Abstract
Background
Alpha1-antitrypsin (AAT) is a serine protease inhibitor that serves as a counterbalance to the activity of elastases, e.g., neutrophil elastase in lung tissue. AAT deficiency (AATD) is a rare disorder usually arising from mutations to the SERPINA1 gene that codes for AAT. The most common AATD alleles are S and Z which produce ~ 40% and ~ 90% reductions in serum AAT, respectively. Rare genetic variants (> 500 identified) can also be associated with mild to severe AATD.
Results
This report describes a novel mutation of SERPINA1 producing AATD, which we have designated, Q0RİZE. This mutation was identified in a 44-year-old woman admitted with massive hemoptysis and treated with bronchial artery embolization. Computed tomography revealed centriacinar and panacinar emphysema with prominent air entrapment, atelectasis, and localized bronchiectasis. Serum AAT was < 0.27 g/L (below detection limit). Genetic analysis showed homozygous deletion of exons I to III.
Conclusions
Although many SERPINA1 variants have been identified, variants with large deletions and identified in a homozygous individual, as seen in this case with Q0RIZE, are uncommon. AATD is an underdiagnosed and undertreated disease. Wider screening of COPD patients could result in earlier diagnosis and treatment that could preserve lung function.
Similar content being viewed by others
Background
Under normal physiologic conditions, alpha1-antitrypsin (AAT), a serine protease inhibitor, acts to counteract the activity of elastases, primarily neutrophil elastase (NE) in the lungs. AAT is a serum anti-protease synthesized primarily in the liver and transported to the lungs through the bloodstream. When AAT levels are deficient (AAT deficiency (AATD)) the regulation of NE activity is disrupted, and NE activity is increased potentially leading to destruction of lung tissue and impaired gas exchange [1].
AATD is a relatively rare inherited disorder that increases the propensity for lung and liver disease. The SERPINA1 gene codes for the AAT protein. Genetic analyses have revealed that the M-allele of this gene is associated with normal serum levels of AAT and is the most common allele. At present, the pathogenic alleles most often associated with reduced serum AAT (90% of AATD cases) are the S- (40% reduction in serum AAT) and Z-alleles (90% reduction in serum AAT) [2, 3]. More than 500 additional variants of the SERPINA1 gene have been identified [4, 5]. Approximately 40% of these variants are associated with some degree of AATD [6] and may be responsible for up to 17% of clinical cases of AATD [7, 8].
Despite the large amount of research identifying SERPINA1 variants and the clear clinical significance early diagnosis and treatment, AATD is underdiagnosed and consequently undertreated. This case report summarizes the clinical findings for a patient with a previously undescribed SERPINA1 genetic variant.
Case presentation
This 44-year-old female patient was hospitalized in the pulmonary clinic due to massive hemoptysis in September 2022. The interventional radiology department was consulted, and embolization of her hypertrophied bronchial arteries was performed the same day. Chest computed axial tomography (CAT) scans were performed and widespread emphysema was detected. The CAT scan showed centriacinar and panacinar emphysematous air entrapment in both lungs - more prominent in the lower lobes (Figs. 1 and 2). Atelectasis and volume loss were observed in the right middle and lower lobes and in the left lingular segment. Localized tractional bronchiectasis was seen in the right middle lobe, lingula inferior and both lower lobes. In the posterior segment of the right upper lobe, an area of cystic bronchiectasis was seen that appeared to merge with an area in the central lobe (Fig. 3).
For medical history, the patient reported fatigue and dyspnea with exercise since her teenage years and was diagnosed with asthma at that time. Her father is the only first degree relative with confirmed chronic obstructive pulmonary disease (COPD). She also has a fraternal cousin (male) that was diagnosed to be homozygous ZZ and is on AAT augmentation therapy.
The patient is currently employed and reports occupational exposure to dust in her job at a local tea factory. She reported that she is a former smoker (starting at 18 years old) with a 10 pack-year exposure prior to quitting in 2014. She also reported that her husband is a heavy smoker and therefore she has ongoing secondhand and thirdhand exposure to cigarette smoke.
