Abstract
Background
Due to individual differences and lack of objective biomarkers, only 30-40% patients with major depressive disorder (MDD) achieve remission after initial antidepressant medication (ADM). We aimed to employ radiomics analysis after ComBat harmonization to predict early improvement to ADM in adolescents with MDD by using brain multiscale structural MRI (sMRI) and identify the radiomics features with high prediction power for selection of selective serotonin reuptake inhibitors (SSRIs) and serotonin norepinephrine reuptake inhibitors (SNRIs).
Methods
121 MDD patients were recruited for brain sMRI, including three-dimensional T1 weighted imaging (3D-T1WI)and diffusion tensor imaging (DTI). After receiving SSRIs or SNRIs for 2 weeks, the subjects were divided into ADM improvers (SSRIs improvers and SNRIs improvers) and non-improvers according to reduction rate of the Hamilton Depression Rating Scale, 17 item (HAM-D17) score. Then, sMRI data were preprocessed, and conventional imaging indicators and radiomics features of gray matter (GM) based on surface-based morphology (SBM) and voxel-based morphology (VBM) and diffusion properties of white matter (WM) were extracted and harmonized with ComBat harmonization. Two-level reduction strategy with analysis of variance (ANOVA) and recursive feature elimination (RFE) was utilized sequentially to decrease high-dimensional features. Support vector machine with radial basis function kernel (RBF-SVM) was used to integrate multiscale sMRI features to construct models for early improvement prediction. Area under the curve (AUC), accuracy, sensitivity, and specificity based on the leave-one-out cross-validation (LOO-CV) and receiver operating characteristic (ROC) curve analysis were calculated to evaluate the model performance. Permutation tests were used for assessing the generalization rate.
Results
After 2-week ADM, 121 patients were divided into 67 ADM improvers (31 SSRIs improvers and 36 SNRIs improvers) and 54 ADM non-improvers. After two-level dimensionality reduction, 8 conventional indicators (2 VBM-based features and 6 diffusion features) and 49 radiomics features (16 VBM-based features and 33 diffusion features) were selected. The overall accuracy of RBF-SVM models based on conventional indicators and radiomics features was 74.80% and 88.19%. The radiomics model achieved the AUC, sensitivity, specificity, and accuracy of 0.889, 91.2%, 80.1% and 85.1%, 0.954, 89.2%, 87.4% and 88.5%, 0.942, 91.9%, 82.5% and 86.8% for predicting ADM improvers, SSRIs improvers and SNRIs improvers, respectively. P value of permutation tests were less than 0.001. The radiomics features predicting ADM improver were mainly located in the hippocampus, medial orbitofrontal gyrus, anterior cingulate gyrus, cerebellum (lobule vii-b), body of corpus callosum, etc. The radiomics features predicting SSRIs improver were primarily distributed in hippocampus, amygdala, inferior temporal gyrus, thalamus, cerebellum (lobule vi), fornix, cerebellar peduncle, etc. The radiomics features predicting SNRIs improver were primarily located in the medial orbitofrontal cortex, anterior cingulate gyrus, ventral striatum, corpus callosum, etc.
Conclusions
These findings suggest the radiomics analysis based on brain multiscale sMRI after ComBat harmonization could effectively predict the early improvement of ADM in adolescent MDD patients with a high accuracy, which was superior to the model based on the conventional indicators. The radiomics features with high prediction power may help for the individual selection of SSRIs and SNRIs.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Background
Major depressive disorder (MDD) is the leading cause of disability in adolescents and the incidence increases dramatically during adolescence [1]. Approximately 70% of adolescents with MDD will relapse within five years, and early-onset depression is also associated with high recurrence rates, poor functional outcomes, and refractory depression in later years [2, 3]. Therefore, there is no doubt about the importance of early and proactive treatment for adolescent MDD patients. Antidepressant medication (ADM) remains the preferred treatment strategy for MDD, and clinical guidelines recommend the selective serotonin reuptake inhibitors (SSRIs) or 5-hydroxytryptamine and serotonin norepinephrine reuptake inhibitors (SNRIs) as the first-line antidepressant agents [4, 5]. However, due to individual differences in patients with MDD, less than 50% of patients benefit from ADM and only 30-40% achieve clinical remission after initial antidepressant treatment [4, 5]. Currently, in clinical practice, the selection of drugs for MDD is mainly relied on the measurements of symptoms and the experience of psychiatrists owing to lack of objective biomarkers for selecting antidepressants [4]. This “trial-and-error” approach takes approximately 4–8 weeks to find out whether antidepressant drugs are effective and then to develop the follow-up treatment program, which not only results in a lower response rate to subsequent medications and a waste of medical resources, but also prolongs rehabilitation time and increases suicide risk [4,5,6,7]. Therefore, it is of great significance for optimizing the way of drug selection if the response prediction of individualized treatment can be achieved prior to initial ADM [6, 7]. Previous studies suggested that early improvement in MDD signified clinical “turning point”, stable remission and good prognosis [8,9,10,11]. To clarify the earliest time point when improvement occurred, a meta-analysis and an observational study both stated that the response to ADM could occur as early as 2 weeks after initial ADM [10, 11]. With the help of early response prediction, psychiatrists can make decisions to continue or change treatment regimens earlier, especially to be able to cease ineffective treatment and reduce side effect and therapy-related risks. In the past few decades, numerous neuroimaging studies have focused on the exploration of the pathogenesis of MDD, while little is known about the pathophysiological substrate underlying the response of some patients to antidepressant medication while others with difficulty in rehabilitation [9,10,11].
