Abstract
Background
Xylanase inhibitors have been confirmed to be involved in plant defence. OsXIP is a XIP-type rice xylanase inhibitor, yet its transcriptional regulation remains unknown.
Results
Herbivore infestation, wounding and methyl jasmonate (MeJA) treatment enhanced mRNA levels and protein levels of OsXIP. By analyzing different 5’ deletion mutants of OsXIP promoter exposed to rice brown planthopper Nilaparvata lugens stress, a 562 bp region (–1451 – −889) was finally identified as the key sequence for the herbivores stress response. Using yeast one-hybrid screening, coupled with chromatin immunoprecipitation analysis, a basic helix-loop-helix protein (OsbHLH59) and an APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor OsERF71 directly binding to the 562 bp key sequence to activate the expression of OsXIP were identified, which is further supported by transient expression assay. Moreover, transcriptional analysis revealed that mechanical wounding and treatment with MeJA resulted in an obvious increase in transcript levels of OsbHLH59 and OsERF71 in root and shoot tissues.
Conclusions
Our data shows that two proteins as direct transcriptional activators of OsXIP responding to stress were identified. These results reveal a coordinated regulatory mechanism of OsXIP, which may probably be involved in defence responses via a JA-mediated signaling pathway.
Similar content being viewed by others

Background
Xylanase inhibitors (XIs) are a kind of plant-produced proteinaceous inhibitor that inhibits the activity of xylanase [1]. Recently, RIXI, riceXIP, OsXIP and OsHI-XIP xylanase inhibitors have been identified in rice plants [2–5], which all belong to XIP-type XIs. XIs have been thought to be involved in plant defence mainly for the reason that XIs only inhibit xylanases of microbial origin but not of plant origin. And many data provide evidence that XIs do indeed participate in plant defense [1].
Xylanase inhibitor genes act as defence-responsive genes in stress-induced signal transduction pathways. Taxi-Ia expression was induced 2.5 times and the transcripts of Taxi-Ib/III and Taxi-IIb/IV rose up to 20-fold by F. graminearum infection of wheat lemma, palea and ovary [6]. Infestation of wheat leaves by the powdery mildew fungus B. graminis induced the expression of Taxi-Ib/III and Taxi-IIb/IV [6]. The transcripts of OsXIP and riceXIP were drastically induced by wounding and methyl jasmonate (MeJA) treatment in the root [2]. Our previous study also revealed that pathogens can induce the expression of the rice xylanase inhibitor gene RIXI [7]. In planta direct evidence for this role has not been reported until Moscetti et al. [8] found that constitutive expression of the xylanase inhibitor TAXI-III delayed Fusarium head blight symptoms. Furthermore, overexpression of the RIXI xylanase inhibitor improved disease resistance of rice to the fungal pathogen, Magnaporthe oryzae [9]. In addition, overexpression of OsHI-XIP enhanced resistance in rice to herbivores, which is also the first time that a xylanase inhibitor has been demonstrated to play a role in resistance among rice herbivores [5]. However, the molecular basis underlying the regulation of XIs in plant defense is poorly understood.
A number of biotic and abiotic stress-responsive elements were observed by comparative analysis of cis-elements of xylanase inhibitors gene promoter by bioinformatics softwares PLACE and PlantCARE. The promoter region of a gene can provide valuable information about the factors inducing expression. For instance, cis-acting elements implicated in pathogen- and wound-inducible gene expression, i.e., GCC-box and W-box sequences could be recognized in the promoter region of TAXI-III [6]. Also investigation of the durum wheat Xip-II upstream region revealed the presence of a number of cis-acting elements controlling the expression of defense-related genes such as several W-boxes and a Myb-binding element, supporting its role in plant defense against pathogens [10]. The importance of these promoters regions has not yet been confirmed by promoter deletion analyses.
OsXIP is a XIP-type rice xylanase inhibitor, which was induced by various stresses such as MeJA treatment and wounding. And the expression patterns of OsXIP and riceXIP resemble each other and the induction of their expression by wounding may occur via a JA-mediated signaling pathway [2]. However, whether OsXIP plays an important role in resistance to invaders via a JA-mediated signaling pathway remains unclear.
Despite all these observations, there have been no reports on in planta functional characterization of the promoter region of xylanase inhibitor gene and its transcriptional regulation pattern so far. In this study, the promoter of OsXIP was cloned and analyzed, and a 562 bp region (−1451 to −889) was identified as the key sequence for the herbivores stress response by promoter deletion analyses. Using this 562 bp sequence as the bait, OsbHLH59 [11] and OsERF71 [12] proteins as direct transcriptional regulators of OsXIP responding to stress were identified. Collectively, our results, for the first time, reveal a transcriptional regulatory mechanism of OsXIP involved in defence responses.
Methods
Plant materials, growth conditions and stress treatments
The rice genotypes used in this study were Nipponbare wild-type (WT) and transgenic lines (see below). Rice seeds were sown in water and grown in normal culture solution in a greenhouse with natural day length extended to light/dark cycle of 14/10 h using high-pressure sodium lamp, with heating or ventilation used to maintain temperature at 28 °C and 18 °C during day and night respectively.
