J–3J correlations between protons and carbons.
The HMBC spectrum of compound 1 showed correlations between the H-19 protons and C-2 and C-6. It can be assumed that there was a methyl group (C-19) attached at C-4 of the A ring. There were 2 sp2 methylene groups (C-18 and C-21) in compound 1. The 3J correlation between the H-18 protons and C-16 and the 2J correlation between the H-21 protons and C-8 helped elucidate the position of the double bond in compound 1. The 3J correlations between the H-14 protons and C-23 and between the H-24 protons and C-22 suggested a gem dimethyl group was present at the C-13 position. The presence of a gem dimethyl fragment was supported by the absorption bands at 1384.90 and 1450.96 cm− 1 in the infrared spectrum. There were also 2 double bonds in compound 1 (δC 123.66 ppm, 136.55 ppm, 146.84 ppm, and 118.45 ppm). The double bond between C-11 and C-14 was confirmed by the correlations between H-11 and C-24, H-11 and C-23, and H-14 and C-23. The 3J correlations between H-25 and C-27 and between H-27 and C-25 indicated the second double bond was between C-25 and C-26.
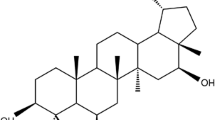
Based on the connectivity of the fragments of compound 1 determined from analysis of the two-dimensional NMR data, the structure of compound 1 was proposed to be that of a diterpenoid derivative. This compound has a molecular formula of C31H50O3 with seven degrees of unsaturation. The proposed structure of compound 1 is shown in Fig. 2.
Based on the results of the mass spectrometric measurements, compound 1 had a molecular weight of m/z 469.99. This result supported our proposed structure of compound 1, which has a molecular formula of C31H50O3 and a molecular weight of 470.
Based on a comparison of the NMR data of compound 1 to those of a reference compound, it was clear that compound 1 has signals that similar to those of the reference compound; thus, compound 1 was determined to be a diterpenoid derivative [22]. Furthermore, compound 1 was a terpenoid.
Inhibition of S. mutans biofilm formation by a terpenoid from M. pendans bulbs
The MIC value of the isolated terpenoid towards S. mutans was 40 ppm. The MBIC value of the terpenoid towards S. mutans biofilm formation was 50 ppm (Table 2). However, with three repetitions, the standard deviation for each measurement increased. This result was probably caused by the tendency of S. mutans to aggregate, which altered the amount of bacteria present in the initial culture preparation in each batch, which in turn impacted each measurement.
Results of the ability of the terpenoid from M. pendans to eradicate S. mutans biofilm
The MBEC value of the terpenoid towards the Streptococcus mutans biofilm for 1 min of treatment tended to increase; thus, the eradication percentage, or the amount of biofilm damaged also increased over time. At concentrations above 100 ppm, the relationship between activity and concentration was no longer linear, and those data were not included in the graph. The terpenoid from M. pendans bulbs was able to eradicate approximately 40% of the biofilms that had formed in 1 min at the highest concentrations tested (Table 2). After 30 min of treatment, a discrepancy was found between different experiments. Only one plate showed results consistent with those found for the 1 min treatment.
Discussion
The results of the NMR experiments conducted in this study showed that compound 1 was a diterpenoid derivative called a terpenoid with a molecular formula of C31H50O3. Previous research conducted by Widyawati [23] also found an active compound, which was a diterpenoid derivative, with the molecular formula of C25H40O4. However, there was a different substituent present at C11, and the number of carbons was different (25 carbons in the prior study compared to 31 carbons in this study). Although the core of the compounds was the same, the overall characteristics showed that they were different compounds. The MIC value of the C25H40O4 terpenoid towards S. mutans was 78.125 ppm [23], while the MIC of the terpenoid isolated in this study was 40 ppm. Perumal’s study [15] showed that the most active compound of Euphorbia hirta L. was a terpenoid. Terpenoids are the largest group of natural plant products known for having potential antimicrobial activities.
The MBIC value of the terpenoid from M. pendans bulbs towards S. mutans biofilm was 50 ppm, and its MIC value was 40 ppm. The concentration of a compound required to inhibit the growth of a bacterial biofilm was higher than that required for a planktonic biofilm. This is because the exopolysaccharides contained in the biofilms make penetration more difficult [24]. The active compounds of the natural substances show bactericidal and bacteriostatic effects by preventing bacterial attachment to the surface of the pharynx, skin, and tooth mucosa; inhibition of glycolytic enzymes; pH reduction; reduction of biofilm and plaque formation; and reduction of the hydrophobicity of the cell surface [25].
