Abstract
Purpose
Osteosarcoma (OS) is a primary bone tumor with high malignancy and poor prognosis. Ferroptosis plays a crucial role in OS. This study aimed to evaluate the effects of Ankyrin 1 (ANK1) on OS and to investigate its specific mechanisms.
Methods
Microarray datasets related to “osteosarcoma” were selected for this study. Relevant hub genes in OS were identified through bioinformatics analysis. Transfected U-2OS and MG-63 cells were used for in vitro experiments. The effects of ANK1 overexpression on cell viability, migration, and invasion were determined through CCK-8, wound healing, and transwell assays. An OS mouse model was established for the in vivo experiments. Hematoxylin-eosin staining and immunohistochemistry were conducted to observe the histological effects of ANK1 overexpression on mouse tumors. TUNEL staining was performed to evaluate apoptosis in mouse.
Results
There were 159 common differentially expressed genes in the GSE16088 and GSE19276 datasets. The hub genes ANK1, AHSP, GYPB, GYPA, KEL, and ALAS2 were identified. Pan-cancer analysis verified that ANK1 was closely associated with cancer prognosis and immune infiltration. Furthermore, ANK1 overexpression inhibited the proliferation, migration, and invasion of OS cells and promoted ferroptosis, while ferroptosis inhibitor (fer-1) weakened these effects. Moreover, ANK1 overexpression suppressed tumor growth, promoted apoptosis, reduced the number of Ki67 positive cells, and elevated the number of caspase-3 positive cells in vivo.
Conclusions
ANK1 is a prognosis biomarker of OS that can alleviate the progression of OS by promoting ferroptosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor in children and adolescents [1, 2]. It has a tendency for local invasion and metastasis [1, 3] and a high risk of recurrence. The typical symptoms of OS are local pain, swelling, and limited joint activity [4]. Currently, the primary treatments include tumor resection and postoperative multi-drug chemotherapy [2]. Although the progression of OS has greatly improved with continuous improvements at the medical level, the prognosis of OS remains unsatisfactory [1]. Therefore, it is necessary to identify effective strategies for OS treatment.
Ankyrin 1 (ANK1), a member of the ankyrin family, is expressed in various biological systems and is a membrane-associated cytoskeletal protein [5]. The methylation of RHBDF2, ANK1, BIN1, and other loci may play a role in the onset of Alzheimer’s disease [6]. ANK1 methylation regulates microRNA-486-5p expression, which distinguishes lung tumors based on smoking history and histology [7]. Importantly, co-mutations of ANK1 and IFITM2 are detected in all patients with multiple myeloma, and the interaction between malignant plasma cells and the microenvironment during clonal evolution is well understood [8]. However, studies on the mechanism of action of ANK1 in OS are scarce.
Ferroptosis is an iron-dependent cell death process with complex mechanisms mainly involving iron, amino acid, and lipid metabolism [9, 10]. It is of vital importance in many diseases, particularly in the progression of tumors such as colorectal cancer, gastric cancer, and triple-negative breast cancer [11,12,13,14]. The induction of ferroptosis has become an effective therapeutic target for inhibiting tumor development [14]. The inhibition of ferroptosis inhibitory protein 1 promotes ferroptosis and plays an anti-tumor role in liver cancer [15]. Apolipoprotein L3 enhances anti-tumor immunity in colorectal cancer by promoting LDHA-mediated ferroptosis [16]. In addition, recent studies have shown that ferroptosis has great therapeutic potential for OS treatment [17]. Differentiation therapy promotes ferroptosis and amplifies apoptosis, effectively combating OS and inhibiting its progression [18]. In OS, gambogenic acid, a bioactive ingredient isolated from gamboge, induces oxidative stress that causes ferroptosis and apoptosis in OS cells through the P53/SLC7A11/GPX4 axis while slowing tumor growth [19]. Nonetheless, the role of ANK1 in ferroptosis remains elusive.