At present, research with different neuroimaging modalities of magnetic resonance imaging (MRI) have identified several potential biomarkers associated with treatment response to specific therapies in MDD. For example, findings have shown that changes of cortex thickness and volume of gray matter (GM) in multiple brain regions including, but not limited to, the prefrontal cortex, hippocampus, anterior cingulate cortex at baseline can serve as imaging biomarkers to predict remission after medication and cognitive behavioral therapy (CBT) [12,13,14]. A recent study found that the anisotropy fraction (FA) of the white matter (WM), the most used metric for diffusion tensor imaging (DTI), could be utilized to predict antidepressant response. Increase of FA in the superior corona radiata and external capsule was correlating with drug response [15]. FA in the right amygdala, cingulate gyrus and terminal fasciculus could also predict antidepressant remission at 4–12 weeks [16,17,18]. One study identified the hippocampus and amygdala centered implicit emotion regulation circuitry as a sensitive biomarker in predicting early efficacy of SSRIs treatment [19]. FA tracts connecting hippocampus and amygdala have been reported to predict remission following SSRIs treatment [20]. The role of hippocampus in predicting 4-week SSRIs treatment response was also emphasized in functional research [21]. For SNRIs treatment, a neuroimaging study highlighted the importance of orbital superior frontal gyrus (ORBsup) and putamen centered neural circuitry as a biomarker [19]. Positron emission tomography / computed tomography (PET/CT) study also suggested that the activity of the anterior medial orbitofrontal cortex was associated with the remission of MDD after treatment with SNRIs [22, 23]. In addition, SSRI acts by blocking 5-hydroxytryptamine reuptake, whereas SNRI works by blocking the reuptake of 5-hydroxytryptamine, norepinephrine and dopamine at dendrites and axons, and the SSRI and SNRI neurofibrillary projection areas have been reported to be different [24]. All these evidence indicate that there may be some brain regions in a variety of spatially diverse GM and WM in associated with drug efficacy, so it is reasonable to explore imaging markers for predicting ADM response and assisting drug selection by combining multiscale structural imaging features of gray and white matter. Moreover, structural MRI (sMRI) is usually part of brain routine examination, and structural imaging data are easily available and relatively stable. However, most of the previous studies used only a single imaging modality and the results were often inconsistent and difficult to compare [12,13,14,15,16,17]. Also, these researches mainly focused on medium to long-term outcomes (4–12 weeks) [10, 11], with less involvement in predicting the earlier response to ADM in MDD patients.
Radiomics, a framework that combines machine learning with medical imaging to quantitatively reveal macroscopic heterogeneity that is unrecognizable to the human eyes, has greatly expanded the applications of conventional imaging in clinical practice [25, 26] and has been employed to investigate imaging biomarkers for neuropsychiatric disorders, such as Alzheimer’s disease [27], bipolar disorder [28] and attention deficit hyperactivity disorder [29]. By extracting and selecting high-weighted radiomics features, diagnostic models outperform those classifiers based on the routine imaging indicators, and the discriminant radiomics features can be used as potential markers. However, properties of MRI scanners, such as manufacturer, field strength, nonlinear gradient fields, and longitudinal drift, increase the bias and variability of brain sMRI [26, 30], thereby impacting the consistency and reproducibility of downstream analyses [31, 32], which will hinder the exploration of radiomics models and their transformation into diagnostic or predicting tools [30]. Like the “batch effect” in genomics, the term “scanner effect” is used in neuroimaging to refer to such abiotic variation [30]. Several methods have been proposed to harmonize CT and PET-CT conventional imaging indicators, such as hybrid white stripe and histogram matching, in previous studies on breast phantom [32], brain tumor [33], and prostate cancer [30]. Only ComBat technique has been validated in radiomics studies [31, 32], which could effectively address the issues of poor reproducibility and stability of the radiomics features.
To date, there has been few radiomics studies on the prediction of early improvement to ADM in adolescents with MDD based on harmonized multiscale sMRI [34]. As such, in present study, we constructed a prediction model for early improvement (2 weeks) to ADM in adolescent MDD patients with radiomics analysis after ComBat harmonization based on multiscale sMRI, including shape parameters of GM based on surface-based morphology (SBM) and voxel-based morphology (VBM) from the three-dimensional T1 weighted imaging (3D-T1WI), and diffusion properties of WM from the diffusion tensor imaging (DTI), and compared it with the model based on conventional imaging indicators. We also identified radiomics features that might be helpful for the objective first-line drugs selection of SSRIs and SNRIs in clinical practice.
Methods
Participants
138 patients with MDD, aged between 13 and 18 years, all right-handed according to their self-report, were prospectively and consecutively recruited from the Department of Psychiatry of the Second Affiliated Hospital of Kunming Medical University from December 2021 to June 2022. The study was approved by the Ethics Committee according to the principles of the Declaration of Helsinki with approval number KYCS202107. The written informed consent was obtained from participants and from their parents or legal guardians for subjects under 16 years old before enrollment. All participants were diagnosed by psychiatrists based on structured interviews and neuropsychological measurements and met the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-V) criteria, including only unipolar and first-episode depression without any medication and physical therapy such as repetitive transcranial magnetic stimulation (rTMS) and electroconvulsive therapy (ECT) prior to hospitalization. 17 item Hamilton Depression Rating Scale (HAM-D17) was used to assess the severity of depressive symptoms at baseline and after 2-week ADM treatment. Exclusion criteria for MDD patients were: ①accompanied with any other mental disorders; ②any history of cranial injury; ③significant physical diseases undergoing treatment; ④substance/alcohol abuse or dependence; ⑤MRI scanning contraindicated.
All patients received SSRIs or SNRIs according to clinical guideline for the treatment of MDD patients (Chinese version) [5]. The doses were as follows: sertraline 100 ~ 200 mg/day, escitalopram 10 ~ 20 mg/day, fluvoxamine 100 ~ 300 mg/day, paroxetine 20 ~ 60 mg/day, fluoxetine 20 ~ 60 mg/day, venlafaxine 37.5 ~ 225 mg/day, and duloxetine 60 ~ 120 mg/day. HAM-D17 score was assessed again after 2 weeks of complete ADM. A case was defined as early improvement to ADM if the HAM-D17 score decreased ≥ 20% compared with the baseline [35]. 17 patients were excluded because they failed to complete 2-week ADM, including 13 cases of medication regimen change and 4 cases with additional rTMS. Finally, the remaining 121 MDD patients were divided into ADM improvers (SSRIs improvers and SNRIs improvers) and non-improvers according to reduction rate of HAM-D17 score.
MRI protocol
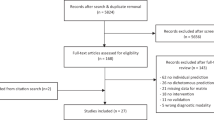
The flowchart of this study is illustrated in Fig. 1. All participants underwent MRI scans within one week prior to ADM after structured interviews and neuropsychological measurements. 3D-T1WI, fluid attenuation inversion recovery (FLAIR), and DTI were performed sequentially using two scanners (Philip 3.0T, GE 3.0T), and the parameters of scanners and sequences are shown in Table S1 and Table S2. Examination of FLAIR was to rule out the presence of a substantial lesion, such as demyelination, brain tumor, vascular malformation, and development abnormalities. All images were reviewed immediately after each sequence completing, and those with motion artifacts needed to be rescanned and those with susceptibility artifacts were excluded. At last, one subject with excessive artifacts in sMRI was excluded. The final sMRI data of 63 subjects acquired at Philip 3.0T scanner and 58 subjects examined on GE 3.0T scanner were used for preprocessing, feature extraction and ComBat harmonization.