The patient has seen an increase in the severity of her pulmonary symptoms over the past three years. Her recent symptoms (May 2023) include dyspnea (primarily with exertion) and fatigue. Her dyspnea score on the Modified Medical Research Council (mMRC) scale was 2 on a 0–4 scale [9,10,11]. Her Global Initiative for Chronic Obstructive Lung Disease (GOLD) COPD status was determined to be Stage B [12]. Her oxygen saturation was 98% and lung auscultation was normal. Her pulmonary function tests showed forced expiratory volume in one second (FEV1) of 1.11 L (37% of predicted value), forced vital capacity (FVC) of 2.23 L (64% of predicted) and FEV1/FVC ratio of 50%. Her liver enzymes were within the normal range (alanine transaminase (ALT) 9 U/L (reference range 0–35 U/L and aspartate transaminase (AST) 12 U/L (reference range 0–35 U/L).
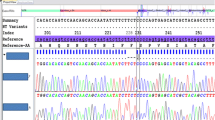
The patient’s serum AAT level was < 0.27 g/L, below the limit of detection (Goldsite Diagnostics, Inc., Shenzhen, China; normal values 0.9-2.0 g/L). A dried blood sample (DBS) was obtained and sent to Progenika Biopharma for genetic analysis (see Methods for details). Sequencing results showed normal coverage for amplicons corresponding to exons IV and V, but extremely low coverage (similar to the negative control) for all other amplicons. This could be interpreted as a homozygous deletion of exons I to III. This was confirmed by multiplex ligation-dependent probe amplification (MLPA) (Fig. 4). Using an exon-based description, this variant is genetically described as c.(?_-573)_(917 + 1_918-1)del. Large deletions are extremely infrequent in SERPINA1 and this is the first description of this variant.
MLPA results for the index patient (A) and a relative (B). The SERPINA1 exon numbering used in this P459-A2 SERPINA1 product is the exon numbering from the RefSeq transcript NM_001127701.1 (transcript variant 5), so that non-coding exon (Ia, Ib and Ic) are numbering as 1, 2 and 3, respectively, and exon II, III, IV and V are indicated as 4, 5, 6 and 7, respectively. Calculated probe ratios normalized to the reference samples are indicated on the y-axis. Arranging probes by chromosomal location reveals the homozygous deletion (probe ratio 0) in the patient’s sample (A) and heterozygous deletion (probe ratio around 0.5) in the relative’s sample (B)
The patient is currently being treated with a long-acting beta agonist, formoterol, and a long-acting muscarinic antagonist, tiotropium. These medications are supplemented with a short-acting beta agonist, salbutamol, and a short-acting muscarinic antagonist, ipratropium bromide, as needed. AAT replacement therapy was initiated in this patient but was discontinued after the second dose due to an allergic skin reaction. The serum IgA level on this patient was normal (1.81 g/L).
DBS samples were obtained from several members of the patient’s family for genetic screening. SERPINA1 gene sequencing and MLPA analysis were performed for these relatives. The results are shown in the family tree in Fig. 5. The patient presented here (index case) is homozygous for the Q0RIZE variant having received a copy of the null variant from both parents. Her father found to be PiZ-Q0RIZE is her only first degree relative diagnosed with COPD. Her cousin has been found to be homozygous ZZ and is on AAT augmentation therapy. Her sisters, brother and sons all have the normal M allele in combination with the Z or null allele and have not been diagnosed with lung or liver disease.
Methods
DBS were obtained from the patient and family members and sent to Progenika for analysis. The first analysis was performed using the A1AT genotyping Test (Progenika Biopharma, SA, Derio, Spain) on the Luminex 200 (Luminex, Austin, TX, USA). This multiplex PCR (polymerase chain reaction) and hybridization test allows the simultaneous identification of 14 genetic variants, corresponding to the most frequent alleles found in AATD patients. If inconclusive, results were obtained with the A1AT Genotyping Test, full sequencing of the SERPINA1 gene was performed using Next Generation Sequencing. The amplicon library was prepared covering the full gene (three non-coding and four coding exons) and sequenced on MiSeq (Illumina, Inc, San Diego, CA, USA). Results were confirmed by multiplex ligation-dependent probe amplification (MLPA) analysis using SALSA MLPA P459-A2 SERPINA1 kit (MRC Holland, Amsterdam, Netherlands). MLPA is based on the amplification of a set of probes that each detect a specific DNA sequence of approximately 60 nucleotides in length. The SALSA MLPA Probemix P459-A2 SERPINA1 contains 21 MLPA probes: nine probes for the SERPINA1 gene and 12 reference probes that detect autosomal chromosomal locations. MLPA is a relative technique: only relative differences can be detected by comparing the MLPA peak patterns of DNA samples. The relative intensity of each individual probe, as compared to the relative probe intensity in various reference DNA samples, reflects the relative copy number of the corresponding target sequence in a sample. A deletion of one or more target sequences is visible as a relative decrease in intensity.