In our study, bioinformatics analysis was used to investigate the hub genes involved in OS, and pan-cancer analysis of the key gene ANK1 was conducted. Subsequently, an OS mouse model was constructed to evaluate the effect of ANK1 on tumor growth. The role of ANK1 in the proliferation, migration, and invasion of OS cells was explored in vitro. In addition, owing to the importance of ferroptosis, the effect of ANK1 on this process was investigated, offering a new potential option for the treatment of OS.
Materials and methods
Screening of differentially expressed genes (DEGs)
The gene expression profiles analyzed in this study were downloaded from the GEO database of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/). Two microarray datasets related to “osteosarcoma,” GSE16088 and GSE19276, were selected and analyzed using GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r). The GSE16088 and GSE19276 datasets were analyzed to identify DEGs (Tumor vs. Control) according to p < 0.05 and |logFC|≥1.25. The DEGs were visualized using volcano plots, and the datasets were normalized and corrected utilizing box plots. Pathway enrichment analysis of DEGs was performed employing GSEA. In addition, a Venn diagram of the DEGs in the two datasets and a heat map of the common DEGs in the datasets were produced.
Functional analysis of common DEGs
Gene Ontology (GO) analysis (molecular function, MF, biological process, BP, and cell component, CC) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed through annotation, visualization, and integration of the screened overlapping DEGs using DAVID (https://david.ncifcrf.gov/summary.jsp), and GO-KEGG data were obtained. Statistical significance was set at p < 0.05. GO and KEGG enrichment analysis of DEGs were analyzed with R language package, and the p-value minimum was selected to display the most significant enrichment. Histograms and bubble diagrams of the enrichment analyses were generated.
Protein-protein interaction (PPI) network
STRING (https://www.string-db.org/) database was used to construct the PPI network, which was employed to analyze common DEGs and predict interactions between proteins encoded by genes that may be involved in OS. Regarding the significance criteria, the confidence score was set to 0.4. Subsequently, the PPI network was visualized using Cytoscape (www.cytoscape.org/), and the hub gene module was identified from the PPI network of common DEGs with MCODE. Target genes were screened comprehensively.
Analysis of hub genes
Enrichment chord and ridgeline diagrams of the hub genes were drawn using R. The expression of hub genes in the original sample data was selected as a variable for principal component analysis. After processing in R, two principal component variables, PC1 and PC2, were obtained. The correlation between genes in the expression profile matrix was visualized using a matrix heat map. A receiver operating characteristic (ROC) curve was drawn employing gene expression profile interaction analysis (http://gepia.cancer-pku.cn/), and the diagnostic accuracy of the selected genes was evaluated using the corresponding area under the ROC curve.
Analysis of target gene in pan-cancer
Sangerbox (http://sangerbox.com/home.html) was utilized to analyze the differences in the target genes in pan-cancer. The standardized pan-cancer dataset was downloaded from UCSC (https://xenabrowser.net/), and samples with expression levels > 0 were retained. Log2 (x + 0.001) transformation was performed on the expression values to screen for cancer species with more than three samples in a single cancer species. R language was used to calculate the differences in expression between normal and tumor samples in each tumor.
TIMER (https://cistrome.shinyapps.io/timer/) was employed to re-evaluate the B cells, neutrophils, macrophages, T cell CD8+, T cell CD4+, and DC infiltration scores of each patient for each tumor according to gene expression. R language was used to analyze the relationship between gene expression and prognosis in each tumor, and the log-rank test was performed to obtain prognostic significance.
Cell culture and transfection
The human osteoblast cell line hFOB1.19 and human OS cell lines MG-63 and U-2OS were purchased from the National Collection of Authenticated Cell Cultures (Shanghai, China). hFOB1.19, MG-63, and U-2OS cells were grown in DMEM containing 1% penicillin-streptomycin (Beyotime, Shanghai, China) and 10% fetal bovine serum (Biosera, France). All cells were grown in an incubator with 5% CO2 and 95% humidity at 37 °C.