Flowchart of the study. Firstly, sMRI (3D-T1WI and DTI) was performed with two scanners in adolescent MDD subjects. FreeSurfer/ANTs hybrid segmentation toolkit of Mindboggle software, CAT 12 suite of SPM software and Diffusion Toolbox of FSL software were used for preprocessing. After that, conventional indicators and radiomics features of shape parameters of GM based on SBM and VBM analysis and diffusion properties of WM were extracted and then harmonized with ComBat technique. After receiving SSRIs or SNRIs for 2 weeks, the subjects were divided into ADM improvers (SSRIs improvers and SNRIs improvers) and non-improvers according to reduction rate of the HAM-D17 score. Finally, the features were decreased and filtered based on ANOVA and RFE successively and those with high prediction power were employed to construct models based on RBF-SVM. The performance was estimated using LOO-CV and ROC curve. sMRI, structural MRI. 3D-T1WI, three-dimensional T1 weighted imaging. DTI, diffusion tensor imaging. MDD, major depressive disorder. SPM, statistical parametric mapping. FSL, FMRIB’s software library. SBM, surface-based morphology. VBM, voxel-based morphology. GM, gray matter. WM, white matter. ANOVA, analysis of variance. RFE, recursive feature elimination. SVM, support vector mechanism. RBF, radial basis function kernel. LOO-CV, leave-one-out cross-validation. ROC, receiver operator characteristic. ADM, antidepressant medication. SSRIs, selective serotonin reuptake inhibitors. SNRIs, serotonin norepinephrine reuptake inhibitors. HAM-D17, Hamilton Depression Rating Scale, 17 item
SBM-based 3D-T1WI preprocessing and GM features extraction
3D-T1WI preprocessing based on SBM was accomplished using the FreeSurfer/ANTs hybrid segmentation pipeline [29] with the Desikan-Killiany-Tourville (DKT) atlas [36]. After performing the steps of motion correction, skull stripping, bias field correction, intensity normalization, expansion and smoothing, spherical mapping and alignment, and cortical surface reconstruction, the hybrid segmentation was achieved by combining the FreeSurfer recon-all toolkit (https://surfer.nmr.mgh.harvard.edu/) and the ANTsXNet CorticalThickness toolbox (http://stnava.github.io/ANTs/) introducing new gray-white matter boundaries to reduce segmentation errors, which has been proven to have superior performance over the FreeSurfer package [37]. Then, the following measures were automatically calculated using the feature extraction toolkit of Mindboggle (http://www.mindboggle.info/) [29] software: ①surface area of labeled brain; ②surface morphometric measures of cortical mesh vertex within labeled brain region and sulci; ③statistical metrics for each morphometric measure. In DKT atlas, there were thirty-eight cortical/subcortical regions and twenty-four sulci in each hemisphere. Therefore, based on the SBM method, 344 conventional indicators and 2,338 radiomics features representing GM surface morphometry were extracted from 3D-T1WI of each subject.
VBM-based 3D-T1WI preprocessing and GM features extraction
3D-T1WI preprocessing based on VBM was performed using the CAT 12 toolkit [38] of the Statistical Parametric Mapping (SPM 12, https://www.fil.ion.ucl.ac.uk/spm/) software. The main steps were as follows: ①images were registered from the original brain space to a standard space coordinate system established by the Montreal Neurological Institute (MNI); ②bias field was estimated using the N4-Bias-Field-Correction algorithm [39] and then overlayed onto the original images to achieve bias field correction, thereby reducing the intensity difference of the same tissue and facilitating tissue segmentation; ③after skull stripping, the spatially normalized images were segmented into GM, WM, and cerebrospinal fluid (CSF). According to the Automated Anatomical Labeling Atlas (AAL) atlas, the preprocessed images of each subject were labeled into one hundred and sixteen brain regions, including ninety cerebral regions and twenty-six cerebellar regions. A neuroradiologist with 7-year experience of neuroimaging used ITK-SNAP 4.0 software (http://www.itksnap.org/pmwiki/) to examine and correct areas of brain tissue that were segmented incorrectly. Then, the segmented data were imported into LIFEx software (https://www.lifexsoft.org), and a total of sixty-three radiomics features of three categories, namely, five shape features, twelve first-order histogram features, and forty-six texture features were extracted from each brain label according to the image biomarkers standardization initiative (IBSI) guidelines [40] and radiomics quality scoring (RQS) tool [41]. Finally, based on the VBM analysis, 206 conventional indicators and 10,044 radiomics features representing GM density morphometry were extracted from 3D-T1WI images of each subject.
DTI preprocessing and WM diffusion features extraction
DTI was preprocessed using Diffusion Toolbox v2.0 [42] of FMRIB’s Software Library (FSL, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) for susceptibility-induced distortions, eddy-current and head motion correction, tensor fitting and registration to standard space. Diffusion tensors data from all subjects were used to build a template specific to this study by using an iterative tensor-based registration algorithm combined with local WM fiber bundle alignment. Each fraction map of the Johns Hopkins ICBM-DTI-81 white matter atlas was registered to the study-specific template through deformable alignment. Through nearest neighbor interpolation and alignment from each subject to the template, the label of the WM region in the map was aligned and warped into a single space, generating a partial anisotropic map, and labeling forty-eight WM regions. The following measures were computed, including the anisotropy fraction (FA), mean diffusivity (MD), axial diffusivity (AD), radial diffusivity (RD) and their corresponding statistical metrics for each WM label. Therefore, 192 conventional indicators and 768 radiomics features were extracted from each DTI data.
ComBat harmonization
The extracted features were harmonized between the two scanners using the ComBat technique [31,32,33], which was designed to adjust any abiotic differences, i.e., scanner effects, that might be caused by the scanners, coils, and/or protocols parameters. The final ComBat-harmonized radiomics features were defined as:
In the above function, \({\widehat{\alpha }}_{v}\) is the overall value at voxel \(v\), such as cortical thickness, volume, or FA value; \({X}_{ij}\) is the matrix of covariates (e.g., age, sex); \({\widehat{\beta }}_{v}\) represents the voxel-based regression coefficients corresponding to the \(X\). \({\widehat{\alpha }}_{v}\) and \({\delta }_{iv}^{\text{*}}\) denote respectively the additivity and multiplication effects of site \(i\) on voxel \(v\). The above ComBat function assigned a specific transformation to each measure extracted from T1WI and DTI, respectively.