For wounding stress, 14-day-old seedlings were cut into 5–10 mm width and floated on distilled water. For phytohormone treatment, 14-day-old rice seedlings were submerged in 200 μM MeJA solution for 0, 2, 6, 12 and 24 h, and then shoots and roots were harvested separately. For BPH treatment, plants were individually infested with 20 adult BPH confined in a glass cylinder [diameter 4 cm, height 8 cm, with 48 small holes (diameter 0.8 mm)], the top of which was covered with a piece of sponge. One empty cylinder was used for control plants (non-infested).
Construction of the OsXIP promoter vectors and rice transformation
The full-length OsXIP gene promoter named OP1 (−2070 bp to +52 bp) was amplified from the genomic DNA of WT with OP1-U (forward primer) and OP1-L (reverse primer). Then a series of nested 5’ deletions of OP1 fragments OP2 (−1451 bp to +52 bp), OP3 (−889 bp to +52 bp), OP4 (−569 bp to +52 bp), OP5 (−380 bp to +52 bp), OP6 (−172 bp to +52 bp), OP7 (−90 bp to +52 bp) were amplified by PCR from pMD19T-OP1 using the common reverse primer OP1-L and either the forward primers OP2-U, OP3-U, OP4-U, OP5-U, OP6-U, or OP7-U, respectively. The primers are shown in Additional file 1: Table S1. The full-length promoter and 5’-deletion derivatives were cloned into the pBI101.3-GUS upstream of GUS (β-glucuronidase). Empty vector pBI101.3-GUS was used as a negative control (VC). All constructs were mobilized into Agrobacterium tumefaciens EHA105 and transformed into calli derived from mature seeds of rice according to a previously described protocol [13, 14]. Approximately 90 calli were co-cultured for each vector, with the number of putative independent transformed plants being regenerated being 25, 19, 20, 16, 22, 13, 17 and 10 respectively for OP1-OP7 and a vector control. Primary transformants (T0) were raised, transferred to soil and allowed to grow in a greenhouse. Seeds were harvested and used for analysis in the next generation.
RNA extraction and quantitative RT-PCR
Total RNA was isolated from roots and shoots of rice seedlings using the RNAprep pure plant kit (Tiangen) according to the manufacturer’s protocol. RNA (1 μg) was used to synthesize the first strand complementary DNA (cDNA) with an oligo (dT) primer according to the instruction of the PrimeScript first-strand cDNA synthesis kit (Takara). The qRT-PCR assay was performed on LightCycler480 instrument (Roche) using a SYBR® Premix Ex TaqTM kit (Takara). A rice actin gene Osactin (GenBank: Os03g50885) was used as an internal standard to normalize cDNA concentrations. The primers for qRT-PCR are listed in Additional file 2: Table S2. The relative quantification of gene expression was analyzed by the comparative method (2−ΔΔCt) [15] with some modifications. Using the 2−ΔΔCt method, data were presented as the fold-change in mRNA expression normalized to the endogenous reference gene (Osactin) and relative to the control.
Western blot analysis
OsXIP-specific polyclonal antibody was produced against a 15-residue synthetic peptide sequence of OsXIP (CGGRRNGVYRPFGDA) by GenScript USA Inc (China). Anti-OsXIP rabbit polyclonal antibody or anti-β-actin mouse monoclonal antibody (Beijing ComWin Biotech Co.,Ltd, CW0096) was used as the primary antibody. Immunoblot analysis was performed as described by Akulinkina et al. [16]. Samples were prepared from leaves of 2-weeks old rice plants treated with BPH for 24 h, wounding for 12 h and MeJA for 12 h, respectively. Protein fractions were separated by SDS-PAGE, and then transferred onto nitrocellulose membrane. Finally, immune complexes on a membrane were detected with BCIP/NBT. The reaction was stopped after 3 minutes of incubation by rinsing the membrane with water.
Quantitative GUS analysis
Quantitative GUS activity was measured according to the method described by Jefferson et al. [17] with some modifications. Briefly, the shoots or roots of 14-days seedlings that carried different fragments of OP1 were homogenized in GUS extraction buffer (50 mM PBS, pH 7.0, 10 mM EDTA, pH 8.0, 20% methanol, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mM β-mercaptethanol). Crude protein extract (50 μl) was added to 450 μl of extraction buffer containing 2 mM 4-methylumbelliferyl-β-D-glucuronide (MUG) at 37 °C for 30 min or 60 min, and thereafter 200 μl of the reaction mixture was added to 800 μl of stop buffer (0.2 M Na2CO3). The 4-methylumbelliferone fluorescence was measured using a spectrofluorophotometer (RF-5301PC, Shimadzu) at 460 nm with excitation at 355 nm. Protein concentration was quantified by methods described by Bradford [18]. GUS activity was calculated as pmol of 4-methylumbelliferon (4-MU) min per minute and per milligram of total soluble proteins and presented as GUS activity relative to the VC.