In this study, when the highest concentrations of the M. pendans terpenoid were tested for 1 min, they could destroy approximately 40% of the biofilms that had been formed. Cowan [26] stated that the target of active compounds for bacterial eradication could be reached through disruption of the mechanisms of cell wall biosynthesis and permeability of the cell membranes, surface adsorption of compound components, inhibition of protein synthesis or nucleic acid metabolism, or inhibition of the enzyme activity through oxidation. Terpenoids can influence the release of planktonic cells from biofilms. Terpenoids can also influence the membrane integrity of all organisms and eradicated most biofilm cells [27].
This study was the first that successfully showed the antimicrobial effect of a terpenoid from M. pendans under both the planktonic conditions and against S. mutans biofilms. Thus, natural compounds can be considered potential molecules for the prevention of dental plaque [28]. Natural substances were shown to have a direct effect on the formation of cariogenic biofilms by inhibiting the expression of glucosyltransferase activity in S. mutans and S. sobrinus [3, 29].
Conclusions
A terpenoid with a molecular formula of C31H50O3 extracted from M. pendans has potential to be developed as an antibacterial agent, particularly with to prevent the formation of biofilms with an MBIC value of 50 ppm and to eradicate approximately 40% of S. mutans biofilms.
Abbreviations
- ATCC:
-
American type culture collection
- CFU:
-
Colony forming unit
- COSY:
-
Correlation spectroscopy
- DEPT:
-
Distortionless enhancement by polarization transfer
- ES-MS:
-
Electrospray mass spectrometry
- HMBC:
-
Heteronuclear multiple bond correlation
- HMQC:
-
Heteronuclear multiple quantum coherence
- IR:
-
Infrared
- NMR:
-
Nuclear magnetic resonance
- TLC:
-
Thin-layer chromatography
- UPLC:
-
Ultra-performance liquid chromatography
- UV-VIS:
-
Ultraviolet-visible
References
Klein M, Duarte S, Xiao J, Mitra S, Foster T, Koo H. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Applied and Environmental Microbiology. 2009;75(3):837–41.
Yoshida A, Kuramitsu H. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl Environ Microbiol. 2002;68(12):6283–91.
Jeon J, Rosalen P, Falsetta M, Koo H. Natural products in caries reasearch: current (limited) knowledge, challenges and future perspective. Caries Res. 2011;45:243–63.
Vrani E, La-Evi A, Mehmedagi A, Uzunovi A. Formulation ingredients for toothpastes and mouthwashes. Bosnian Journal of Basic Medical Sciences. 2004;4(4):51–8.
Gold J. The role of chlorhexidine in caries prevention. Oper Dent. 2008;33(6):710–1.
Supriatno DR. Antitumor activity of Papua’s Myrmecodia pendens in human oral tongue squamous cell carcinoma cell line through induction of cyclin-dependent kinase inhibitor p27Kp1 and suppression of cyclin E. J of Cancer Res and Ther. 2014;2(3):48–53.
Soeksmanto A, Subroto M, Wijaya H, Simanjuntak P. Anticancer activity test for extracts of sarang semut (Myrmecodia pendens) to HeLa and MCM-B2 cells. Pak J Biol Sci. 2010;13(3):148–51.
Soeksmanto A, Simanjuntak P, Subroto MA. Uji toksisitas akut ekstrak air tanaman sarang semut (Myrmecodia pendans) terhadap histologi organ hati mencit. Jurnal Nature Indonesia. 2012;12(2):152–5.
Abdolshahi A, Mojteba HH, Jared SR, Mehrdad T, Aliakbar S, Jaime ATS. Choice of solvent extraction technique affects fatty acid composition of pistachio (Pistacia vera L.) oil. J Food Sci Technol. 2015;52(4):2422–7.
Kustrin SA, David WM, Ahmad PY. Thin-layer chromatography-bioassay as powerful tool for rapid identification of bioactive compounds in botanical extracts. Mod Chem Appl. 2013;3:1–2.
Gartika M, Inne SS, Mieke HS, Alex C, Danny H. Antibacterial activities of papain against Streptococcus mutans ATCC 25175. Int Journal of Development Research. 2014;4(10):2075–7.
Eloff JN. A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Plant Med. 1998;64:711–3.
Gupta A. Biofilm quantification and comparative analysis of MIC (minimum inhibitory concentration) and MBIC (minimum biofilm inhibitory concentration) value for different antibiotics against E. Coli. Int J Curr Microbiol App Sci. 2015;4(2):198–224.