MG-63 and U-2OS cells were divided into Control, pcDNA-NC (transfected with the empty vector pcDNA3.1-NC), and pcDNA-ANK1 (transfected with pcDNA3.1-ANK1 to overexpress ANK1) groups. The vectors (600 ng/µL) were transfected into the cells using Lipo6000™ (Invitrogen, USA). Additionally, MG-63 and U-2OS cells were treated with the ferroptosis inhibitor ferrostatin-1 (Fer-1, 2 µM) for 48 h.
Real-time fluorescence quantitative PCR (RT-qPCR)
The TRIZOL reagent (Invitrogen) was used to extract total RNA. RNA sample (5 µL) was extracted and diluted with RNase free water, and the concentration of RNA and the absorption values at 260 nm and 280 nm were measured through a UV spectrophotometer. Hiscript II QRT Supermix (Vazyme, Nanjing, China) was employed to synthesize cDNA). Subsequently, RT-qPCR was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) with an ABI7500 quantitative PCR (Applied Biosystems, Inc., USA) under the following conditions: pre-denaturation at 95 °C for 30 s, denaturation at 95 °C for 10 s, and annealing at 60 °C for 30 s for 40 cycles; GAPDH was used as an internal reference, and this experiment was repeated 3 times. The primer sequences are listed in Table 1.
Cell counting kit-8 (CCK-8) assay
MG-63 and U-2OS cells (1.2 × 104/well) were inoculated in 96-well plates and incubated for 24 h. Subsequently, the proliferation of MG-63 and U-2OS cells was detected through the CCK-8 assay. After incubation with 10% CCK-8 solution in the dark for 2 h at 37 °C, the optical density (OD) value was measured in each well every 1 h. Optimal incubation time was determined using OD. The OD of the samples was measured at 450 nm using a microplate reader.
Wound healing assay
Migration of MG-63 and U-2OS cells was assessed using a wound healing assay. Transfected cells (1 × 105/well) were seeded in 6-well plates and cultured until they reached 90% confluence. Subsequently, a wound gap was created using a pipette tip. Cells were then washed with PBS to remove cell debris and were continuously cultured for 24 h at 37℃, 5% CO2. The wound distance was photographed and measured under a microscope (Olympus, Tokyo, Japan) at 0 and 24 h post-wounding. ImageJ software was used to analyze the migration ability.
Transwell assay
The invasion of MG-63 and U-2OS cells was detected utilizing a transwell assay. Briefly, 100 µL transfected cells (1 × 105/mL) were plated into the upper chamber of 24-well plate, and DMEM (containing 20% FBS) was filled in the lower chamber. After incubation for 24 h, the cells were fixed in paraformaldehyde (4%) for 30 min and stained with crystal violet (0.1%) for 20 min. The stained cells were washed with PBS and photographed under a microscope (Olympus, Japan).
Western blotting (WB)
MG-63 and U-2OS cells were subjected to RIPA to isolate the total protein samples. Proteins were separated on a 10% SDS-PAGE gel and transferred onto a PVDF membrane. Subsequently, the membrane was sealed in skimmed milk (5%) and then incubated with primary antibodies at 4 °C for a night, including anti-ANK1 (PA5-114853, 1:1000, Thermo Fisher scientific, Waltham, MA, USA), anti-Caspase-3 (ab32351, 1:1000, Abcam, Shanghai, China), anti-PTGS2 (ab179800, 1:1000, Abcam), anti-GPX4 (ab125066, 1:1000, Abcam), anti-FTH (ab183781, 1:1000, Abcam), and anti-GAPDH (ab9485, 1:1000, Abcam). Subsequently, the cells were incubated with a goat anti-rabbit antibody (1:5000, Abcam) for 1 h at 20℃-25℃. Finally, the membrane was placed on a Tanon 5200 Chemiluminescent Imaging System incubation plate, and ECL solution was added for development. The results were calculated using the ImageJ software.
Enzyme linked immunosorbent assay (ELISA)
The levels of MDA, GSH, and Fe in transfected OS cells were evaluated using the corresponding ELISA kit according to the manufacturer’s instructions (Esebio, Shanghai, China). Total 50 µL of diluted samples and standards were added to the wells of the microtiter plate. Subsequently, 100 µL of HRP-labeled antibody was added to each well, and the plates were incubated for 60 min at 37℃. The OD of each well was determined at 450 nm within 15 min. The sample levels were calculated using a standard curve.