Yeast one-hybrid (Y1H) screening
The Y1H screening used the Matchmaker Gold One-Hybrid Library Screening System (Clontech, Cat. Nos. 630491). The bait sequence (562 bp fragment) was cloned into the pAbAi vector that harbors the AUR1-C gene, conferring resistance to Aureobasidin A (AbA, a cyclic depsipeptide antibiotic used as a yeast selection marker). The resulting pAbAi-Bait construct was then linearized and integrated into the genome of the Y1HGold yeast strain by homologous recombination to generate a bait-specific reporter strain. The minimal inhibitory concentration of Aureobasidin A for the bait-specific reporter strain was determined. And the strain was used to screen a cDNA library generated from the leaves of WT treated by BPH for 24 h. The transformants were initially screened on selective medium (SD/−Leu/AbA100) and the positive colonies were identified by PCR and DNA sequencing.
For the re-transformation assay, the full-length CDSs of candidate genes were amplified from cDNA using the primers 59-AD-U/L and 71-AD-U/L as listed in Additional file 1: Table S1. The PCR products were then cloned into the pGADT7 vector and the resulting constructs were transferred into the bait reporter yeast strain mentioned above, respectively. The cells were grown on SD/−Leu and (SD/−Leu/AbA100) plates at 30 °C for 3 days, and resuspended in liquid media to OD600 of 0.1 (10−1) and diluted in a 10× dilution series (10−2 to 10−3). Of each dilution, 7 μl was spotted on media selecting for both plasmids (SD/−Leu) and selecting for interaction (SD/−Leu/AbA100), supplemented with 100 ng ml−1 to suppress background growth. The empty vector pGADT7 was used as a negative control.
Chromatin immunoprecipitation (ChIP)-PCR analysis
The 35Sp::OsbHLH59:GFP and 35Sp::OsERF71:GFP expression vectors were constructed by subcloning the full-length CDSs without terminators of OsbHLH59 and OsERF71 into the pCAMBIA1300-sGFP vector under the control of the 35S promoter [19], respectively. The primers 59-GFP-U/L and 71-GFP-U/L are listed in Additional file 1: Table S1. The resulting constructs were then introduced into Agrobacterium tumefaciens strain EHA105, and transformed into WT rice.
2–3 g of 3–weeks-old 35S:OsbHLH59-GFP or 35S:OsERF71-GFP transgenic seedlings were used for ChIP-PCR experiments as described in Haring et al. [20]. In brief, transgenic rice was fixed with 60 mL of 1.0% formaldehyde by vacuuming for 10 min. The chromatin DNA was sheared to 200–500 bp fragments by sonicating. Sheared DNA was incubated with GFP antibody (Biogot) (ChIP). Chromatin before immunoprecipitation was used as an input control. The primers for PCR of the target DNA are listed in Additional file 3: Table S3.
Transient expression in Nicotiana benthamiana leaves
The construction of vectors and transient expression in Nicotiana benthamiana leaves were performed as described by Ding et al. [21]. Briefly, for construction of effector vectors, the full length ORFs of OsbHLH59 and OsERF71 were amplified and cloned into the pCAMBIA1300-sGFP vector under the control of the 35S promoter. The 5’–deleted OP2 and OP3 promoters were constructed into the reporter vector pGreenII0800-LUC [22]. The recombinant plasmids were transferred into the Agrobacterium EHA105 lines. Then the EHA105 lines were co-infiltrated into the N. benthamiana leaves as described previously [23]. The Firefly and Renilla luciferase activities were quantified using a Dual Luciferase assay kit (Promega, http://www.promega.com/).
Subcellular localization
The 35Sp::OsbHLH59:GFP and 35Sp::OsERF71:GFP expression vectors were infiltrated into the N. benthamiana leaves by Agrobacterium-mediated transformation [24]. The two constructs were also transfected into rice protoplasts according to the protocol of Yoo et al. [25] with some modifications. The rice protoplasts were isolated from stems of 12-day-old WT seedlings by enzyme hydrolysis. Then 10 μg plasmid DNA was polyethylene glycol/calcium-transfected into these protoplasts. Empty vector was used as a control. The cells were observed with a confocal microscope (Zeiss LSM 710).
Results
Herbivore infestation, wounding and methyl jasmonate (MeJA) treatment enhanced mRNA levels and protein levels of OsXIP
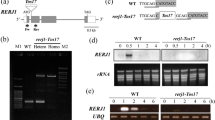
qRT-PCR analysis revealed herbivore infestation, mechanical wounding, and MeJA treatment, especially wounding, resulted in an obvious increase in transcript levels of OsXIP (Fig. 1a). OsXIP expression was induced 3.5 times by BPH infestation and five-fold by wounding and MeJA treatment. To determine whether these stress also affected the protein levels of OsXIP, we analyzed the protein levels of OsXIP by quantitative GUS activity of transgenic rice that carried the full-length OsXIP gene promoter named OP1 (−2070 bp to +52 bp) and western blotting. The expression levels of GUS all significantly rose up (Fig. 1b). Immunoblot analysis showed that the level of OsXIP also increased upon these stress (Fig. 1c). These results demonstrate that OsXIP is a stress-responsive gene.
