Abstract
Objective:
The aim of this study was to assess associations of PART1 rs27565 and DEFB1 rs11362 polymorphisms with the prevalence of dental caries in twelve-year-old children in Nandan County, Guangxi, China.
Methods:
A total of 1,061 children were included in this cross-sectional study and divided into two groups based on the Decayed, Missing and Filled teeth (DMFT) index: caries-free children (DMFT score = 0) and children with caries (DMFT score ≥ 1). Demographic characteristics, oral hygiene behaviour and dietary habits were collected through household records and questionnaires. Genomic DNA was extracted from buccal cells, and PART1 rs27565 and DEFB1 rs11362 polymorphisms were genotyped using a custom-designed 48-Plex single nucleotide polymorphism-scan kit.
Results:
Carriers of the PART1 rs27565 C allele (odds ratio [OR] = 1.338, 95% confidence interval (CI) = 1.015–1.764, P value = 0.039) and carriers of the DEFB1 rs11362 T allele (OR = 1.364, 95% CI = 1.056–1.762, P value = 0.017) had a higher risk of caries. Carriers of the PART1 rs27565 TC or CC genotype who ate sugary food more than once a week had a 1.6-fold higher risk of caries than TT carriers who ate sugary food at most once a week (OR = 1.579, 95% CI = 1.032–2.414, P value = 0.035). Carriers of the DEFB1 rs11362 CT or TT genotype who ate sugary food more than once a week had a 2.1-fold higher risk of caries than CC carriers who ate sugary food at most once a week (OR = 2.057, 95% CI = 1.438–2.940, P value < 0.001).
Conclusion:
PART1 rs27565 and DEFB1 rs11362 polymorphisms were associated with caries in 12-year-old children in Nandan County, Guangxi, China. Carriers of the PART1 rs27565 TC or CC genotype and the DEFB1 rs11362 CT or TT genotype who ate sugary food more than once a week had a high probability of having caries.
Similar content being viewed by others
Introduction
Dental caries is a major public health problem worldwide. Untreated caries in permanent teeth was the most prevalent health condition, affecting 35% of the global population; untreated caries in deciduous teeth was the tenth most prevalent health condition, affecting 9.0% of the global paediatric population [1]. Caries may cause toothache, masticatory dysfunction and psychological disorders and impact people’s quality of life [1,2,3,4]. Overall, the global burden of untreated caries in deciduous and permanent teeth has remained constant over the past 30 years, despite various measures implemented to prevent and control caries [1]. Thus, studying the factors affecting the prevalence of caries is very important.
Dental caries is likely influenced by multiple factors, such as the oral microbiota, sugars, tooth structure, salivary and fluoride exposure, socioeconomic status, oral hygiene habits and genetic factors [2, 5]. Among these factors, the oral microbiota plays a key role in the aetiology of caries. The ecological plaque hypotheses, which are now generally accepted as the most plausible explanations for the microbial aetiology of caries, consider that caries is a consequence of an unfavourable shift in the balance of the resident microbiota driven by changes in the dental environment [2, 6]. Caries will not occur in the absence of a cariogenic dental biofilm composed of complex microbial communities [2].
Studies have shown that the oral microenvironment and the development of dental biofilms are affected not only by diet and oral hygiene habits but also by natural immune factors. For example, individuals with very low levels of saliva antibodies or proteins are more sensitive to plaque accumulation and subsequent caries formation [7, 8]. The structures, functions and levels of these antibodies or proteins might be influenced by genetic variants [9, 10].