Establishment of OS mouse model
Twelve female BALB/c nude mice (6–8 weeks old, 18–20 g) were purchased from SiPeiFu Biotechnology Co., Ltd (Beijing, China) and randomly assigned to the oe-NC and oe-ANK1 groups (n = 6). Mice were subcutaneously inoculated with 0.1 mL of transfected MG-63 cells (1 × 107/mL), individually. After 4 weeks, the mice were euthanized by isoflurane inhalation followed by cervical dislocation. Subsequently, the tumor tissues were obtained and weighed. The tumor volume was measured weekly and calculated as (length × width2)/2. All animal experiments were approved by Scientific Research Ethics Committee of the First Affiliated Hospital of Gannan Medical University (No. LLSC-2024-236). Humanitarian endpoint: Tumor weight did not exceed 10% of the body weight of the mice.
Hematoxylin-eosin (HE) staining
HE staining was performed to observe pathological changes in the tumor tissues of the mice. The tissues were fixed in 4% paraformaldehyde solution and dehydrated using an ethanol gradient. The tissues were embedded in paraffin and cut into 4 μm thick slices. The paraffin slices were dewaxed in xylene and ethanol (Sigma, USA). The slices were then stained with hematoxylin and eosin. Finally, the slices were dehydrated and sealed with neutral gum. The staining results were observed under an optical microscope.
TUNEL staining
The TUNEL apoptosis assay kit (Beyotime, Shanghai, China) was used to assess apoptosis. The paraffin sections were deparaffinized in xylene and washed with PBS. Sections were then incubated with proteinase K at 37 °C for 30 min. The sections were washed and incubated with H2O2 at 20℃-25℃ for 10 min. The TUNEL reagents were supplemented and incubated at 37 °C in dark for 60 min. Finally, DAB was applied for development. The sections were stained with hematoxylin and sealed. A fluorescence microscope (Olympus) was used to observe apoptotic cells.
Immunohistochemistry (IHC)
The paraffin-embedded sections were dewaxed, rehydrated, and then incubated with anti-Ki67 (ab15580, 1:400, Abcam) and anti-caspase-3 (ab184787, 1:400, Abcam) antibodies overnight at 4 °C. The slices were then incubated with HRP-labeled secondary antibody at 37 °C for 1 h. They were then stained with DAB and hematoxylin and photographed under a microscope.
Statistical analysis
All data were processed using GraphPad Prism 9.0 statistical software and are expressed as means ± SD. Comparisons between multiple groups were performed using a one-way ANOVA and Tukey’s test. A t-test was applied for comparisons between the two groups. Statistical significance was set at p < 0.05.
Results
Screening of DEGs
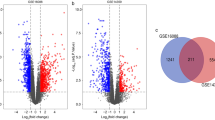
A total of 4271 DEGs were screened from GSE16088, which included six normal samples (Control) and 14 OS samples (Tumor). There were 606 DEGs in GSE19276, including five normal samples (Control) and 44 OS samples (Tumor). The DEGs in the two datasets were analyzed, and a volcano map of the DEGs was obtained (Fig. 1A-B). After GEO2R analysis, the data correction results of 20 samples from the GSE16088 dataset and 49 samples from the GSE19276 dataset were obtained. The standardization and cross-comparability of the data were evaluated, showing that most of the selected samples were centered, and the numerical distribution met the standard, indicating cross-comparability in the microarray data (Fig. S1). The top three pathways with the most significant p values were selected for display (Fig. 1C-D). The top 15 DEGs with the smallest p-values were selected (Tables S1 and S2) and visualized using R language (Fig. 1E-F). Finally, 159 common DEGs were identified in the GSE10688 and GSE19276 dataset (Fig. S2).