Inducible expression of OsXIP after different treatments. a Transcript levels of OsXIP in rice leaves after different treatments were analyzed by qRT-PCR. Two-weeks-old WT seedlings were treated with BPH for 24 h, wounding for 12 h and MeJA for 12 h, respectively. The data represent means ± SD of three independent replicates. Asterisks indicate statistically significant differences compared with CK (0 h) (**P < 0.01; Student’s t test). b GUS activity of OP1 transgenic rice after different treatments was measured by quantitative fluorescence method. Two-weeks-old transgenic rice seedlings were treated with BPH for 24 h, wounding for 12 h and MeJA for 12 h, respectively. The data represent means ± SD of three independent replicates. Asterisks indicate statistically significant differences compared with CK (**P < 0.01; Student’s t test). c Western blot analysis of OsXIP. Protein fractions were isolated from leaves of WT treated with BPH for 24 h, wounding for 12 h and MeJA for 12 h, respectively and subjected to immunoblot with anti-OsXIP antibody (top panel). M, markers of proteins, the sizes of the markers are indicated at the left of the picture. Fractions corresponding to 10 μg were loaded into each lane and equal loading was confirmed by anti-actin antibody (bottom panel)
Identification of herbivore-responsive promoter region
Given the observation that the expression of OsXIP was induced by different stress (Fig. 1), we used the PLACE and PlantCARE to analyse the promoter sequence of OsXIP. And as expected, a series of biotic and abiotic stress-responsive cis-regulatory elements exist in the promoter region (Additional file 4: Figure S1), such as W-box (TGACY) element, ABRE (CACGTG) element, MYB-binding (CGGTCA) site. To further determine the key sequences of the OsXIP promoter responding to herbivory, the transgenic plants that carried a set of 5’ deletion promoter reporter constructs, OP2, OP3, OP4, OP5, OP6 or OP7 were obtained (Fig. 2) and their quantitative GUS activity was measured.
Schematic representation of 5’ deletion promoter constructs and their GUS expression in shoot tissues. TSS, transcriptional start site. For every construct, three independent T2 transgenic lines were measured, and similar results were obtained. The data represent means ± SD of three independent replicates
Without BPH treatment (Fig. 2), about 110- and 54-fold higher expression of GUS was detected with OP1 and OP2 in the shoots, respectively, compared with vector control (VC). It was interesting to observe a sudden drop in expression with OP3, which was 18-fold higher compared with VC and similar to OP4. Surprisingly, the relative GUS expression for OP5 and OP6 are both three-fold greater than OP3 and OP4, despite having a greater deletion in the promoter region. Both OP5 and OP6 retain restore the expression level observed in OP2, whereas OP7 with the greatest promoter deletion has very minimal expression.
After infestation with BPH for 24 h (Fig. 3), GUS expression increased significantly in both the shoots and roots of transgenic lines OP1 and OP2. Elevated expression of GUS was also observed in the shoots of OP5, OP6 and OP7 and root of OP6. However, no significant change in expression was observed in the shoots of OP3 and OP4 and roots of OP4, OP5 and OP7. A sudden decrease in GUS expression was observed in the root of OP3. As mentioned above, the GUS expression of OP1 and OP2 was induced after BPH infestation, but OP3 and OP4 were not induced. Comparing the promoter regions upstream from the GUS gene in the constructs OP2 and OP3, OP2 had 562 bp region [−1451 bp to −889 bp relative to the transcriptional start site (TSS)] that was deleted from OP3. And the 562 bp fragment contains some stress-responsive cis-acting elements, such as W-box, ARR1AT, MBS. So we speculated that the 562 bp fragment was the key sequence involved in herbivore stress response. While the 290 bp fragment (−380 bp to −90 bp) was involved in herbivore stress response, this 562 bp fragment was then used for further research in this paper.
GUS expression of 5’ deletion promoter constructs infested by BPH in T2 transgenic plants. Relative (to VC) GUS activity in shoots (a) and roots (b) of different transgenic lines (OP1–OP7) infested by BPH for 24 h. For every construct, three independent T2 transgenic lines were measured, and similar results were obtained. The data represent means ± SD of three independent replicates. Asterisks indicate statistically significant differences compared with control (Non-infested) (**P < 0.01; Student’s t test)
Proteins bound to the herbivores-responsive promoter region
To determine the proteins bound to the herbivores-responsive promoter region, we used the 562 bp fragment mentioned above as a bait to screen transformants from a cDNA library generated from the leaves of rice plants infested with BPH for 24 h by yeast one-hybrid (Y1H) screening system. Through initial screening, DNA sequencing and BLAST analysis, some proteins including functional proteins and regulatory proteins showing interaction with OsXIP promoter were obtained. The functional proteins included fructose 1, 6-bisphosphatase, HSP, photosystem, Tify domain containing protein. While only two genes LOC_Os02g02480 and LOC_Os06g09390, according to the rice genome annotation of The Institute for Genomic Research (TIGR; http://rice.plantbiology.msu.edu/), were the candidates of regulatory proteins. Based on BLAST analysis and literatures, LOC_Os02g02480 is a basic helix-loop-helix protein (OsbHLH59) [11]; LOC_Os06g09390 is an APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor (OsERF71) [12].