Discussion
In this article, we report the case of a patient with severe emphysema and AATD. Genetic analysis for this patient showed a homozygous deletion of exons I to III, a variant we are calling Q0RIZE. Large deletions such as this are uncommon as null mutations are most commonly caused by single base deletions [5]. However, the null mutations Q0isola de procida [13] and Q0riedenburg [14] were found to be due to deletions of exons II to V. It is even more unusual that this patient appears to be homozygous for this allele.
In this case, a novel null variant was identified in a homozygous patient. This patient had serum AAT levels below the limit of detection and significant emphysema likely complicated by a 10 pack-year history of smoking and on-the-job exposure to dust. AATD was identified in this patient at 44 years of age and with advanced disease. The patient is currently being treated with inhaled dual bronchodilators and with short acting bronchodilators as needed.
AATD is an underdiagnosed disease due to the lack of widespread testing. Due to the lack of a diagnosis in affected individuals, it is consequently undertreated. Earlier detection of AATD could lead to earlier treatment, including assistance with reduction of environmental hazards (e.g., smoking cessation therapy for the patient and household members and workplace risk reduction) and AAT augmentation therapy (if feasible and warranted), which could help improve or maintain the patient’s lung function and quality of life.
As previously noted, a large number of variants of SERPINA1 have been identified. The clinical characteristics of some of these variants have not fully described. When previously undescribed SERPINA1 variants are tentatively identified, full characterization of the patient’s medical history, lab values and genetic profile could be helpful in expanding the knowledge base of AATD and in providing guidance for selecting the best course of treatment.
Conclusions
-
We report a patient with severe emphysema hospitalized for massive hemoptysis. Genetic analysis revealed the patient was homozygous for a previously undescribed SERPINA1 variant: Q0RIZE.
-
This patient had a delayed diagnosis due to inadequate access to advanced diagnostic technology and genetic analysis.
-
Reporting this and other novel mutations and their clinical profiles is important to advance our understanding of AATD and improving its diagnosis and treatment.
Data availability
The data that support the findings of this study are not openly available due to reasons of confidentiality and are available from the corresponding author upon reasonable request. Data are located in controlled access data storage at Recep Tayyip Erdoğan University, School of Medicine, Department of Chest Diseases.
Abbreviations
- AAT :
-
Alpha1-antitrypsin
- AATD:
-
Alpha1-antitrypsin deficiency
- ALT:
-
Alanine transaminase
- AST:
-
Aspartate transaminase
- CAT:
-
Computed axial tomography
- COPD:
-
Chronic obstructive pulmonary disease
- FEV1:
-
Forced expiratory volume in one second
- FVC:
-
Forced vital capacity
- GOLD:
-
Global Initiative for Chronic Obstructive Lung Disease (GOLD) COPD status
- mMRC:
-
Modified Medical Research Council dyspnea score
- NE:
-
Neutrophil elastase
References
Bashir A, Shah NN, Hazari YM, Habib M, Bashir S, Hilal N, et al. Novel variants of SERPIN1A gene: interplay between alpha1-antitrypsin deficiency and chronic obstructive pulmonary disease. Respir Med. 2016;117:139–49.
Lopez-Campos JL, Casas-Maldonado F, Torres-Duran M, Medina-Gonzálvez A, Rodriguez-Fidalgo ML, Carrascosa I, et al. Results of a Diagnostic Procedure based on Multiplex Technology on dried blood spots and buccal swabs for subjects with suspected Alpha1 Antitrypsin Deficiency. Arch Bronconeumol (Engl Ed). 2021;57(1):42–50.