Reduction of features
In order to eliminate extraneous features which affected the fitting efficiency and accuracy of models, a two-level selection strategy was used sequentially in this study. Firstly, analysis of variance (ANOVA) was applied to filter out features with P value less than 0.05. Then, recursive feature elimination (RFE) was applied to further reduce dimensionality of features by ranking them according to their relevance to each other and importance to the model in the reverse elimination process [43].
Identification of radiomics features with high prediction power
In present study, because of small sample size, LOO-CV was used to validate the prediction model to avoid overfitting while being able to maximize the inclusion of samples during model training and testing. In each LOO-CV iteration of n samples, n-1 samples were used for training and one sample was left for testing. RFE process was conducted on the training dataset and selected the desired features by recursively and continuously reducing the size of the feature datasets. Each feature’s prediction power was estimated quantitatively by its weight vector, which was the distance from the feature to the separation hyperplane or decision boundary in the vector mapping space. Then, the prediction power in all LOO-CV iterations were averaged and ranked, and the top 30% were identified as the most powerful features.
Prediction model construction and evaluation
When obtaining feature datasets with high prediction power, we used selected conventional indicators and harmonized radiomics features to construct support vector machine with radial basis function kernel (RBF-SVM) model on the LIBSVM toolbox (https://www.csie.ntu.edu.tw/~cjlin/libsvm) for prediction ADM non-improvers, SSRIs improvers and SNRIs improvers. All participants were labeled into three types (Label 1 = SSRIs-improver, Label 0 = ADM-non-improver, Label-1 = SNRIs-improver) to identify decision boundaries in the input space. To ensure the accuracy of parameter tuning, the grid search method was used to determine the optimal regularization parameter C and the kernel function parameter gamma for RBF-SVM model. After the tuning range and tuning step were given, the possible values of each parameter were calculated. Then all combination situations were traversed, and finally the best parameters were returned. Area under the curve (AUC), accuracy, sensitivity, and specificity based on LOO-CV results of the SVM were computed to evaluate the performance of the models. Permutation tests were used for assessing the generalization rate.
Statistical analysis
Statistical analyses were conducted using R 3.5.1 software (Comprehensive R Archive Network). Quantitative data were summarized as the mean ± standard deviation or median with interquartile range, and categorical variables were described as numbers and percentages. ANOVA or Chi-square test was used for the univariate comparison as appropriate between the groups. AUC, sensitivity, and specificity were calculated by analysis of receiver operating characteristic (ROC) curve at cut-off score corresponding to the highest Youden index. Pairwise and multiple AUCs comparisons were performed using the DeLong tests. P value less than 0.05 was statistically significant.
Results
Demographic data and clinical information of participants
Results of demographic data and clinical information of participants are listed in Tables 1 and 2. There was no significant difference between ADM improvers and non-improvers, SSRIs improvers and SNRIs improvers in terms of age, gender, years of education, and baseline HAM-D17 score (P > 0.05).
Features selection and identification of radiomics features with high prediction power
After preprocessing, a total of 742 conventional indicators (344 SBM-based features, 206 VBM-based features, and 192 DTI diffusion features) (Table S3) and 13,150 radiomics features (2,338 SBM-based features, 10,044 VBM-based features, and 768 DTI diffusion features) were extracted from 3D-T1WI and DTI (Table S4). These features were harmonized with ComBat technique. Then, the two-level dimensionality reduction strategy was performed and only 8 conventional indicators (2 VBM-based features, 6 diffusion features) and 49 radiomics features (16 VBM-based features, 33 diffusion features) were selected (Fig. 2).
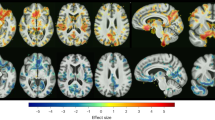
Prediction power of 49 radiomics features for predicting early improvement of MDD patients to ADM with SSRIs and SNRIs for 2 weeks. The grayscale bar represents the power, the darker the color, the greater the power; the lighter the color, the smaller the power. MDD, major depressive disorder. ADM, antidepressant medication. SSRIs, selective serotonin reuptake inhibitors. SNRIs, serotonin norepinephrine reuptake inhibitors
A 49-dimensional features dataset with different prediction power was obtained after each LOO-CV iteration and 121 49-dimensional feature datasets were achieved after 121 iterations, in which the best feature dataset was determined when the model reached its highest values after feature search completing (Fig. 3, Table S5). The prediction power in all LOO-CV iterations were averaged and ranked in descending order, and the top 30% were defined as the most powerful features for predicting ADM improvers, SSRIs improvers and SNRIs improvers. The radiomics features predicting ADM improver were mainly located in the hippocampus, medial orbitofrontal gyrus, anterior cingulate gyrus, amygdala, superior frontal gyrus, cerebellum (lobule vii-b), middle temporal gyrus, body of corpus callosum, anterior limb of internal capsule and anterior corona radiata. The radiomics features predicting SSRIs improver were primarily distributed in hippocampus, amygdala, inferior temporal gyrus, thalamus, cerebellum (lobule vi), fornix, and cerebellar peduncle. The radiomics features predicting SNRIs improver were primarily located in the medial orbitofrontal cortex, anterior cingulate gyrus, ventral striatum, accumbency area, knee of corpus callosum, internal capsule and anterior corona radiata.
Learning curve for selection of ComBat-harmonized radiomics feature based on RFE. The X- / Y-axes represents the number of ComBat-harmonized radiomics features selected and the prediction accuracy, respectively. The highest overall accuracy (88.19%) of the radiomics prediction model was achieved when based on a minimum number (49) of radiomics features, and then the prediction performance did not improve as the number of features increased. RFE, recursive feature elimination
Prediction models performance
RBF-SVM models were constructed by using selected conventional imaging indicators and radiomics features. The optimal values of the regularization parameter C and the kernel function parameter gamma for RBF-SVM by the process of tuning and searching were 102 and 10− 1, respectively. The detailed results of the SVM based on the two types of features are illustrated in Table 3. The overall accuracy of SVM based on conventional indicators and radiomics features after harmonization were 74.80% and 88.19%. The SVM model using ComBat-harmonized radiomics features based on the results of LOO-CV and ROC analysis had the better performance for predicting ADM improvers, SSRIs improvers and SNRIs improvers with the AUC, accuracy, sensitivity, and specificity of 0.889, 91.2%, 80.1% and 85.1%, 0.954, 89.2%, 87.4% and 88.5%, 0.942, 91.9%, 82.5% and 86.8%, respectively, which was superior to the model based on the conventional imaging indicators (Fig. 4). P value of permutation tests were less than 0.001.