PART1 and DEFB1 are two of the immune-related genes [11, 12]. This study focused on PART1 rs27565 and DEFB1 rs11362 polymorphisms due to their previous potential association with dental caries. PART1 rs27565 is located on chromosome 5q13.3, which was reported to be associated with a lower caries susceptibility by Vieira using a genome-wide linkage scan [13]. mRNA expression of PART1 has been detected in saliva, salivary glands, and the prostate [12]. The PART1 rs27565 T allele was associated with low caries susceptibility in Filipino families [12]. The study by Kelly also reported a higher frequency of the C allele in the caries group [14], while another study showed that the association between the PART1 rs27565 polymorphism and the risk of caries was not significant after the Bonferroni adjustment of the P value [15]. DEFB1, consisting of two exons and one intron, encodes β-defensin 1, which is synthesized as a 64–68 amino acid prepropeptide and is expressed in the major salivary glands, tongue, gingiva, and buccal mucosa [16, 17]. β-Defensin 1 has broad-spectrum antibacterial (against gram-positive and gram-negative bacteria), antiviral and antifungal activities, protecting individuals from pathogens [18]. The rs11362 is located in the 5′ untranslated region (5`-UTR) of the reference sequence of DEFB1 (NM_005218) at the − 20 (c.-20G > A) position [19]. A relationship between the DEFB1 rs11362 polymorphism and caries was observed in some populations. Carriers of the DEFB1 rs11362 T allele have a higher risk of caries than carriers of the C allele, as documented in studies of children from Ribeirão Preto in Brazil [20] and Gansu Province in China [19], as well as in adults from Turkey [21] and North America [22]. In contrast, studies examining Italian adults and Latvia children with cleft lip suggested that CC genotype carriers have a higher risk of caries [23, 24]. In addition, some studies did not report any association between the rs11362 polymorphism and caries [25, 26].
Although the relationship between gene polymorphisms and caries has been studied for decades and genetic factors play a well-established role in an individual’s risk of caries, the results of studies are inconsistent, potentially due to differences in the study populations [27]. Therefore, research on the relationship between gene polymorphisms and caries prevalence in diverse populations with various geographic origins and different ages will advance our understanding of the role of genetics in caries development and/or progression [27]. Nandan County, located in northwestern Guangxi Province in southern China, is an area containing ethnic minority groups and thus has a population that exhibits large differences in genetic backgrounds and living habits compared with those of other populations. Nandan is a remote mountainous area, and people live in relative isolation because of the poor construction of infrastructure facilities and the traffic inconvenience. Although the inhabitants belong to different ethnic subgroups, they share the same ancestry and have relatively consistent genetic backgrounds. Studies assessing the correlation between gene polymorphisms and dental caries in this area are still scarce. Therefore, the purpose of this study was to explore the relationships of PART1 rs27565 and DEFB1 rs11362 polymorphisms with caries prevalence in 12-year-old children in Nandan County.
Subjects and methods
Subjects
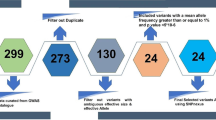
Twelve-year-old children in Nandan County, Guangxi (water and milk are not fluoridated) were selected as the study population. The expected prevalence of caries in 12-year-old children residing in Guangxi was 43.8% [28]. The sample size was calculated by using Quanto program (Version 1.2.3, https://bio.tools/QUANTO) based on the association between the prevalence of caries and the frequency of the risk allele in the population. The frequencies of the PART1 rs27565 C allele and DEFB1 rs11362 T allele, which are potentially associated with high caries risk, are 0.48 and 0.40 in Asian populations, respectively (HaploReg v4.1, https://broadinstitute.org). It was calculated that at least 974 participants would be required with α = 0.05, 1-β = 0.8 and expected odds ratio = 1.5. The expected odds ratio was determined by the results from previous studies [13, 20]. This calculated sample size also met the statistical requirements of binary logistic regression analysis in this study. Then, fifteen of 55 primary schools in Nandan County were selected using a random number generator software program (Microsoft Office Excel 2010, USA). Twelve-year-old children from the selected schools who were free of chronic diseases and unrelated were recruited. Finally, 1,061 children were included after excluding subjects with incomplete data.
This study was approved by the Institutional Research Ethics Committee of Guangxi Medical University. All the children assented to participate in this survey on-site, and guardians provided written informed consent. All methods were conducted in accordance with relevant guidelines and regulations.
Data collection
Clinical examinations were performed by three dentists to collect data on caries experience. The dentists were trained by an experienced dental epidemiologist. The inter- and intra-examiner consistency test was conducted following the protocol suggested by the World Health Organization (WHO) [29]. Thirty participants were assessed and the kappa test was used to observe the consistency of measurements with each examiner.
The experience of dental caries in permanent teeth was recorded using the Decayed, Missing and Filled teeth (DMFT) index according to the standard recommended by the WHO in 2013 [29]. Children were divided into two groups based on the DMFT index: caries-free children (DMFT score = 0) and children with caries (DMFT score ≥ 1).