The interaction between herbivores-responsive promoter region and the corresponding complete encoding products of the two genes were re-tested by Y1H assay. The analysis showed that the two proteins interacted specially with the promoter region (Fig. 4a). Meanwhile, we performed ChIP-PCR analysis by transforming 35S:OsbHLH59-sGFP and 35S:OsERF71-sGFP into rice to determine whether OsbHLH59 and OsERF71 regulate gene expression by binding to the 562 bp region in vivo. Our results showed that the promoter fragments of OsXIP were detected in the ChIP assays (Fig. 4b), further confirming that OsbHLH59 and OsERF71 can directly bind to the promoter motifs in vivo.
OsbHLH59 and OsERF71 bind to OsXIP promoter in vitro and in vivo. a TFs (OsbHLH59, and OsERF71) bind to OsXIP promoter in yeast. Bait strain Y1HGold[pBait-AbAi] yeast cells was transformed with a prey vector, containing OsbHLH59 and OsERF71 fused to a GAL4 activation domain, respectively. Cells were grown in liquid media to OD600 of 0.1 (10−1) and diluted in a 10× dilution series (10−2 to 10−3). Of each dilution, 7 μl was spotted on media selecting for both plasmids (SD/−Leu) and selecting for interaction (SD/−Leu/AbA100), supplemented with 100 ng ml−1 AbA to suppress background growth. b ChIP-PCR analysis. The ChIP of TFs (OsbHLH59, and OsERF71) assays was performed using transgenic rice expressing the 35S:OsbHLH59-GFP fusion or 35S:OsERF71-GFP fusion. Products of ChIP assays were amplified using five specific primers (listed in Additional file 1: Table S1)
Transcriptional activation of the OsXIP promoter by OsbHLH59 and OsERF71
We also performed the tobacco transient expression assay to further clarify the DNA-binding activities of the two proteins. The dual luciferases vector was used as a reporter system following Hellens et al. [22]. The full length ORFs of OsbHLH59 and OsERF71 were amplified and cloned into the effector vector. The OP2 and OP3 promoters were constructed into the reporter vector as P1 and P2 (Fig. 5a), which infiltrated N. benthamiana leaves alone or co-expressed with the corresponding effector vectors, respectively. It was obvious that co-expression of OsbHLH59 remarkably increased LUC expression driven by the OP2 promoter, as did expression of OsERF71 (Fig. 5b), suggesting OsbHLH59 and OsERF71 proteins promote the transcription of OsXIP. These results indicated that the two proteins may function as positive transcriptional regulators of OsXIP expression.
Tobacco transient transactivation assay for the interaction between OsbHLH59, OsERF71 and OsXIP promoter. a Characterization of OsXIP promoter and structures of vectors. Full length of the promoter from the translational start site (ATG) was indicated. OP2 and OP3 promoters were constructed into the report vector, and TFs (OsbHLH59, and OsERF71) were cloned into the effect vector, respectively. b Transient expression assay in N. benthamiana. LUC, Firefly luciferase activity; REN, Renilla luciferase activity (used as control). Data show ratios of LUC to REN and represent means ± SD of three independent replicates
Subcellular localization of OsbHLH59 and OsERF71
To further evaluate the role of the OsbHLH59 and OsERF71 proteins, their subcellular localization were determined. We constructed 35Sp::OsbHLH59:GFP and 35Sp::OsERF71:GFP fusion genes, and transiently expressed the constructs in N. benthamiana leaves (Additional file 5: Figure S2) and rice protoplasts (Fig. 6), respectively. Fluorescence analysis revealed that the proteins localized only in the nucleus (Fig. 6). Cells infiltrated with GFP construct (control) yielded fluorescence both in the cytosol and the nucleus. These results further indicate that the two proteins may be transcriptional factors that function in the nucleus to regulate OsXIP expression.
Subcellular localization of OsbHLH59 and OsERF71 in rice protoplasts. The rice protoplasts were transformed with 35Sp::OsbHLH59:GFP, 35Sp::OsERF71:GFP or pCAMBIA1300-GFP. The transformed cells were observed under a confocal microscope. The photographs were taken under detecting GFP fluorescence, bright field, chloroplast auto-fluorescence, and merged microscope images, respectively. Empty vector (pCAMBIA1300-GFP) was used as a control. Bars, 10 μm
Influence of abiotic stress on the expression of transcription factors
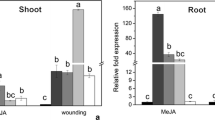
The expression patterns of the two genes that encode transcription factors bound to the rice herbivore-responsive cis-elements were examined by qRT-PCR in wild-type rice plants treated with MeJA or wounding at varying time intervals (2–24 h). The expression of these genes appeared to be responsive upon abiotic stress and was similar (Fig. 7). In shoot tissues, transcript expression of OsbHLH59 and OsERF71 increased concomitantly with time under MeJA treatment to 6 h, while decreased at 2 h and thereafter increased under wounding stress. The expression levels of OsbHLH59 and OsERF71 in root tissues were maximally induced approximately six- and seven-fold, and seven- and eight-fold after MeJA and wounding treatment, respectively. These results further suggest that the genes encoding these transcription factors may be involved in defence responses against herbivores by a JA- mediated pathway.