Discussion
Precision medicine is a long-term goal pursued in psychiatry clinics [44, 45]. Nevertheless, antidepressant drug selection is a complex and challenging issue in clinical practice [4, 5]. Gradually, neuroimaging studies based on radiomics and machine learning are demonstrating an important subsidiary role in the personalized diagnosis and treatment of psychiatric disorders [15,16,17,18, 27,28,29]. In this study, by integrating imaging features of brain multiscale sMRI after harmonization and selection, the radiomics model performed significantly better than the model based on the conventional imaging indicators, indicating that the integration and selection of high-dimensional features improved the predictive efficacy of the features, and the outstanding ability to select features was just the advantage of machine learning [25,26,27]. This radiomics framework confirmed that there were indeed differences in gray and white matter of important brain regions involved in early improvement to ADM of MDD patients by quantitatively estimating the power of each feature contributing to prediction model, which also made the results more interpretable.
The predictive ability of imaging markers represents the outcome of specific ADM agents to alleviate or eliminate structural or functional pathological changes in gray and white matter [25, 26]. The radiomics features identified in this study for the prediction of early ADM efficacy may help to choose SSRIs or SNRIs, thereby improving early response and long-term remission rates in patients with MDD. Features of the medial orbitofrontal gyrus, hippocampus, cerebellum lobule vii-b, corpus callosum, anterior limb of internal capsule and anterior corona radiata were extremely important in predictive models of 2-week ADM improvement. These brain regions are essential components of the brain’s emotion network and closely related to emotional processing, memory, and regulation [46, 47]. Previous studies have demonstrated that antidepressants could reverse impaired neuroplasticity and neurogenic changes in the hippocampus, such as loss of dendritic spines and synapses [48, 49]. According to the recent findings on hippocampal physiological function [50], the synergy between the hippocampus and other relevant brain regions at baseline could be integrated into a holistic feature to predict the beneficial effects of antidepressants on emotional memory and regulation. However, this needs to be further validated by neuropharmacological and psychological experiments after medication administration.
The present study showed that radiomics features of some brain regions for implicit emotion regulation such as hippocampus and amygdala play a prominent role in predicting 2-week improvement of SSRIs treatment. Several neurophysiological studies revealed that SSRIs had an acute neurological effect on emotion regulation by affecting 5-hydroxytryptamine reuptake and thus increasing 5-hydroxytryptamine concentrations in the synaptic space to regulate activity in relevant brain regions, especially in the hippocampus and amygdala [50,51,52]. There were also imaging studies showing that FA values of the WM fiber tract conjoining hippocampus and amygdala could predict long-term remission after receiving SSRIs [53], and the fornix connecting the hippocampus was an important predictor of early response to SSRIs [16, 17]. SSRIs are known to mediate hippocampal responsiveness to treatment by increasing neural progenitor cells, and a functional MRI study predicting 4-week efficacy also highlighted the hippocampal response to SSRIs therapy [54]. As for the amygdala, one study showed higher FA values in the amygdala of remitters compared to non-remitters after treatment with SSRIs [15, 16], which might be related to the 5-hydroxytryptamine-mediated amygdala response pattern as a key pathway for the antidepressant effects of SSRIs. MDD is a kind of psychiatric disorder with impaired emotion regulation, and habitual use of emotion regulation strategies plays an important role in MDD episodes [46, 47]. Therefore, based on our results and the known mechanisms of action of SSRIs, it is reasonable to use imaging features of emotional regulation-localized brain regions as markers to predict the early efficacy of SSRIs in MDD patients.
The present study also identified the radiomics features of some brain regions such as the medial orbitofrontal cortex, anterior cingulate gyrus, ventral striatum, accumbency area, knee of corpus callosum, internal capsule and anterior corona radiata as imaging markers for 2-week improvement of SNRIs. This was consistent with the results of previous neuropharmacological studies showing that the brain regions associated with SNRIs response were mainly located in the emotional regulation and reward circuits [50, 52]. The medial orbitofrontal cortex and anterior cingulate gyrus are involved in learning, motivation, and reward behavior in MDD patients [55, 56], while the ventral striatum is engaged in reward signaling in the neural system, and abnormalities in these regions can lead to anhedonia [46, 55,56,57]. SNRIs increase activity in the medial orbitofrontal cortex and ventral striatum by affecting norepinephrine reuptake and dopamine release to alleviate the pleasure deficit in MDD patients [52]. Positron emission tomography (PET) study also suggested that the activity of the anterior medial orbitofrontal cortex was associated with the remission of MDD after treatment with SNRIs [22, 23]. Meanwhile, our study indicated that DTI-related diffusion features of white matter in brain regions associated with emotional regulation and reward circuits were important biomarkers for predicting early improvement to SNRIs. Therefore, the results of this study were consistent with the above theoretical perspective.
Previous neuroimaging studies revealed that morphological features of the brain surface based on SBM analysis had a high weight in the diagnostic model of MDD [34] and could be used as predictive markers of response to antidepressant treatment [12,13,14]. For example, structural abnormalities accompanied by decreased functional connectivity at baseline in the hippocampus, superior frontal gyrus, middle temporal gyrus, cingulate gyrus, and amygdala associated with processing emotional processing were better predictors of antidepressant response [12,13,14, 16, 17]. However, the results of the present study showed that none of SBM-based features were selected in the radiomics prediction model, which might be related to the selecting strategy of imaging features by ranking them according to their relevance to each other and importance to the model in the reverse elimination process during machine learning, so the relationship between SBM-based imaging indicators and MDD drug efficacy needs to be further investigated.
Several limitations are certainly needed to be considered in our study. First, due to small sample size, we used the leave-one-out cross-validation and failed to use external samples for validation, which might affect the reliability of the prediction model. Second, the brain multiscale sMRI were obtained using different scanners, and although the radiomics features were harmonized by using the ComBat technique, it might also affect the model prediction performance. Third, the MDD patients were simply grouped using only scale scores and medication types, without considering the heterogeneity associated with different subtypes of depression and various biological entities, which may affect the effectiveness of the model.
Conclusions
In summary, our findings suggest that the radiomics model based on brain multiscale sMRI after ComBat harmonization could effectively predict the early improvement of ADM in adolescent MDD patients with a high accuracy, which was superior to the model based on the conventional imaging indicators. The radiomics features with high prediction power may help for the individual SSRIs and SNRIs selection.