Demographic characteristics, such as sex, ethnicity (defined by both parents being of the same ethnic group), age and parental education level (the highest education level attained by the parents) were obtained from household records. Data on oral hygiene behaviour and dietary habits were collected using a structured questionnaire designed with reference to the Fourth Chinese National Oral Health Survey [30]. Dietary habits were assessed, specifically whether the children regularly ate at school and the frequency of sugary food intake. Sugary food was defined as sweet foods, sweetened milk/yoghurt/tea/coffee and other sweet drinks [30]. The frequency of sweet food intake was recorded in six categories (0 = seldom, 1 = one to three times per month, 2 = once per week, 3 = two to six times per week, 4 = once per day, and 5 = more than twice per day) [30] and then divided into two groups: at most once a week (with scores of 0, 1 and 2) and more than once a week (with scores of 3, 4 and 5). Structured questionnaires were completed by children and reviewed by research assistants.
DNA isolation and genotyping
Subjects were asked to gargle and do not eat or drink within 30 min before sampling to ensure that the sample was not contaminated by food or drink. Four disposable swabs were used to scrape both buccal mucosae back and forth (at least 20 times) without touching the teeth to obtain buccal mucosal cells. The heads of the swabs were sealed in cryovials, immediately frozen in dry ice, transported to the laboratory and stored in a -80 °C freezer until needed. Genomic DNA was extracted from buccal mucosal cells with the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions.
The genotyping work was performed using a custom-designed 48-Plex single nucleotide polymorphism-scan kit (Genesky Biotechnologies, Inc., Shanghai, China). The kit was developed according to a patented genotyping technology by Genesky Biotechnologies, Inc., which was based on double ligation and multiplex fluorescence polymerase chain reaction (PCR). The genotyping processes were as follows: (1) a 1% agarose gel was used for quality inspection and to estimate the concentration. The concentration of the DNA sample ranged from 30 to 50 ng/µl. (2) Next, 2.5 µl of 4X DNA Lysis Buffer were added to 4 µl of DNA samples, the volume was increased to 10 µl with water, and samples were mixed and centrifuged. Then, the reaction was performed at 98 °C for 5 min and then immediately placed on ice. (3) Ten microliters of linking reaction premix were added to the frozen, degraded DNA sample, centrifuged for 30 s (3000 rpm), and immediately placed in the PCR instrument and cycled using the following program: 98℃ for 2 min, 5 cycles of 95 °C for 1 min, 58 °C for 3 h) and 94 °C for 2 min, followed by an incubation at 72 °C until subsequent reactions. The ligated products were then amplified by PCR using fluorescent primers using the following program: a hold cycle of 95 °C for 2 min, followed by 9 touchdown amplification cycles of 94 °C for 20 s, 62 °C (-0.5 °C/cycle) for 40 s, 72 °C for 1.5 min; 25 amplification cycles of 94 °C for 20 s, 57 °C for 40 s, and72°C for 1.5 min; and finally, an extension step at 68 °C for 1 h. (4) One microlitre of PCR product (after a 10-fold dilution) was mixed with 0.5 µl of Liz500 SIZE STANDARD and 8.5 µl of hi-di. After denaturation at 95 °C for 5 min, an ABI 3730XL sequencer was used to obtain the original data. (5) GeneMapper 4.1 (Applied Biosystems, USA) was used to analyse the data and record the fluorescence of markers and length of PCR products, as well as the corresponding single-nucleotide polymorphism (SNP) loci/allele information.
Statistical analysis
Sex, ethnicity, parental education level, frequency of tooth brushing, frequency of sugary food intake, regular eating at school and genotype were the categorical variables and are presented as percentages. Categorical variables were compared between the two groups using Chi-square tests. Binary logistic regression analysis was performed to determine the associations between caries and the risk factors. Caries status was the dependent variable, whereby DMFT greater than zero is denoted as “1” and DMFT equal to zero as “0”. Chi-square tests and binary logistic regression analyses were performed using the Statistical Package for Social Sciences 25.0 software (SPSS, Chicago, IL, USA). Hardy–Weinberg equilibrium (HWE) was calculated using PLINK 1.90. A P value < 0.05 was considered statistically significant.
Results
Demographic characteristics and environmental factors
The kappa values for inter- and intra-examiner data during calibration were greater than 0.85. The demographic characteristics of the participants and environmental factors are shown in Table 1. A total of 1,061 children were enrolled in this study, including 586 caries-free children and 475 children with caries. The prevalence of caries was 44.8%. The proportion of female children with caries (51.0%) was higher than that of male children (39.3%) (P value < 0.001). Children who brushed their teeth more than once a day, ate sugary food more than once a week and did not regularly eat at school had higher rates of dental caries (P value < 0.05).