Expression patterns of the genes encoding transcription factors under abiotic stress analysed by qRT-PCR. Two-weeks-old WT seedlings were treated with 200 μM MeJA or wounding. Samples were collected immediately after treatment (0 h) and at 2 to 24 h after treatment. The data represent means ± SD of three independent replicates. Letters indicate significant differences between means using Duncan’s multiple range mean comparisons (5% α)
Discussion
The phytohormone jasmonic acid (JA) play a vital role in plant defense when plants are exposed to invaders [26]. In general, herbivorous insects and necrotrophic pathogens are more sensitive to JA-induced defenses [27, 28]. Wounding that caused by mechanical injury or insect feeding, leads to the accumulation of JA. Subsequently, the JA pathway is activated, which induces JA-responsive gene expression [29]. Our results revealed that herbivore infestation, mechanical wounding and MeJA treatment enhanced the expression of OsXIP at the transcriptional and protein levels (Fig. 1), which is consistent with OsHI-XIP [5]. To date, there is one report that the xylanase inhibitor XIP-I could inhibit a xylanase from the digestive tract of the coffee berry borer [30]. OsXIP is a wound stress-responsive gene in rice [31]. When the herbivore feeds on the plants, wounding generates and then induces the accumulation of jasmonic acid (JA), which in turn activates wounding-associated defense pathways [32]. In our study, the expression level of OsXIP was up-regulated by BPH and the N. lugens induction process was very similar to that caused by wounding. When the rice was infested by BPH, wounding generated first and then activated the wound-responsive gene OsXIP, thus enhancing resistance against herbivores.
Bioinformatic analysis of the promoter of OsXIP revealed that it was a stress-induced promoter. OsXIP promoter contained stress-responsive cis-acting elements, such as ARR1AT, W-box, G-box and TGACG-motif. The W-box (TGACY), has been confirmed to be present upstream of salicylic acid or wound signal responsive genes; The G-box (CACGTG) and G-box-like (CANNTG, also called the E-box) are known to the binding sites of basic helix-loop-helix (bHLH) transcription factors [33]; TGACG motif has been reported to be essential for responsiveness to MeJA; ARR1AT and WRKY71OS exist in the promoter of growth regulators responsive genes. Thus, OsXIP gene responded to different stress (Fig. 1).
OsXIP promoter deletion analyses showed that GUS expression of OP3 reduced sharply compared with OP2, while OP4 slightly enhanced and OP5 reached a maximum (Fig. 2). This suggested there were at least an enhancer (e) and a repressor (r) motif between OP2 and OP3 (−1451 bp to −889 bp) and OP4 and OP5 (−569 bp to −380 bp), respectively. So a regulation model of the OsXIP gene was proposed: an enhancer site (e) may exist in −1451 bp to −889 bp, and a repressor site (r) may be located between −569 bp and −380 bp in the promoter. GUS expression in OP2 was high due to the binding of enhancer (E) to its site (e). The enhancer might preferentially bound to the promoter and facilitate the gene expression on the grounds that binding of enhancer E at e site prevents the repressor R from binding to r site by either binding with R or obstructing r site. GUS expression of OP3 and OP4 decreased because of deletion of enhancer site (e). GUS expression of OP5 and OP6 increased after deletion of the r site. Serial deletion promoter constructs between −1451 and −889 bp and between −569 and −380 bp can be constructed to determine the locations and sequences of e and r by transient expression assay and electrophoretic mobility shift assay (EMSA).
After BPH stress, a 562 bp region (−1451 to −889) in the OsXIP promoter were finally identified as the key sequences involved in the herbivores stress response (Fig. 3). By Y1H screening, positive DNA-protein binding in vitro and in vivo (Fig. 4) and nuclear localization (Fig. 6), OsbHLH59 and OsERF71 proteins were confirmed as the direct regulators of OsXIP expression. The two proteins activated the expression of OsXIP gene by transient transactivation assay (Fig. 5). Moreover, the two genes encoding corresponding OsbHLH59 and OsERF71 proteins showed differential expression upon MeJA and wounding stress (Fig. 7), further suggesting that they may be involved in positively regulating OsXIP expression via JA-mediated defence responses.
In the present study, two proteins (OsbHLH59, and OsERF71) as transcription activators of OsXIP were found. OsbHLH59 is a member of bHLH transcription factors, which are known to bind to G-box or E-box [11]. We analyzed the bait sequence and found that several E-box elements were present, including CAGTTG, CACTTG and CAATTG. And whether OsbHLH59 binds to these E-box elements of the bait can be further determined by EMSA. Some bHLH transcription factors in rice are involved in stress responses. For instance, RERJ1 (OsbHLH006) responded to wound and drought [34, 35]; OsBP-5 (OsbHLH102) is related to transcriptional regulation of the rice Wx gene [36]; OsbHLH094 forms a complex with RSS3 and JASMONATE ZIM-DOMAIN (JAZ) proteins to modulate the expression of JA-responsive genes [37]; DPF (OsbHLH25) positively regulates the accumulation of diterpenoid phytoalexins [38].
