Abstract
Background
Epilepsy is a complex disorder caused by various factors, including genetic aberrance. Recent studies have identified an essential role of the sodium channel Nav1.6, encoded by the gene SCN8A, in epileptic encephalopathy.
Case presentation
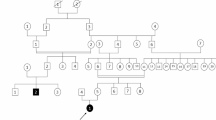
Using parent-offspring trio targeted-exome sequencing, we identified a de novo heterozygous missense mutation c.3953A > G (p.N1318S) in SCN8A in a 3-year-and-9-month Chinese female patient with early infantile epileptic encephalopathy and a normal magnetic resonance imaging of the brain.
Conclusions
This de novo mutation was only detected in the patient but not in her parents. Bioinformatic analysis indicates the pathogenicity of this mutation. Administration of the sodium channel blocker well controlled seizures in the patient. Therefore, we recommend trio targeted-exome sequencing as a routine method for pathogenic variant screening in patients with intractable epilepsy and a normal MRI.
Similar content being viewed by others
Background
Epilepsy is one of most common pediatric neurologic disorders. The prevalence rate is estimated 12/1000 in pediatric patients. One third of them shows pharmacoresistance, and 40% of patients who are younger than 3 years are related to the epileptic encephalopathy [1]. Epileptic encephalopathy (EE) refers to a heterogenous group of epileptic disorders, characterized by intractable seizure, impairment and regression of cognitive and behavioral functions [1, 2]. The causes of EE include structure anomalies, inborn errors of metabolism and genetic insults. The exploration of numerous genetic variants is attributed to wide-spreading applications of next-generation sequencing [3], including ion channel mutations. Voltage-gated sodium channels are responsible for the initiation and propagation of action potentials. Malfunctions of sodium channels are involved in epileptic seizures [4]. Here we reported a patient with infantile EE probably caused by a de novo missense mutation of SCN8A (c.3953A > G, p.N1318S).
Case presentation
The 3-year-and-9-month old girl was born at term with uneventful maternal pregnancy, delivery and family history. Her birth weight and body length were both within normal limits. She had the first afebrile seizure at night sleep presented as generalized tonic-clonic seizure lasting for 1–2 min while she held the normal developemtal milestone of social smiling at her age of 2 months. Topiramate was prescribed, but seizures still happened, until oxcarbazepine monotherapy employed that seizure temporarily resolved for half a year. Later, afebrile seizure flared up and the frequency was around 1–2 fits per month, even several convulsive status epilepticus. She came to our attention 1 year later. Physical and neurologic examinations, including the head circumference, muscle tone and deep tendon reflex, and the laboratory data were all unremarkable. Metabolic survey of amino and organic acids, brain magnetic resonance imaging (MRI) and fluorodeoxyglucose (FDG)-positron emission tomography (PET) demonstrated unremarkable findings (Fig. 1). The test of Wechsler Preschool and Primary Scale of Intelligence (WPPSI-IV) showed mild cognitive impairment (full scale IQ = 66). Interictal electroencephalogram (EEG) revealed normal background activity without obvious epileptiform discharge. Ictal video EEG recorded the seimology of generalized tonic, tonic-clonic seizures, and generalized electrical decrement with superimposed fast activity at EEG ictal onset (Fig. 2). Phenytoin (5 mg/kg/day), valproic acid (20 mg/kg/day), levetiracetam, clonazepam were ever tried, but there was little benefit. Meanwhile, short-term seizures that were provoked by low-grade fever developed then.
Genetic counselling was recommended because her seizures were poorly controlled. High-throughput sequencing of exons of disease-causing genes were performed on the patient and her parents. Genomic DNA extraction and library preparation followed the standard Illumina protocols (Illumina, San Diego, USA) with minor adaptation provided by Joy Orient (Joy Orient Translational Medicine Research Center Co. Ltd., Beijing, China). Agilent Bioanalyzer 2100 (Agilent Technologies, USA) was used for quality control of DNA size distribution and enrichment. Target capturing was performed using Roche (Roche AG., Basel, Switzerland) product customized by Joy Orient, which uses 91,867 probes to capture total 7,465,978 bp of exons regions of 3372 genes that are potentially associated with 4213 known Mendelian genetic diseases. A HiSeq2500 sequencer was used to perform high-throughput sequencing. Exon-enriched DNA was sequenced by the Illumina hiseq2500 platform following the manufacturer’s instructions (Illumina). Raw image files were processed by the BclToFastq (Illumina) for base calling and generating the raw data. The low-quality variations were filtered out using the quality score ≥ 20 (Q20). The sequencing reads were aligned to the NCBI human reference genome version hg19 using BWA. Samtools and Pindel were used to screen SNP and indel of the sequence. All genetic variants were screened by pathogenicity, mode of inheritance and clinical phenotypes (Table 1).