A, B. Volcano map of differentially expressed genes (DEGs) in GSE10688 and GSE19276 dataset. The abscissa is log2FoldChange, and the ordinate is -log10(p-value). The red dot indicates the up-regulated gene in this group, and the blue dot indicates the down-regulated gene in this group. C, D. GSEA enrichment analysis. The related pathway enrichment analysis was performed using the total differential genes in the GSE16088 and GSE19276 datasets, respectively. E, F. Heat map of DEGs in GSE10688 and GSE19276 dataset. Genes are represented horizontally, and each column is a sample. Red indicates high expression genes, and blue indicates low expression genes
GO and KEGG enrichment analysis
GO and KEGG functional analyses were performed on 159 common DEGs. The top five GO terms with the smallest p-values in each GO classification were selected for display (Table S3). The DEGs were mainly enriched in response to axon injury and hypoxia in BP, extracellular exosomes and specific granule lumen in CC, and protein binding and calcium ion binding in MF (Fig. 2A-B).
Functional enrichment analysis of common DEGs. A. Histogram of GO enrichment analysis. The abscissa is GO term, and the ordinate is the number of genes enriched for each term. B. Bubble diagram of GO enrichment analysis. The color depth of the node represents the corrected p-value, and the size of the node refers to the number of genes involved. C. Histogram of KEGG enrichment analysis. D. Bubble diagram of KEGG enrichment analysis
KEGG enrichment was assessed using -log10 (p-value), count value, and the number of genes enriched in each term. Based on the change in the p-value, the 10 most significantly enriched KEGG terms were selected for display (Table S4). The results showed that DEGs were mainly enriched in small cell lung cancer, cancer pathways, and amino acid biosynthesis (Fig. 2C-D).
PPI network
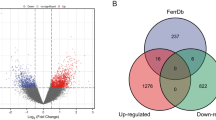
A PPI network based on DEGs was constructed using STRING, and a PPI network panorama was obtained (Fig. S3A). The Cytoscape software was used to visualize the PPI network model (Fig. S3B), and MCODE was used to identify a highly interconnected cluster as a potential functional molecular complex of OS, including ANK1, OSBP2, AHSP, GYPB, CA1, GYPA, KEL, ALAS2, RHAG (Fig. S3C).
Analysis of hub genes
Based on the gene expression in the original data samples of GSE16088, an enrichment string diagram was drawn using R language. The results showed that the DEGs were mainly enriched in protein binding, plasma membrane, and membrane biological processes (Fig. 3A). The ridgeline map was drawn using the R language, and the peak represented the concentration of the data distribution (Fig. 3B). Based on the expression of the selected genes in the original sample data of GSE16088, principal component analysis was performed as a variable. After processing with the R language function, PC1 and PC2 were obtained. These components provide a 95% explanation rate and can be used to distinguish between control and OS samples (Fig. 3C). Based on expression profile data in GSE16088, a matrix heat map was constructed for visualization (Fig. 3D).
Analysis of hub genes. A. String diagram. LogFC is the fold change of the gene, which is used for sorting and gene color. The remaining columns are signaling pathways, and different links of the gene indicate whether the gene is in the pathway. B. Ridgeline map. The abscissa is the amount of gene expression, the shape of the peak represents the dispersion between a set of data, and its height is the number of samples corresponding to the amount of gene expression. C. Principal component analysis of hub genes. The coordinate axes PC1 and PC2 are the first and second principal components (i.e., the explanatory rate of potential variables to the difference). Points represent samples, and different colors represent different groups. D. Matrix correlation analysis. Matrix analysis is used to perfectly display the correlation of genes between matrices and visualize them
ROC curve
After a literature review and screening, ANK1, AHSP, GYPB, GYPA, KEL, and ALAS2 were selected as hub genes for further research, and the key gene, ANK1, was chosen as the research target gene. To verify the classification effect of ANK1 expression between OS samples and healthy controls, the ROC curves of ANK1, AHSP, GYPB, GYPA, KEL, and ALAS2 were drawn using the original sample data of GSE16088 with the R language (Fig. S4). The results showed that the false-positive rate of ANK1 was 1.1%, while the true-positive rate was 98.9%, demonstrating that ANK1 is an excellent indicator for distinguishing OS patients from healthy controls.