Dickens JA, Lomas DA. Why has it been so difficult to prove the efficacy of alpha-1-antitrypsin replacement therapy? Insights from the study of disease pathogenesis. Drug Des Devel Ther. 2011;5:391–405.
Seixas S, Marques PI. Known mutations at the cause of Alpha-1 Antitrypsin Deficiency an updated overview of SERPINA1 variation spectrum. Appl Clin Genet. 2021;14:173–94.
Wiesemann GS, Oshins RA, Flagg TO, Brantly ML. Novel SERPINA1 alleles identified through a large Alpha-1 Antitrypsin Deficiency Screening Program and Review of known variants. Chronic Obstr Pulm Dis. 2023;10(1):7–21.
Silva D, Oliveira MJ, Guimaraes M, Lima R, Gomes S, Seixas S. Alpha-1-antitrypsin (SERPINA1) mutation spectrum: three novel variants and haplotype characterization of rare deficiency alleles identified in Portugal. Respir Med. 2016;116:8–18.
Meira L, Boaventura R, Seixas S, Sucena M. Alpha-1 Antitrypsin Deficiency Detection in a Portuguese Population. COPD. 2018;15(1):4–9.
Ferrarotti I, Baccheschi J, Zorzetto M, Tinelli C, Corda L, Balbi B, et al. Prevalence and phenotype of subjects carrying rare variants in the Italian registry for alpha1-antitrypsin deficiency. J Med Genet. 2005;42(3):282–7.
Mahler DA, Wells CK. Evaluation of clinical methods for rating dyspnea. Chest. 1988;93(3):580–6.
Bestall JC, Paul EA, Garrod R, Garnham R, Jones PW, Wedzicha JA. Usefulness of the Medical Research Council (MRC) dyspnoea scale as a measure of disability in patients with chronic obstructive pulmonary disease. Thorax. 1999;54(7):581–6.
Stenton C. The MRC breathlessness scale. Occup Med (Lond). 2008;58(3):226–7.
Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease (2021 Report). Global Initiative for Chronic Obstructive Lung Disease.; 2021. Available at: https://goldcopd.org/wp-content/uploads/2020/11/GOLD-REPORT-2021-v1.1-25Nov20_WMV.pdf (Accessed 26 January 2024).
Takahashi H, Crystal RG. Alpha 1-antitrypsin null(isola di procida): an alpha 1-antitrypsin deficiency allele caused by deletion of all alpha 1-antitrypsin coding exons. Am J Hum Genet. 1990;47(3):403–13.
Poller W, Faber JP, Weidinger S, Olek K. DNA polymorphisms associated with a new α1-antitrypsin PI Q0 variant (PI Q0riedenburg). Hum Genet. 1991;86(5):522–4.
Acknowledgements
The authors acknowledge Michael K. James, Ph.D., for medical writing assistance and Jordi Bozzo, Ph.D., for editorial assistance.
Funding
The publication of this case was supported by Grifols.
Author information
Authors and Affiliations
Contributions
DK, ÜŞ, BD, LG, and LO participated in the conceptualization of the case report and genetic analysis. DK and ÜS analyzed and interpreted the patient information regarding her pulmonary disease. LO and LG performed the genetic analyses. DK, ÜŞ, BD, LG, and LO participated in the creation of the first draft and subsequent revisions. DK, ÜŞ, BD, LG, and LO read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from the patient described herein for their anonymized information to be published in this article. Ethics committee approval was not sought for this study because Recep Tayyip Erdoğan University does not require ethical approval for reporting individual cases. This study was completed in accordance with the Helsinki Declaration as revised in 2013.
Consent for publication
Written informed consent was obtained from the patient for their anonymized information to be published in this article.
Competing interests
DK and ÜS have no competing interests related to this work. BD is an employee of Grifols Deutschland. LO and LG are employees of Progenika, a Grifols Company.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Karadoğan, D., Şahin, Ü., Dreger, B. et al. Case report of a novel alpha1-antitrypsin null variant in Türkiye: Q0RİZE. BMC Pulm Med 24, 91 (2024). https://doi.org/10.1186/s12890-024-02900-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-024-02900-6