Data Availability
The data used and analyzed in this study are available from the corresponding author on reasonable request.
Abbreviations
- MDD:
-
Major depressive disorder
- ADM:
-
Antidepressant medication
- sMRI:
-
Structural magnetic resonance imaging
- SSRIs:
-
Selective serotonin reuptake inhibitors
- SNRIs:
-
Serotonin norepinephrine reuptake inhibitors
- 3D-T1WI:
-
Three-dimensional T1 weighted imaging
- DTI:
-
Diffusion tensor imaging
- GM:
-
Gray matter
- WM:
-
White matter
- CSF:
-
Cerebrospinal fluid
- SBM:
-
Surface-based morphology
- VBM:
-
Voxel-based morphology
- ANOVA:
-
Analysis of variance
- RFE:
-
Recursive feature elimination
- SVM:
-
Support vector machine
- RBF:
-
Radial basis function kernel
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- LOO-CV:
-
Leave-one-out cross-validation
- HAMD:
-
Hamilton depression rating scale
- DSM-V:
-
Diagnostic and statistical manual of mental disorders, fifth edition
- rTMS:
-
Repetitive transcranial magnetic stimulation
- ECT:
-
Electroconvulsive therapy
- FLAIR:
-
Fluid attenuation inversion recovery
- DKT:
-
Desikan-Killiany-Tourville
- SPM:
-
Statistical parametric mapping
- AAL:
-
Automated anatomical labeling atlas
- IBSI:
-
Image biomarkers standardization initiative
- RQS:
-
Radiomics quality scoring
- FSL:
-
FMRIB’s software library
- FA:
-
Anisotropy fraction
References
Hasin DS, Sarvet AL, Meyers JL, et al. Epidemiology of adult DSM-5 major depressive disorder and its specifiers in the United States. JAMA Psychiatry. 2018;75(4):336–46.
Rice F, Riglin L, Lomax T, Souter E, Potter R, Smith DJ, Thapar AK, Thapar A. Adolescent and adult differences in major depression symptom profiles. J Affect Disord. 2019;243:175–81.
LeMoult J, Humphreys KL, Tracy A, Hoffmeister JA, Ip E, Gotlib IH. Meta-analysis: exposure to early life stress and risk for depression in childhood and adolescence. J Am Acad Child Adolesc Psychiatry. 2020;59(7):842–55.
Walter HJ, Abright AR, Bukstein OG, Diamond J, Keable H, Ripperger-Suhler J, Rockhill C. Clinical practice guideline for the assessment and treatment of children and adolescents with major and persistent depressive disorders. J Am Acad Child Adolesc Psychiatry. 2022;21:S0890-8567(22)01852-4.
Feng Y, Xiao L, Wang WW, Ungvari GS, Ng CH, Wang G, Xiang YT. Guidelines for the diagnosis and treatment of depressive disorders in China: the second edition. J Affect Disord. 2019;253:352–56.
Avenevoli S, Swendsen J, He JP, Burstein M, Merikangas KR. Major depression in the national comorbidity survey-adolescent supplement: prevalence, correlates, and treatment. J Am Acad Child Adolesc Psychiatry. 2015;54(1):37–44.
Sharma A, Barrett MS, Cucchiara AJ, Gooneratne NS, Thase ME. A breathing-based meditation intervention for patients with major depressive disorder following inadequate response to antidepressants: a randomized pilot study. J Clin Psychiatry. 2017;78(1):59–63.
Tadić A, Wachtlin D, Berger M, Braus DF, van Calker D, Dahmen N, Dreimüller N, Engel A, Gorbulev S, Helmreich I, Kaiser AK, Kronfeld K, Schlicht KF, Tüscher O, Wagner S, Hiemke C, Lieb K. Randomized controlled study of early medication change for non-improvers to antidepressant therapy in major depression-the EMC trial. Eur Neuropsychopharmacol. 2016;26(4):705–16.
Henkel V, Seemüller F, Obermeier M, Adli M, Bauer M, Mundt C, Brieger P, Laux G, Bender W, Heuser I, Zeiler J, Gaebel W, Mayr A, Möller HJ, Riedel M. Does early improvement triggered by antidepressants predict response/remission? Analysis of data from a naturalistic study on a large sample of inpatients with major depression. J Affect Disord. 2009;115(3):439–49.
Szegedi A, Jansen WT, van Willigenburg AP, Stassen HH, Thase ME. Early improvement in the first 2 weeks as a predictor of treatment outcome in patients with major depressive disorder: a meta-analysis including 6562 patients. J Clin Psychiatry. 2009;70(3):344–53.
Kim JM, Kim SY, Stewart R, Yoo JA, Bae KY, Jung SW, Lee MS, Yim HW, Jun TY. Improvement within 2 weeks and later treatment outcomes in patients with depressive disorders: the CRESCEND study. J Affect Disord. 2011;129(1–3):183–90.
Maller JJ, Broadhouse K, Rush AJ, Gordon E, Koslow S, Grieve SM. Increased hippocampal tail volume predicts depression status and remission to anti-depressant medications in major depression. Mol Psychiatry. 2018;23(8):1737–44.
Saricicek Aydogan A, Oztekin E, Esen ME, Dusmez S, Gelal F, Besiroğlu L, Zorlu N. Cortical thickening in remitters compared to non-remitters with major depressive disorder following 8-week antidepressant treatment. Acta Psychiatr Scand. 2019;140(3):217–26.
Bartlett EA, DeLorenzo C, Sharma P, Yang J, Zhang M, Petkova E, Weissman M, McGrath PJ, Fava M, Ogden RT, Kurian BT, Malchow A, Cooper CM, Trombello JM, McInnis M, Adams P, Oquendo MA, Pizzagalli DA, Trivedi M, Parsey RV. Pretreatment and early-treatment cortical thickness is associated with SSRI treatment response in major depressive disorder. Neuropsychopharmacology. 2018;43(11):2221–30.
Grieve SM, Korgaonkar MS, Gordon E, Williams LM, Rush AJ. Prediction of nonremission to antidepressant therapy using diffusion tensor imaging. J Clin Psychiatry. 2016;77(4):e436–43.
Hoogenboom WS, Perlis RH, Smoller JW, et al. Limbic system white matter microstructure and long-term treatment outcome in major depressive disorder: a diffusion tensor imaging study using legacy data. World J Biol Psychiatry. 2014;15(2):122–34.