Gordya N, Andrey Y, Anastasia K, et al. Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: Calliphora vicina medicinal maggots. Plos One. 2017;12(3)
Perumal S, Roziahanim M. Chemical analysis, inhibition of biofilm formation and biofilm eradication potential of Euphorbia hirta L. against clinical isolates and standard strains. BMC Complementary & Alternative Medicine. 2013;13:346.
LaPlante KL, Sarkisian SA, Woodmansee S, Rowley DC, Seeram NP. Effects of cranberry extracts on growth and biofilm production of Escherichia coli and Staphylococcus species. Phytother Res. 2012;26:1371–4.
Patra JK, Kim ES, Oh K, Kim HJ, Dhakal R, Kim Y, Baek KH. Bactericidal effect of extract and metabolites of Robinia pseudoacacia L. on Streptococcus mutans and Porphyromonas gingivalis causing dental plaque and periodontal inflammatory diseases. Molecules. 2015;20:6128–39.
Dong WW, Jiao W, Deng MC, Yang CB, Yue JM, Lu RH. A new steroid glycoside derivative from Acorus calamus L. J of The Chinese Chemical Society. 2008;55:1277–9.
Khatun M, Billah M, Quader MA. Sterols and sterol glucoside from Phyllanthus species. Dhaka Univ. J. Sci. 2012;60:5–10.
Seebacher W, Simic N, Weis R, Saf R, Kunert O. Complete assignments of 1H and 13C NMR resonances of oleanolic acid, 18α-oleanolic acid, ursolic acid and their 11-oxo derivatives. Magn Reson Chem. 41:636–8. https://doi.org/10.1002/mrc.1214.
Govenalp Z, Kilik N, Kazaz C, Kaya Y, Demiezer LO. Chemical constituen of Galium tirtunerte. Turk J. 2006;30:515–23.
Filho ER, Magnani RF, Xie W, Mirocha CJ, Pathre SJ. Hidroxiliation of labdane diterpene cupresic acid by Fusarum graminearum. J Braz Chem. 2002;13(2):266–9.
Widyawati. Potential of terpenoid isolated from Myrmecodia pendans as antibacterial against Streptococcus mutans. Int Journal of Development. Research. 2016; 6(10):10350–4.
Suzana KS, Robert EWH. Mode of action the new antibiotic for gram positive pathogens daptomycin comparison with cationic antimicrobial peptides and lipeptides. Biochim Biophys Acta. 1758;2006:1215–23.
Abachi S, Song L, Rupaisinghe HPV. Molecular mechanism of inhibition of Streptococcus species by phytochemicals. Molecules. 2016;21:215.
Cowan MM. Plant product as antimicrobial agents. Clin Microbiol Rev. 1999;12:564–82.
Mastelic J, Politeo O, Jerkovic I, Radosevic N. Composition and antimicrobial activity of Helichrysum italicum essential oil and its terpene and terpenoid fractions. Chem Nat Comp. 2005;41:35–40.
Sa NC, et al. Antimicrobial and antibiofilm action of Carbane diterpene from Croton nepetaefolius against oral bacteria. SciVerse ScienceDirect, 2011. https://doi.org/10.1016/j.archoralbio.2011.10.016.
Kalesinskas P, Kačergius T, Ambrozaitis A, Peciuliene V, Ericson D. Reducing dental plaque formation and caries development, a review of current methods and implications for novel pharmaceuticals. Stomatologija, Baltic Dental and Maxillofacial Journal. 2014;16(2):44–52.
Acknowledgements
We would like to thank Intan Taufik from the School of Life Sciences and Technology, Institut Teknologi Bandung and Eti Apriyanti from the Department of Chemistry, Faculty of Science, Universitas Padjadjaran who generously assisted with the studies conducted in their respective laboratories.
Funding
This work was supported by Riset Kompetisi Dosen Universitas Padjadjaran 2017 (Contract No. 872/UN6.3.1/LT/2017 LPPM Unpad).
Availability of data and materials
We have submitted additional figures and all supporting data to the Springer Nature repository with submission number: DSS-SPRN-00045.
Author information
Authors and Affiliations
Contributions
MG provided idea for this research and drafted this paper. DK analysed all the chemical structures in this paper. HTP and MHS helped to evaluate and edit this paper. All of the authors have approved this article.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gartika, M., Pramesti, H.T., Kurnia, D. et al. A terpenoid isolated from sarang semut (Myrmecodia pendans) bulb and its potential for the inhibition and eradication of Streptococcus mutans biofilm. BMC Complement Altern Med 18, 151 (2018). https://doi.org/10.1186/s12906-018-2213-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-018-2213-x