Pan-cancer analysis of ANK1
ANK1 was significantly upregulated in 14 tumors and downregulated in 12 tumors (Fig. S5A). ANK1 expression in 44 cancers and the overall survival rates of the corresponding samples were obtained. Finally, according to the log-rank test, high expression in two tumor types was associated with poor prognosis, and low expression in four tumor types was linked to poor prognosis (Fig. S5B). The B cell, neutrophil, macrophage, T cell CD8+, T cell CD4+, and DC infiltration scores of each patient for each tumor were evaluated using TIMER based on gene expression. In addition, ANK1 expression was significantly associated with immune infiltration in 35 tumors (Fig. S5C).
Validation of hub genes via RT-qPCR
RT-qPCR showed that compared with hFOB1.19, the expression of ANK1, AHSP, GYPB, GYPA, KEL, and ALAS2 in human OS cells (MG-63 and U-2OS) was significantly reduced (Fig. 4).
Overexpression of ANK1 inhibits the proliferation, migration, and invasion of OS cells
In OS cells, the expression of ANK1 in the pcDNA-ANK1 group was significantly higher than that in the pcDNA-NC group (Fig. 5A). In addition, CCK-8, wound healing, and transwell assays verified that ANK1 overexpression suppressed the proliferation, migration, and invasion of OS cells (Fig. 5B-D). The protein expression of ANK1 and Caspase-3 was detected through WB, indicating that compared with the pcDNA-NC group, overexpression of ANK1 significantly increased the Caspase-3 level in OS cells, demonstrating that ANK1 promoted cell apoptosis (Fig. 5E).
A. RT-qPCR detection of transfection rate. B. CCK8 assay was used to detect cell viability. C. Wound healing assay was used to detect cell migration ability. D. Transwell assay was used to detect cell invasion. E. Western blot was performed to evaluate the levels of ANK1 and Caspase-3. **p < 0.01 ***p < 0.001 vs. pcDNA-NC (n = 3)
Overexpression of ANK1 promotes ferroptosis in OS cells
WB was performed to detect the expression of ferroptosis-related proteins in MG-63 and U-2OS cells. In OS cells, compared to the pcDNA-NC group, the levels of PTGS2 and FTH in the pcDNA-ANK1 group were significantly increased, while GPX4 was reduced (Fig. 6A). The MDA, GSH, and Fe contents were detected using ELISA. Furthermore, compared to the pcDNA-NC group, ANK1 overexpression significantly elevated the levels of MDA and Fe, whereas GSH was decreased (Fig. 6B). Thus, overexpression of ANK1 promoted ferroptosis in OS cells.
Overexpression of ANK1 suppresses tumor growth and promotes ferroptosis in OS
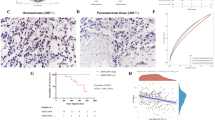
Overexpression of ANK1 retarded tumor growth and significantly reduced tumor volume and weight in OS mice (Fig. 7A-D). HE staining revealed a dense tumor tissue structure in the oe-NC group; conversely, the tissue structure in the oe-ANK1 group was more loosely arranged (Fig. 7E). TUNEL staining demonstrated that ANK1 overexpression significantly promoted the apoptosis of OS cells (Fig. 7F). IHC verified that in comparison with the oe-NC group, oe-ANK1 significantly reduced the number of Ki67 positive cells and increased the number of caspase-3 positive cells in OS (Fig. 7G-H). Furthermore, WB was performed to evaluate the levels of ANK1 and ferroptosis-related proteins, which showed that compared to the oe-NC group, ANK1 overexpression significantly increased the levels of PTGS2 while inhibiting FTH and GPX4 (Fig. 7I).