Analysis of risk factors
The variables (demographic characteristics and environmental factors) with P values < 0.05 were included in the binary logistic regression model (forward LR method) to determine the risk factors for caries. Female children had a 59.5% higher chance of having caries than male children (odds ratio [OR] = 1.595, 95% confidence interval (CI) = 1.246–2.042, P value < 0.001). Baikuyao children had a 35.7% lower chance of having caries than Han children (OR = 0.643, 95% CI = 0.471–0.879, P value = 0.006). Children who ate sugary foods more than once a week had a 34.0% higher probability of having caries than children who ate sugary foods at most once a week (OR = 1.340, 95% CI = 1.021–1.760, P value = 0.035) (Table 2).
Genetic analysis
All DNA samples were successfully genotyped. The minor allele frequencies (MAFs) of rs27565 and rs11362 were 0.472 and 0.280, respectively. The genotyped polymorphisms agreed with Hardy-Weinberg equilibrium (HWE) (Table 3).
Both the genotype and allele distributions of PART1 rs27565 and DEFB1 rs11362 were significantly different between caries-free children and children with caries experience (P value < 0.05) (Table 4).
The demographic and environmental risk factors identified above (sex, ethnicity and frequency of sugary food intake) were included as covariates in the binary logistic regression analyses (Enter method) to identify the association between genotypes and alleles of PART1 rs27565 and DEFB1 rs11362 with dental caries (dependent variable: DMFT score = 0 and DMFT score ≥ 1). The results are presented in Table 5. The codominant model showed that carriers of the PART1 rs27565 CC genotype had an approximately 1.7-fold higher risk of caries than TT genotype carriers (OR = 1.692, 95% CI = 1.194–2.397, P value = 0.003), and carriers of the DEFB1 rs11362 TT genotype also had a 1.7-fold higher risk of caries than CC genotype carriers (OR = 1.713, 95% CI = 1.084–2.708, P value = 0.021) (Table 5). Children who carried the C allele of the rs27565 polymorphism had a higher risk of caries in the dominant, additive and allelic models (OR = 1.338, 95% CI = 1.015–1.764, P value = 0.039; OR = 1.296, 95% CI = 1.089–1.543, P value = 0.003; and OR = 1.303, 95% CI = 1.094–1.911, P value = 0.003;, respectively). Children who carried the T allele of the rs11362 polymorphism also had a higher risk of caries in the dominant, additive and allelic models (OR = 1.364, 95% CI = 1.056–1.762, P value = 0.017; OR = 1.306, 95% CI = 1.072–1.590, P value = 0.008; and OR = 1.307, 95% CI = 1.074–1.592, P value = 0.008;, respectively).
The frequency of sugary food intake was a significant environmental risk factor for caries in the present study. A binary logistic regression model (Enter method) was also used to analyse the effects of different combinations of genetic polymorphisms (PART1 rs27565 and DEFB1 rs11362) and the frequency of sugary food intake on caries. After adjusting for sex and ethnicity, carriers of the PART1 rs27565 TC or CC genotype who ate sugary food more than once a week had a 1.6-fold increase in their risk of dental caries compared to TT carriers who ate sugary food at most once a week (OR = 1.579, 95% CI = 1.032–2.414, P value = 0.035). Carriers of the DEFB1 rs11362 CT or TT genotype who ate sugary food more than once a week had a 2.1-fold increased risk of caries than CC carriers who ate sugary food at most once a week (OR = 2.057, 95% CI = 1.438–2.940, P value < 0.001) (Table 6).
Discussion
This study identified the relationship of PART1 and DEFB1 polymorphisms with caries prevalence among 12-year-old children in Nandan County, Guangxi, China. Our results showed that carriers of the PART1 rs27565 C allele and the DEFB1 rs11362 T allele had higher risks of dental caries.
In this study, Baikuyao children had a lower caries prevalence than Han and Zhuang children, potentially due to their lower frequency of sugary food intake. Sugary food intake is an important risk factor for dental caries [2]. Our previous study reported lower parental education levels for Baikuyao children [31]. A lower parental education level often indicates a lower household income [32]. As most of the Baikuyao population could barely afford their household expenses, they might not have much money to buy sugary food for their children [31]. According to the published literature, the relationship between income and added sugar intake is curvilinear, increasing from very low-income to middle-income households, followed by a decrease among children from high-income households [33]. Moreover, most Baikuyao children often eat regularly at school, which may reduce their access to sugar and lead to their lower frequency of sugary food intake [33]. Nevertheless, other factors may be involved in different caries statuses among this population, and we will expand the sample in future studies.