In Arabidopsis, the bHLH transcription factor MYC2 is a key positive transcriptional regulator of JA signaling pathway, which is inhibited by JAZ transcriptional repressors [39, 40]. Similarly, OsbHLH062 interacted with OsJAZ9 to regulate JA-responsive genes expression in rice [41]. Furthermore, the JA signaling pathway in Arabidopsis composes of the two major branches: the ERF branch and MYC branch [29]. And the ERF branch is controlled by AP2/ERF transcription factors, such as ORA59 [42]. Thus, the OsERF71 protein, an AP2/ERF transcription factor, may belong to the ERF branch to positively regulate the expression of OsXIP. In addition, transcriptional analysis revealed mechanical wounding and MeJA induced transcriptional expression of OsbHLH59 and OsERF71 in rice (Fig. 7). These results reinforce the possibility that the induction expression of OsXIP by wounding may be regulated by the JA-mediated signaling pathway [2]. So OsbHLH59 and OsERF71 may belong to the MYC branch and the ERF branch, respectively, to positively regulate the expression of OsXIP. This speculation is further supported by the findings that OsXIP was independent of growth and development in rice plants [30] and its expression was not regulated by phytohormones associated with growth [2].
Conclusions
In summary, we reveal the transcriptional regulatory mechanism of OsXIP and its involvement in defense response in rice. In response to herbivore infestation, mechanical wounding or MeJA stress, OsbHLH59 and OsERF71 transcription factors promote this process by activating the expression of OsXIP via directly binding to its promoter. Our discovery contributes to clarify the regulatory mechanism of OsXIP and gives us a better understanding of the function of OsXIP in plant defence.
Abbreviations
- BPH:
-
Brown planthopper
- ChIP:
-
Chromatin immunoprecipitation
- GFP:
-
Green fluorescent protein
- GUS:
-
β-glucuronidase
- JA:
-
Jasmonic acid
- JAZ:
-
JASMONATE ZIM-DOMAIN
- MeJA:
-
Methyl jasmonate
- XIs:
-
Xylanase inhibitors
References
Dornez E, Croes E, Gebruers K, De Coninck B, Cammue BPA, Delcour JA, Courtin CM. Accumulated evidence substantiates a role for three classes of wheat xylanase inhibitors in plant defense. Crit Rev Plant Sci. 2010;29:244–64.
Tokunaga T, Esaka M. Induction of a novel XIP-type xylanase inhibitor by external ascorbic acid treatment and differential expression of XIP-family genes in rice. Plant Cell Physiol. 2007;48:700–14.
Goesaert H, Gebruers K, Courtin CM, Delcour JA. Purification and characterization of a XIP-type endoxylanase inhibitor from Rice (Oryza sativa). J Enzyme Inhib Med Chem. 2005;20:95–101.
Durand A, Hughes R, Roussel A, Flatman R, Henrissat B, Juge N. Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J. 2005;272:3227.
Xin ZJ, Wang Q, Yu ZN, Hu LC, Li JC, Xiang CY, Wang BH, Lou YG. Overexpression of a xylanase inhibitor gene, OsHI-XIP, enhances resistance in rice to herbivores. Plant Mol Biol Rep. 2014;32:465–75.
Igawa T, Ochiai-Fukuda T, Takahashi-Ando N, Ohsato S, Shibata T, Yamaguchi I, Kimura M. New TAXI-type xylanase inhibitor genes are inducible by pathogens and wounding in hexaploid wheat. Plant Cell Physiol. 2004;45:1347–60.
Hou CX, Zhan YH, Jiang DA, Weng XY. Functional characterization of a new pathogen induced xylanase inhibitor (RIXI) from rice. Eur J Plant Pathol. 2014;138:405–14.
Moscetti I, Tundo S, Janni M, Sella L, Gazzetti K, Tauzin A, Giardina T, Masci S, Favaron F, D’Ovidio R. Constitutive expression of the xylanase inhibitor TAXI-III delays Fusarium head blight symptoms in durum wheat transgenic plants. Mol Plant Microbe In. 2013;26:1464–72.
Hou CX, Lv T, Zhan YH, Peng YY, Huang YY, Jiang D, Weng XY. Overexpression of the RIXI xylanase inhibitor improves disease resistance to the fungal pathogen, Magnaporthe oryzae, in rice. Plant Cell Tiss Org. 2015;120:167–77.
Elliott G, Durand A, Hughes RK, Kroon PA, D’Ovidio R, Juge N. Isolation and characterisation of a xylanase inhibitor Xip-II gene from durum wheat. J Cereal Sci. 2009;50:324–31.
Li XX, Duan XP, Jiang HX, Sun YJ, Tang YP, Yuan Z, Guo JK, Liang WQ, Chen L, Yin JY, Ma H, Wang J, Zhang DB. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–84.
Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–32.
Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–82.
Mei CS, Qi M, Sheng GY, Yang YN. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe In. 2006;19:1127–37.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–8.