The effect of ANK1 on tumor. A. The weight change of mice. B. Tumor weight of mice. C. Tumor volume of mice. D. Tumor map of mice. E. HE staining was used to observe the changes of tissue morphology. F. TUNEL staining was executed to observe apoptosis level. G. Immunohistochemistry was used to detect the expression of Ki67. H. The expression of caspase-3 was detected by immunohistochemistry. I. Western blot was performed to detect the expression of ferroptosis-related proteins PTGS2, FTH and GPX4 in vivo. **p < 0.01 ***p < 0.001 vs. oe-NC (n = 6)
To further explore the role of ferroptosis in OS, a ferroptosis inhibitor (Fer-1) was applied to OS cells. In OS, ANK1 overexpression significantly suppressed cell viability, migration, and invasion, whereas fer-1 weakened these effects (Fig. 8A-C). Furthermore, fer-1 also significantly attenuated the stimulatory effects of ANK1 on ferroptosis (Fig. 8D).
Fer-1 attenuated the inhibitory effect of ANK1 on OS. A. CCK8 assay was used to detect cell viability. B. Wound healing assay was used to detect cell migration ability. C. Transwell assay was used to detect cell invasion. D. Western blot was performed to evaluate the levels of ferroptosis-related proteins PTGS2, FTH and GPX4. ***p < 0.001 vs. pcDNA-NC, ###p < 0.001 vs. pcDNA-ANK1 (n = 3)
In summary, ANK1 overexpression promoted ferroptosis and significantly inhibited OS progression.
Discussion
OS is the most common malignant bone tumor with a low survival rate [20]. Therefore, it is necessary to identify more effective therapeutic targets for OS to improve the survival rate. In this study, the hub gene ANK1 was identified through bioinformatics analysis, and pan-cancer analysis was performed. ANK1 was found to exhibit a correlation with the prognosis of various cancers. Furthermore, through in vitro and in vivo experiments, we found that ANK1 inhibited the proliferation, migration, and invasion of OS cells; suppressed tumor growth; and promoted ferroptosis in OS.
With the advent of big data, the use of bioinformatics technology to analyze the mechanism of disease in medical research has become increasingly prevalent. Identifying disease-related marker genes provides a direction for targeted disease therapy. The circular RNA circ_001422 was identified using bioinformatics analysis and high-throughput sequencing, revealing that it could interact with miR-195-5p and promote OS progression through the FGF2-PI3K-AKT axis [21]. Chen et al. selected microarray datasets to identify hub genes and found that myofascial fibrosarcoma oncogene homolog B is a key transcriptional regulator in OS [22]. Shi et al. explored hub genes and pathways of OS with lung metastasis using microarray datasets to identify potential therapeutic targets [23]. Herein, DEGs in the microarray datasets were screened through bioinformatics analysis, and hub genes closely related to OS, ANK1, AHSP, GYPB, GYPA, KEL, and ALAS2, which may play a role in OS progression, were identified using the PPI network. AHSP, alpha-hemoglobin-stabilizing protein, is a red blood cell-specific chaperone protein that can be used to stain and identify erythroid precursors in bone marrow biopsies of leukemia, metastatic cancer, and other diseases [24, 25]. GYPA and GYPB belong to a small gene family on the chromosome and are responsible for encoding the glycoproteins GPA and GPB on the erythrocyte membrane [26]. Wang et al. identified that HBG1, SNCA, and GYBP are significantly associated with atrial fibrillation, are involved in the occurrence of atrial fibrillation complicated by stroke, and can be used as early therapeutic targets [27]. KEL is an oncogenic gene that plays a role in the immune escape of tumor cells, thus expanding our understanding of the pathogenesis of acute erythroleukaemia [28]. ALAS1 is a rate-limiting enzyme in haeme biosynthesis [29], and haeme isolation inhibits the development of lung adenocarcinoma and squamous cell carcinoma [30]. ANK1 is associated with malignant progression of Down syndrome in children and acute myeloid leukemia in older patients [31, 32]. Herein, ANK1 was identified as a hub gene for OS. Meanwhile, through pan-cancer analysis, we found that ANK1 was related to various cancers and closely correlated with immune infiltration in 35 tumors. Thus, ANK1 appears to participate in OS progression. Namløs et al. identified that miRr-486-5p, encoded in the last intron of the ANK1 gene, highly correlates with cancers and is regulated by DNA methylation in OS [33]. Another study reported that nine representative genes, including ANnK1, associated with OS (screened through random forest and artificial neural networks) can serve as new targets for the early diagnosis and effective treatment of OS [34]. In this study, in vivo and in vitro assays demonstrated that ANK1 was downregulated in OS and that overexpression of ANK1 inhibited the proliferation, migration, and invasion of OS cells and suppressed tumor growth. Hence, ANK1 plays a pivotal role in alleviating OS progression.