The multivariate analyses indicated that the PART1 rs27565 polymorphism may be a risk factor for caries. PART1 encodes a long noncoding RNA (lncRNA). Transcription factors with immune function, such as nuclear factor interleukin-3 (NFIL3)-regulated, nuclear factor of activated T cells1 (NFATC1) and regulatory factor X1 (RFX1), may bind to PART1 and then be coexpressed, resulting in antibacterial and immune activity [12]. The expression of PART1 in the oral cavity affects the proliferation of oral squamous cell carcinoma [34] and was also speculated to affect dental caries by regulating immunity and oral microbes [12]. We speculated that compared with the rs27565 T allele, the C allele is associated with a lower ability of immune-related transcription factors to bind PART1 and a lower level of coexpressed products. Therefore, individuals with the rs27565 C allele may be more sensitive to plaque accumulation, resulting in an increased risk of caries. Our results were consistent with those reported by Shimizu, who found that carriers of the PART1 rs27565 C allele had a higher risk of caries in the Filipino family [12]. The study by Kelly also revealed that the C allele frequency was higher in the caries group [14]. However, studies explaining the biological mechanism underlying the association between the PART1 rs27565 polymorphism and caries occurrence are insufficient.
The DEFB1 rs11362 T allele was also a risk factor for caries in the current study. DEFB1 rs11362 is a site in the promoter. SNPs in the promoter region may alter transcriptional activity compared with that region in the wild type gene [35]. Therefore, we speculate that the DEFB1 rs11362 T allele may reduce the expression level of β-defensin 1, resulting in lower antibacterial ability and thereby increasing the risk of caries. The studies by Ozturk, Yildiz and Wu supported this hypothesis. They found that carriers of the DEFB1 rs11362 TT genotype had a higher risk of caries [19, 21, 22]. A meta-analysis published in 2020 also showed that individuals with the TT genotype had a seven times higher risk of caries in permanent dentition than individuals with the CC genotype [11]. However, our results are inconsistent with those reported by Krasone and Navarra, who found that CC genotype carriers had a higher risk of caries [23, 24]. Possible explanations for these inconsistencies mainly included differences in genetic heterogeneity. Our results suggested that the DEFB1 rs11362 polymorphism may be a potential biomarker for caries among 12-year-old children in Nandan County, Guangxi, China.
We also found that carriers of the PART1 rs27565 TC + CC genotype and DEFB1 rs11362 CT + TT genotype who ate sweets more than once a week had a higher risk of caries, with ORs of 1.579 and 2.057, respectively. Although this result does not prove the interaction between genes and environmental factors, it suggests a stronger risk effect on caries when the two risk factors are present at the same time.
Our results supported the possibility of DEFB1 rs11362 and PART1 rs27565 polymorphisms as caries risk markers and provided a basis for identifying caries-susceptible populations and developing caries prevention strategies. However, our study has several limitations. First, we did not perform a subgroup analysis stratified by ethnicity because of the small sample size. Second, all independent variables were categorical data. Compared with continuous variable, categorical variables may reduce some available information, and different cut-off points of categorizing continuous data may lead to slightly different results. Third, environmental factors are very complex. However, insufficient environmental factors were included in this study, and thus more of them should be included in future studies.
Conclusion
PART1 rs27565 and DEFB1 rs11362 polymorphisms might be associated with caries in 12-year-old children in Nandan County, Guangxi, China. Carriers of the PART1 rs27565 TC + CC genotype and the DEFB1 rs11362 CT + TT genotype who ate sugary food more than once a week had a higher risk of developing dental caries. PART1 and DEFB1 polymorphisms may thus be potential risk factors for a childhood caries diagnosis. These results must be confirmed in other populations with a larger sample size.
Availability of data and materials
The data and analysis for the current study are available from the corresponding author on reasonable request.