Akulinkina DV, Bolychevtseva YV, Elanskaya IV, Karapetyan NV, Yurina NP. Association of high light-inducible HliA/HliB stress proteins with photosystem 1 trimers and monomers of the cyanobacterium Synechocystis PCC 6803. Biochemistry (Mosc). 2015;80:1254–61.
Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–7.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Zhou G, Qi J, Ren N, Cheng J, Erb M, Mao B, Lou Y. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009;60:638–48.
Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11.
Ding ZJ, Yan JY, Xu XY, Li GX, Zheng SJ. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 2013;76:825–35.
Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13.
Yang YN, Li RG, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000;22:543–51.
Wroblewski T, Tomczak A, Michelmore R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J. 2005;3:259–73.
Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72.
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521.
Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–27.
Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66.
Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Korbes AP, Memelink J, Ritsema T, Van Wees SCM, Pieterse CMJ. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell. 2013;25:744–61.
Padilla-Hurtado B, Florez-Ramos C, Aguilera-Galvez C, Medina-Olaya J, Ramirez-Sanjuan A, Rubio-Gomez J, Acuna-Zornosa R. Cloning and expression of an endo-1,4-beta-xylanase from the coffee berry borer, Hypothenemus hampei. BMC Res Notes. 2012;5:23.
Tokunaga T, Miyata Y, Fujikawa Y, Esaka M. RNAi-Mediated knockdown of the XIP-type endoxylanase inhibitor gene, OsXIP, has no effect on grain development and germination in rice. Plant Cell Physiol. 2008;49:1122–7.
Erb M, Meldau S, Howe GA. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012;17:250–9.
Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–6.
Kiribuchi K, Jikumaru Y, Kaku H, Minami E, Hasegawa M, Kodama O, Seto H, Okada K, Nojiri H, Yamane H. Involvement of the basic helix-loop-helix transcription factor RERJ1 in wounding and drought stress responses in rice plants. Biosci Biotech Bioch. 2005;69:1042–4.
Kiribuchi K, Sugimori M, Takeda M, Otani T, Okada K, Onodera H, Ugaki M, Tanaka Y, Tomiyama-Akimoto C, Yamaguchi T, Minami E, Shibuya N, Omori T, Nishiyama M, Nojiri H, Yamane H. RERJ1, a jasmonic acid-responsive gene from rice, encodes a basic helix-loop-helix protein. Biochem Biophys Res Commun. 2004;325:857–63.
Zhu Y, Cai XL, Wang ZY, Hong MM. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem. 2003;278:47803–11.
Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, Ando T, Sugimoto K, Mitsuda N, Katoh E, Abe K, Miyao A, Hirochika H, Hattori T, Takeda S. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell. 2013;25:1709–25.
Yamamura C, Mizutani E, Okada K, Nakagawa H, Fukushima S, Tanaka A, Maeda S, Kamakura T, Yamane H, Takatsuji H, Mori M. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 2015;84:1100–13.
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–U664.
Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, Howe GA, He SY. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–88.
Wu H, Ye HY, Yao RF, Zhang T, Xiong LZ. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015;232:1–12.
Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–57.
Acknowledgments
We thank Dr. Jian Xue (College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China) for BPH infestation.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 30971702, 31271632 and 31672462), and by research grants from the Science and Technology Department of Zhejiang Province, China (2016C32086).
Availability of data and materials
All the data supporting the conclusions is included within the manuscript and its additional files.
Authors’ contributions
XW, YZ designed the project. YZ, XS, GR, CH and YH performed the experiments and data analysis. YZ wrote the manuscript. DJ guided the experiments. All authors read and approved the final manuscript.
Competing interests
The authors declared that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
List of primers used for gene constructs. (DOCX 18 kb)
Additional file 2: Table S2.
Primers for quantitative real-time (qRT) PCR. (DOCX 14 kb)
Additional file 3: Table S3.
Primers for chromatin immunoprecipitation (ChIP)-PCR. (DOCX 19 kb)
Additional file 4: Figure S1.
The sequence of the putative promoter region (−2,070/+52) of OsXIP. The transcription start site is indicated as +1, and the putative start codon is underlined; All potential cis-acting elements are boxed; All the primers (forward OP1-U- OP7-U and reverse OP-L) are indicated by red arrows. (DOCX 793 kb)
Additional file 5: Figure S2.
Subcellular localization of OsbHLH59 and OsERF71 in N. benthamiana. The full length ORFs without terminators of OsbHLH59 and OsERF71 were cloned into the pCAMBIA1300-sGFP vector under the control of the 35S promoter. Then N. benthamiana cells were transformed with 35Sp::OsbHLH59:GFP, 35Sp::OsERF71:GFP or pCAMBIA1300-GFP. After incubating for 48 h, the transformed cells were observed under a confocal microscope. The photographs were taken under detecting GFP fluorescence, bright field, and in combination (merge), respectively. Empty vector (pCAMBIA1300-GFP) was used as a control. Bars, 10 μm. (DOCX 1068 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhan, Y., Sun, X., Rong, G. et al. Identification of two transcription factors activating the expression of OsXIP in rice defence response. BMC Biotechnol 17, 26 (2017). https://doi.org/10.1186/s12896-017-0344-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12896-017-0344-7