Importantly, a de novo heterozygous mutation c.3953A > G in SCN8A (the coverage of the variant (DP) is 174, allele frequency (AF) = 70/174 = 0.4) was identified and confirmed by Sanger Sequencing (Fig. 3a). The detailed information of this variant is as the followings: SCN8A (NM_014191.4), missense mutation, c.3953A > G (exon22), p.N1318S (de novo), location: chr12, 52,180,336. This missense mutation causes an amino acid substitution of an asparagine residue with a serine residue (p.N1318S) which occurs at a highly conserved LINKER position between the S4 and S5 segments in the third transmembrane domain (DIII) of SCN8A protein (Fig. 3b and c). This mutation site is absent from ClinVar [5] and HGMD Public [6]. It is neither recorded in the Exome Aggregation Consortium (ExAC) [7], which contains exome sequencing data from 60,706 unrelated individuals, nor in our 100 in-house controls. The substitution p.N1318S is predicted to be highly deleterious by bioinformatic tools, which predict possible impact of an amino acid substitution on the structure and function of a human protein, such as Polyphen2 [8] (HumDiv score = 1; HumVar score = 0.998), SIFT [9] (score = 0), Mutation Taster [10] (score = 1) and a comprehensive index CADD phred score (C score = 23.2) [11] (Table 2).
Since the probable causative gene mutation was found, levetiracetam was stopped, but her family reported more seizures. Accordingly, the therapeutic regimen of a sodium channel blocker, lamotrigine (5 mg/kg/day), valproic acid (24 mg/kg/day) and levetiracetam (10 mg/kg/day) were employed, then she had a second temporary seizure-resolved period of 5 months, even under high fever.
Discussion and conclusions
SCN8A is widely expressed in the central and peripheral nerve systems during the neuronal maturation [4, 12]. It is mapped to chromosome 12q13, encoding neuronal voltage-gated sodium channel α 8-subunit Nav1.6, which forms a complex combined with β subunits to modulate current conductance [4, 13, 14]. Nav1.6 consists of four transmembrane domains (DI-DIV), each containing six segments (S1-S6). Four S4 transmembrane segments are responsible for the voltage sense which contain positively charged arginine and histidine residues. There are fast and slow inactivation phases. Fast-inactivation phase is provided by internal DIII-DIV linkers to occlude the ion-conducting pores. Slow-inactivation phase is involved in a collapse of the pore, which is composed of S5-S6 segments of four domains [4].
Missense mutations of SCN8A accounting for 1% of EE are associated with a wide-spectrum phenotype of heterogenous epilepsy, and SCN8A missense mutations are recently recognized to be associated with early infantile epileptic encephalopathy type 13, ref. [15,16,17,18] which displays multiple seizure types, including focal seizures, generalized seizures (tonic, myoclonic, absence) and epileptic spasms. These patients may experience stormy epilepsies [15, 17]. Fever rarely triggers seizures [4, 13, 17]. Severity of psychomotor delay ranges widely after seizure onset [15, 17]. MRI studies are typically normal [15]. There is no clear correlation between phenotypic severity and genetic mutations so far [1, 4]. SCN8A mutations lead to premature channel opening, impaired inactivation and increased persistent current. Stormy seizures are reported resulting from gain-of-function effects [4]. It could explain that some patients respond well to sodium channel blockers in halting seizures [15, 17, 19, 20].
As to our patient, she was categorized as the intermediate phenotype of EE, who manifested mild psychomotor retardation and infrequent seizure episodes, though developing stormy onset of generalized tonic and tonic-clonic seizures concomitant with normal EEG background activity [15, 17, 18]. Fever barely triggered seizures in our patient as previously reported [4, 13, 17]. MRI demonstrated no significant findings. She responds well to the sodium channel blockers. Targeted exome sequencing revealed a heterozygous missense mutation (c.3953A > G, p.N1318S) in SCN8A which is located in the internal S4-S5 linker of the DIII. This mutation could alter the function of the down-stream protein. Since SCN8A (c.3953A > G, p.N1318S) is a de novo missense mutation, it is necessary to further validate its function in the future.
Availability of data and materials
All available data are presented. The mutation information has been submitted to ClinVar, the ClinVar accession SCV000987318.
Abbreviations
- EE:
-
Epileptic encephalopathy
- EEG:
-
Electroencephalogram
- FDG-PET:
-
Fluorodeoxyglucose-positron emission tomography
- MRI:
-
Magnetic resonance imaging
- WPPSI:
-
Wechsler Preschool and Primary Scale of Intelligence
References
Nashabat M, Al Qahtani XS, Almakdob S, Altwaijri W, Ba-Armah DM, Hundallah K, Al Hashem A, Al Tala S, Maddirevula S, Alkuraya FS, et al. The landscape of early infantile epileptic encephalopathy in a consanguineous population. Seizure. 2019;69:154–72.