Lai CH. Fronto-limbic neuroimaging biomarkers for diagnosis and prediction of treatment responses in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2021;107:110234.
Ning L, Rathi Y, Barbour T, Makris N, Camprodon JA. White matter markers and predictors for subject-specific rTMS response in major depressive disorder. J Affect Disord. 2022;299:207–14.
Xue L, Pei C, Wang X, Wang H, Tian S, Yao Z, Lu Q. Predicting neuroimaging biomarkers for antidepressant selection in early treatment of depression. J Magn Reson Imaging. 2021;54(2):551–59.
Davis AD, Hassel S, Arnott SR, Harris J, Lam RW, Milev R, Rotzinger S, Zamyadi M, Frey BN, Minuzzi L, Strother SC, MacQueen GM, Kennedy SH, Hall GB. White matter indices of medication response in major depression: a diffusion tensor imaging study. Biol Psychiatry Cogn Neurosci Neuroimaging. 2019;4(10):913–24.
Chin Fatt CR, Jha MK, Cooper CM, Fonzo G, South C, Grannemann B, Carmody T, Greer TL, Kurian B, Fava M, McGrath PJ, Adams P, McInnis M, Parsey RV, Weissman M, Phillips ML, Etkin A, Trivedi MH. Effect of intrinsic patterns of functional brain connectivity in moderating antidepressant treatment response in major depression. Am J Psychiatry. 2020;177(2):143–54.
Li J, Lu C, Gao Z, et al. SNRIs achieve faster antidepressant effects than SSRIs by elevating the concentrations of dopamine in the forebrain. Neuropharmacology. 2020;177:108237.
Kennedy SH, Konarski JZ, Segal ZV, Lau MA, Bieling PJ, McIntyre RS, Mayberg HS. Differences in brain glucose metabolism between responders to CBT and venlafaxine in a 16-week randomized controlled trial. Am J Psychiatry. 2007;164(5):778–88.
Wattiez AS, Dupuis A, Privat AM, Chalus M, Chapuy E, Aissouni Y, Eschalier A, Courteix C. Disruption of 5-HT2A-PDZ protein interaction differently affects the analgesic efficacy of SSRI, SNRI and TCA in the treatment of traumatic neuropathic pain in rats. Neuropharmacology. 2017;125:308–18.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–77.
Rondina JM, Ferreira LK, de Souza Duran FL, Kubo R, Ono CR, Leite CC, Smid J, Nitrini R, Buchpiguel CA, Busatto GF. Selecting the most relevant brain regions to discriminate Alzheimer’s disease patients from healthy controls using multiple kernel learning: a comparison across functional and structural imaging modalities and atlases. Neuroimage Clin. 2017;17:628–41.
Huang K, Lin Y, Yang L, Wang Y, Cai S, Pang L, Wu X, Huang L. Alzheimer’s Disease Neuroimaging Initiative. A multipredictor model to predict the conversion of mild cognitive impairment to Alzheimer’s disease by using a predictive nomogram. Neuropsychopharmacology. 2020;45(2):358–66.
Wang Y, Sun K, Liu Z, Chen G, Jia Y, Zhong S, Pan J, Huang L, Tian J. Classification of unmedicated bipolar disorder using whole-brain functional activity and connectivity: a radiomics analysis. Cereb Cortex. 2020;30(3):1117–28.
Sun H, Chen Y, Huang Q, Lui S, Huang X, Shi Y, Xu X, Sweeney JA, Gong Q. Psychoradiologic utility of MR imaging for diagnosis of attention deficit hyperactivity disorder: a radiomics analysis. Radiology. 2018;287(2):620–30.
Orlhac F, Lecler A, Savatovski J, Goya-Outi J, Nioche C, Charbonneau F, Ayache N, Frouin F, Duron L, Buvat I. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur Radiol. 2021;31(4):2272–80.
Leithner D, Schöder H, Haug A, Vargas HA, Gibbs P, Häggström I, Rausch I, Weber M, Becker AS, Schwartz J, Mayerhoefer ME. Impact of ComBat harmonization on PET radiomics-based tissue classification: a dual-center PET/MRI and PET/CT study. J Nucl Med. 2022;63(10):1611–16.
Saint Martin MJ, Orlhac F, Akl P, Khalid F, Nioche C, Buvat I, Malhaire C, Frouin F. A radiomics pipeline dedicated to breast MRI: validation on a multi-scanner phantom study. MAGMA. 2021;34(3):355–66.
Fortin JP, Parker D, Tunç B, Watanabe T, Elliott MA, Ruparel K, Roalf DR, Satterthwaite TD, Gur RC, Gur RE, Schultz RT, Verma R, Shinohara RT. Harmonization of multi-site diffusion tensor imaging data. NeuroImage. 2017;161:149–70.
Ma H, Zhang D, Sun D, Wang H, Yang J. Gray and white matter structural examination for diagnosis of major depressive disorder and subthreshold depression in adolescents and young adults: a preliminary radiomics analysis. BMC Med Imaging. 2022;22(1):164.
Lin HS, Lin CH. Early improvement in HAMD-17 and HAMD-6 scores predicts ultimate response and remission for depressed patients treated with fluoxetine or ECT. J Affect Disord. 2019;245:91–7.
Alexander B, Loh WY, Matthews LG, Murray AL, Adamson C, Beare R, Chen J, Kelly CE, Anderson PJ, Doyle LW, Spittle AJ, Cheong JLY, Seal ML, Thompson DK. Desikan-Killiany-Tourville atlas compatible version of M-CRIB neonatal parcellated whole brain atlas: the M-CRIB 2.0. Front Neurosci. 2019;13:34.
Tustison NJ, Cook PA, Klein A, Song G, Das SR, Duda JT, Kandel BM, van Strien N, Stone JR, Gee JC, Avants BB. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. NeuroImage. 2014;99:166–79.
Ghosh A, Kaur S, Shah R, Oomer F, Avasthi A, Ahuja CK, Basu D, Nehra R, Khandelwal N. Surface-based brain morphometry in schizophrenia vs. cannabis-induced psychosis: a controlled comparison. J Psychiatr Res. 2022;155:286–94.
Dovrou A, Nikiforaki K, Zaridis D, Manikis GC, Mylona E, Tachos N, Tsiknakis M, Fotiadis DI, Marias K. A segmentation-based method improving the performance of N4 bias field correction on T2weighted MR imaging data of the prostate. Magn Reson Imaging. 2023;101:1–12.