Abbreviations
- DMFT:
-
Decayed, Missing and Filled Teeth
- Chr:
-
chromosome SNP:single-nucleotide polymorphism
- OR:
-
odds ratio
- CI:
-
confidence interval
- MAF:
-
minor allele frequency
- HWE:
-
Hardy-Weinberg equilibrium
References
Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, et al. Oral diseases: a global public health challenge. Lancet. 2019;394(10194):249–60.
Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, et al. Dental caries. Nat reviews Disease primers. 2017;3:17030.
Watt RG, Daly B, Allison P, Macpherson LMD, Venturelli R, Listl S, et al. Ending the neglect of global oral health: time for radical action. Lancet. 2019;394(10194):261–72.
Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789–858.
Schwendicke F, Dorfer CE, Schlattmann P, Foster Page L, Thomson WM, Paris S. Socioeconomic inequality and caries: a systematic review and meta-analysis. J Dent Res. 2015;94(1):10–8.
Rosier BT, De Jager M, Zaura E, Krom BP. Historical and contemporary hypotheses on the development of oral diseases: are we there yet? Front Cell Infect Microbiol. 2014;4:92.
Mosaddad SA, Tahmasebi E, Yazdanian A, Rezvani MB, Seifalian A, Yazdanian M, et al. Oral microbial biofilms: an update. Eur J Clin Microbiol Infect Dis. 2019;38(11):2005–19.
Meyle J, Dommisch H, Groeger S, Giacaman RA, Costalonga M, Herzberg M. The innate host response in caries and periodontitis. J Clin Periodontol. 2017;44(12):1215–25.
Chisini LA, Cademartori MG, Conde MCM, Costa FDS, Tovo-Rodrigues L, Carvalho RV, et al. Genes and SNPs in the pathway of immune response and caries risk: a systematic review and meta-analysis. Biofouling. 2020;36(9):1100–16.
Polesello V, Zupin L, Di Lenarda R, Biasotto M, Ottaviani G, Gobbo M, et al. Impact of DEFB1 gene regulatory polymorphisms on hBD-1 salivary concentration. Archives of oral biology. 2015;60(7):1054–8.
Hatipoglu O, Saydam F. Association between rs11362 polymorphism in the beta-defensin 1 (DEFB1) gene and dental caries: A meta-analysis. J Oral Biosci. 2020;62(3):272–9.
Shimizu T, Deeley K, Briseno-Ruiz J, Faraco IM Jr, Poletta FA, Brancher JA, et al. Fine-mapping of 5q12.1-13.3 unveils new genetic contributors to caries. Caries Res. 2013;47(4):273–83.
Vieira AR, Marazita ML, Goldstein-McHenry T. Genome-wide scan finds suggestive caries loci. J Dent Res. 2008;87(5):435–9.
Kelly AM, Kallistova A, Kuchler EC, Romanos HF, Lips A, Costa MC, et al. Measuring the microscopic structures of human dental enamel can predict caries experience. Journal of personalized medicine. 2020; 10(1).
Weber ML. Examining the relationship between the genotype and redefined phenotype of dental caries. Doctor of Philosophy degree thesis. United States: University of Pittsburgh. 2016.
Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, et al. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67(6):2740–5.
Huttner KM, Bevins CL. Antimicrobial peptides as mediators of epithelial host defense. Pediatr Res. 1999;45(6):785–94.
Jurevic RJ, Chrisman P, Mancl L, Livingston R, Dale BA. Single-nucleotide polymorphisms and haplotype analysis in beta-defensin genes in different ethnic populations. Genet Test. 2002;6(4):261–9.
Wu L, Li Z, Zhou J, Ma B, Yu F, Zheng X, et al. An association analysis for genetic factors for dental caries susceptibility in a cohort of Chinese children. Oral Dis. 2022;28(2):480–94.
Oliveira DSB, Segato RAB, Oliveira S, Dutra ALT, Santos ASD, Praxedes ADN, et al. Association between genetic polymorphisms in DEFB1 and microRNA202 with caries in two groups of Brazilian children. Archives of oral biology. 2018;92:1–7.
Yildiz G, Ermis RB, Calapoglu NS, Celik EU, Turel GY. Gene-environment interactions in the etiology of dental caries. J Dent Res. 2016;95(1):74–9.
Ozturk A, Famili P, Vieira AR. The antimicrobial peptide DEFB1 is associated with caries. J Dent Res. 2010;89(6):631–6.