Hussain SA. Epileptic Encephalopathies. Continuum (Minneap Minn). 2018;24(1, Child Neurology):171–85.
Chambers C, Jansen LA, Dhamija R. Review of commercially available epilepsy genetic panels. J Genet Couns. 2016;25(2):213–7.
Meisler MH, Helman G, Hammer MF, Fureman BE, Gaillard WD, Goldin AL, Hirose S, Ishii A, Kroner BL, Lossin C, et al. SCN8A encephalopathy: research progress and prospects. Epilepsia. 2016;57(7):1027–35.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–5.
Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–2.
Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5.
O'Brien JE, Meisler MH. Sodium channel SCN8A (Nav1.6): properties and de novo mutations in epileptic encephalopathy and intellectual disability. Front Genet. 2013;4:213.
Pons L, Lesca G, Sanlaville D, Chatron N, Labalme A, Manel V, Arzimanoglou A, de Bellescize J, Lion-Francois L. Neonatal tremor episodes and hyperekplexia-like presentation at onset in a child with SCN8A developmental and epileptic encephalopathy. Epileptic Disord. 2018;20(4):289–94.
Calhoun JD, Isom LL. The role of non-pore-forming beta subunits in physiology and pathophysiology of voltage-gated sodium channels. Handb Exp Pharmacol. 2014;221:51–89.
Gardella E, Marini C, Trivisano M, Fitzgerald MP, Alber M, Howell KB, Darra F, Siliquini S, Bolsterli BK, Masnada S, et al. The phenotype of SCN8A developmental and epileptic encephalopathy. Neurology. 2018;91(12):e1112–24.
Vaher U, Noukas M, Nikopensius T, Kals M, Annilo T, Nelis M, Ounap K, Reimand T, Talvik I, Ilves P, et al. De novo SCN8A mutation identified by whole-exome sequencing in a boy with neonatal epileptic encephalopathy, multiple congenital anomalies, and movement disorders. J Child Neurol. 2014;29(12):NP202–6.
Denis J, Villeneuve N, Cacciagli P, Mignon-Ravix C, Lacoste C, Lefranc J, Napuri S, Damaj L, Villega F, Pedespan JM, et al. Clinical study of 19 patients with SCN8A-related epilepsy: two modes of onset regarding EEG and seizures. Epilepsia. 2019;60(5):845–56.
Johannesen KM, Gardella E, Encinas AC, Lehesjoki AE, Linnankivi T, Petersen MB, Lund ICB, Blichfeldt S, Miranda MJ, Pal DK, et al. The spectrum of intermediate SCN8A-related epilepsy. Epilepsia. 2019;60(5):830–44.
Moller RS, Johannesen KM. Precision medicine: SCN8A encephalopathy treated with Sodium Channel blockers. Neurotherapeutics. 2016;13(1):190–1.
Boerma RS, Braun KP, van den Broek MP, van Berkestijn FM, Swinkels ME, Hagebeuk EO, Lindhout D, van Kempen M, Boon M, Nicolai J, et al. Remarkable phenytoin sensitivity in 4 children with SCN8A-related epilepsy: a molecular Neuropharmacological approach. Neurotherapeutics. 2016;13(1):192–7.
Acknowledgments
We thank Bing Zhu from Dongfang Affliated Hospital of Xiamen University for the constructive discussion. We also thank Xu Ke, Yang Shuanghao, Quan Xiaofang, Lin cuiping and Chen zhiqiang from Chigene Translational Medical Research Center Co. Ltd. for their constructive discussion.
Funding
The study was supported in part by National Natural Science Foundation of China (81802823 to Y.Z., 81571176, 31871077 and 81822014 to X.W.,), Natural Science Foundation of Fujian Province of China (2018 J01054 to Y.Z., 2017 J06021 to X.W.), the Education and Research Foundation for Young Scholars of Education Department of Fujian Province, China (JAT170004 to Y.Z.), the Nanjing Military funding (No. ZD26 to YY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
YY, XW and YZ conceived and designed the study. KML, GS and YZ wrote the manuscript. YY, FW, KML and XW supervised and made corrections. YY, FW, XZ, JR, YW and XW contributed to the discussion of the results, revisions and approval of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Ethics approval was obtained from the ethics committee of Xiamen Humanity Hospital. The written consent to participate in the genetic testing was obtained from both the patient (the parents signed the form on behalf of their child) and the parents. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Consent for publication
Written informed consent was obtained from the patient’s parents for publication of this Case Report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lin, KM., Su, G., Wang, F. et al. A de novo SCN8A heterozygous mutation in a child with epileptic encephalopathy: a case report. BMC Pediatr 19, 400 (2019). https://doi.org/10.1186/s12887-019-1796-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-019-1796-9