Bettinelli A, Marturano F, Avanzo M, Loi E, Menghi E, Mezzenga E, Pirrone G, Sarnelli A, Strigari L, Strolin S, Paiusco M. A novel benchmarking approach to assess the agreement among radiomic tools. Radiology. 2022;303(3):533–41.
Spadarella G, Stanzione A, Akinci D’Antonoli T, Andreychenko A, Fanni SC, Ugga L, Kotter E, Cuocolo R. Systematic review of the radiomics quality score applications: an EuSoMII radiomics auditing group initiative. Eur Radiol. 2023;33(3):1884–94.
Yang S, Ghosh K, Sakaie K, Sahoo SS, Carr SJA, Tatsuoka C. A simplified crossing fiber model in diffusion weighted imaging. Front Neurosci. 2019;13:492.
Chen C, Qin Y, Chen H, Cheng J, He B, Wan Y, Zhu D, Gao F, Zhou X. Machine learning to differentiate small round cell malignant tumors and non-small round cell malignant tumors of the nasal and paranasal sinuses using apparent diffusion coefficient values. Eur Radiol. 2022;32(6):3819–29.
Quinlan EB, Banaschewski T, Barker GJ, Bokde ALW, Bromberg U, Büchel C, Desrivières S, Flor H, Frouin V, Garavan H, Heinz A, Brühl R, Martinot JL, Paillère Martinot ML, Nees F, Orfanos DP, Paus T, Poustka L, Hohmann S, Smolka MN, Fröhner JH, Walter H, Whelan R, Schumann G, IMAGEN Consortium. Identifying biological markers for improved precision medicine in psychiatry. Mol Psychiatry. 2020;25(2):243–53.
Goldstein-Piekarski AN, Holt-Gosselin B, O’Hora K, Williams LM. Integrating sleep, neuroimaging, and computational approaches for precision psychiatry. Neuropsychopharmacology. 2020;45(1):192–204.
Salzman CD, Fusi S. Emotion, cognition, and mental state representation in amygdala and prefrontal cortex. Annu Rev Neurosci. 2010;33:173–202.
McRae K, Gross JJ. Emotion regulation. Emotion. 2020;20(1):1–9.
Valdés-Tovar M, Estrada-Reyes R, Solís-Chagoyán H, Argueta J, Dorantes-Barrón AM, Quero-Chávez D, Cruz-Garduño R, Cercós MG, Trueta C, Oikawa-Sala J, Dubocovich ML, Benítez-King G. Circadian modulation of neuroplasticity by melatonin: a target in the treatment of depression. Br J Pharmacol. 2018;175(16):3200–08.
Serafini G. Neuroplasticity and major depression, the role of modern antidepressant drugs. World J Psychiatry. 2012;2(3):49–57.
Watson TC, Obiang P, Torres-Herraez A, Watilliaux A, Coulon P, Rochefort C, Rondi-Reig L. Anatomical and physiological foundations of cerebello-hippocampal interaction. Elife. 2019;8:e41896.
Jannini TB, Lorenzo GD, Bianciardi E, Niolu C, Toscano M, Ciocca G, Jannini EA, Siracusano A. Off-label uses of selective serotonin reuptake inhibitors (SSRIs). Curr Neuropharmacol. 2022;20(4):693–712.
Leshem R, Bar-Oz B, Diav-Citrin O, Gbaly S, Soliman J, Renoux C, Matok I. Selective serotonin reuptake inhibitors (SSRIs) and serotonin norepinephrine reuptake inhibitors (SNRIs) during pregnancy and the risk for autism spectrum disorder (ASD) and attention deficit hyperactivity disorder (ADHD) in the offspring: a true effect or a bias? A systematic review & meta-analysis. Curr Neuropharmacol. 2021;19(6):896–906.
Jiang J, Zhao YJ, Hu XY, Du MY, Chen ZQ, Wu M, Li KM, Zhu HY, Kumar P, Gong QY. Microstructural brain abnormalities in medication-free patients with major depressive disorder: a systematic review and meta-analysis of diffusion tensor imaging. J Psychiatry Neurosci. 2017;42(3):150–63.
Oh SJ, Cheng J, Jang JH, Arace J, Jeong M, Shin CH, Park J, Jin J, Greengard P, Oh YS. Hippocampal mossy cell involvement in behavioral and neurogenic responses to chronic antidepressant treatment. Mol Psychiatry. 2020;25(6):1215–28.
Bekhbat M, Li Z, Mehta ND, Treadway MT, Lucido MJ, Woolwine BJ, Haroon E, Miller AH, Felger JC. Functional connectivity in reward circuitry and symptoms of anhedonia as therapeutic targets in depression with high inflammation: evidence from a dopamine challenge study. Mol Psychiatry. 2022;27(10):4113–21.
Zhang FF, Peng W, Sweeney JA, Jia ZY, Gong QY. Brain structure alterations in depression: psychoradiological evidence. CNS Neurosci Ther. 2018;24(11):994–1003.
Wang S, Leri F, Rizvi SJ. Anhedonia as a central factor in depression: neural mechanisms revealed from preclinical to clinical evidence. Prog Neuropsychopharmacol Biol Psychiatry. 2021;110:110289.
Acknowledgements
Not applicable.
Funding
This study was not funded.
Author information
Authors and Affiliations
Contributions
Huan Ma: Conceptualization, Investigation, Methodology, Data curation, Formal analysis, Writing ? original draft, Writing ? review & editing. Dafu Zhang: Methodology, Validation, Visualization, Data curation. Yao Wang: Resources, Formal analysis. Yingying Ding: Conceptualization, Methodology, Supervision, Data curation. Jianzhong Yang: Investigation, Methodology, Resources, Supervision, Writing ? review & editing. Kun Li: Project administration, Writing ? review & editing. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Third Affiliated Hospital of Kunming Medical University according to the principles of the Declaration of Helsinki with approval number KYCS202107. The written informed consent was obtained from participants and from their parents or legal guardians for subjects under 16 years old before enrollment.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1:
Radiomics features and parameters of MR scanners.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ma, H., Zhang, D., Wang, Y. et al. Prediction of early improvement of major depressive disorder to antidepressant medication in adolescents with radiomics analysis after ComBat harmonization based on multiscale structural MRI. BMC Psychiatry 23, 466 (2023). https://doi.org/10.1186/s12888-023-04966-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12888-023-04966-8