Krasone K, Lace B, Akota I, Care R, Deeley K, Kuchler EC, et al. Genetic variation in the promoter region of beta-defensin 1 (DEFB1) is associated with high caries experience in children born with cleft lip and palate. Acta Odontol Scand. 2014;72(3):235–40.
Navarra CO, Robino A, Pirastu N, Bevilacqua L, Gasparini P, Di Lenarda R, et al. Caries and innate immunity: DEFB1 gene polymorphisms and caries susceptibility in genetic isolates from north-eastern Italy. Caries Res. 2016;50(6):589–94.
Lips A, Antunes LS, Antunes LA, Abreu JGB, Barreiros D, Oliveira DSB, et al. Genetic polymorphisms in DEFB1 and miRNA202 are involved in salivary human beta-defensin 1 levels and caries experience in Children. Caries Res. 2017;51(3):209–15.
Abbasoğlu Z, Tanboğa İ, Küchler EC, Deeley K, Weber M, Kaspar C, et al. Early childhood caries is associated with genetic variants in enamel formation and immune response genes. Caries Res. 2015;49(1):70–7.
Chapple IL, Bouchard P, Cagetti MG, Campus G, Carra MC, Cocco F, et al. Interaction of lifestyle, behaviour or systemic diseases with dental caries and periodontal diseases: consensus report of group 2 of the joint EFP/ORCA workshop on the boundaries between caries and periodontal diseases. J Clin Periodontol. 2017;44(Suppl 18):39–51.
Chen W, editor. Survey report on oral health and medical service capacity of Guangxi residents. 1 ed. Guangxi: Guangxi Science and Technology Publishing House; 2019.
Petersen PE, Baez RJ, World Health O. Oral health surveys: basic methods. 5th ed. edn. Geneva: World Health Organization; 2013.
Quan JK, Wang XZ, Sun XY, Yuan C, Liu XN, Wang X, et al. Permanent teeth caries status of 12- to 15-year-olds in China: findings from the 4th national oral health survey. Chin J Dent Res. 2018;21(3):181–93.
Ma F, Chen SY, Yu XT, Liu QL, Zeng XJ. Comparison of the status and analysis of the impact factors of caries in children aged 12 years old between Baikuyao and Zhuang as well as Han population in Guangxi [in Chinese]. Chin J Practical Stomatology. 2019;12(12):729–34.
Reiss F. Socioeconomic inequalities and mental health problems in children and adolescents: a systematic review. Soc Sci Med. 2013;90:24–31.
Chi DL, Scott JM. Added sugar and dental caries in children: a scientific update and future steps. Dent Clin North Am. 2019;63(1):17–33.
Yu Q, Du Y, Wang S, Zheng X. LncRNA PART1 promotes cell proliferation and inhibits apoptosis of oral squamous cell carcinoma by blocking EZH2 degradation. J Biochem. 2021;169(6):721–30.
Milanese M, Segat L, Crovella S. Transcriptional effect of DEFB1 gene 5’ untranslated region polymorphisms. Cancer Res. 2007;67(12):5997. author reply.
Acknowledgements
We sincerely appreciate Centers for Disease Control and Prevention of Nandan town of Guangxi Province and the Nandan Department of Education for their help in conducting this study. We also sincerely appreciate the administrators, teachers, and the children for their cooperation.
Funding
The study was funded by the National Natural Science Foundation of China (No. 81660181) and Natural Science Foundation of Guangxi Province (No. 2016GXNSFAA380284) of China. The study was supported by the Department of dental publish health, College of Stomatology, Guangxi Medical University.
Author information
Authors and Affiliations
Contributions
Xiaojuan Zeng conceived of the study and was responsible for training and supervising the project investigators. Shaoyong Chen, Xueting Yu, Qiulin Liu and Fei Ma performed the research and were responsible for the data collection. Fei Ma, Haoyu He and Xiaojuan Zeng were responsible for the data analysis, interpretation of the findings, and manuscript preparation. All authors discussed the results and revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Research Ethics Committee of Guangxi Medical University (date, February 29, 2016; approval number, 20160229-7). All the children assented to participate in this survey on-site, and their guardians provided written informed consent. All methods were conducted in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ma, F., He, H., Chen, S. et al. Associations of PART1 and DEFB1 polymorphisms with Dental Caries in twelve-year-old children in Southern China: a cross-sectional study. BMC Pediatr 23, 6 (2023). https://doi.org/10.1186/s12887-022-03678-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-022-03678-4