Reportedly, ferroptosis is of vital significance in OS. FANCD2 silencing can suppress the viability, invasion, migration, and tumor growth of OS cells by regulating the JAK2/STAT3 axis and induce ferroptosis [35]. Good results have been achieved in the exploration of drugs that target ferroptosis to alleviate OS. Baicalin promotes ferroptosis and exerts anti-OS activity via the Nrf2/xCT/GPX4 regulatory axis [36]. β-Phenethyl Isothiocyanate induces oxidative stress to trigger ferroptosis in OS cells, retarding tumor growth in OS [37]. Zoledronic acid promotes ferroptosis by reducing ubiquinone and promoting HMOX1 in OS cells while inhibiting their growth [38]. Similar to the above studies, herein, overexpression of ANK1 significantly suppressed the proliferation, migration, and invasion of OS cells and promoted ferroptosis, while the ferroptosis inhibitor (fer-1) weakened these effects, demonstrating that ANK1 can alleviate OS progression by accelerating ferroptosis.
However, this study had some limitations. We did not explore the effect of fer-1 on ANK1 overexpression in OS mice, which should be explored further in the future. Furthermore, to prove the specificity of ANK1 overexpression-induced antitumor effects, we will conduct in vitro assays in the future to show that ANK1 inhibition can result in increased proliferation, migration, and invasion of OS cells.
Conclusion
In this study, the hub genes ANK1, AHSP, GYPB, GYPA, KEL, and ALAS2 were identified using bioinformatics analysis. ANK1 was proved to be associated with prognosis and immune cell infiltration. Overexpression of ANK1 can promote ferroptosis and alleviate OS progression, offering a novel prognostic biomarker and therapeutic option for OS.
Data availability
The datasets GSE16088 and GSE19276 in the study can be downloaded from GEO of NCBI (https://www.ncbi.nlm.nih.gov/geo/).
References
Chen C, et al. Immunotherapy for osteosarcoma: fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021;500:1–10.
Ritter J, S. Bielack S. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–5.
Corre I et al. The Osteosarcoma Microenvironment: a Complex but Targetable Ecosystem. Cells. 2020. 9(4).
Wittig JC, et al. Osteosarcoma: a multidisciplinary approach to diagnosis and treatment. Am Fam Physician. 2002;65(6):1123–32.
Bennett V, Chen L. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr Opin Cell Biol. 2001;13(1):61–7.
De Jager PL, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17(9):1156–63.
Tessema M, et al. ANK1 methylation regulates expression of MicroRNA-486-5p and discriminates lung tumors by histology and smoking status. Cancer Lett. 2017;410:191–200.
Liang Y, et al. Malignant clonal evolution drives multiple myeloma cellular ecological diversity and microenvironment reprogramming. Mol Cancer. 2022;21(1):182.
Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73(11–12):2195–209.
Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–82.
Yang F, et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023;35(1):84–e1008.
Li D, et al. CST1 inhibits ferroptosis and promotes gastric cancer metastasis by regulating GPX4 protein stability via OTUB1. Oncogene. 2023;42(2):83–98.
Wei R, et al. Tagitinin C induces ferroptosis through PERK-Nrf2-HO-1 signaling pathway in colorectal cancer cells. Int J Biol Sci. 2021;17(11):2703–17.
Gong C, et al. Ferroptosis in tumor immunity and therapy. J Cell Mol Med. 2022;26(22):5565–79.
Cheu JW, et al. Ferroptosis Suppressor Protein 1 inhibition promotes Tumor Ferroptosis and Anti-tumor Immune responses in Liver Cancer. Cell Mol Gastroenterol Hepatol. 2023;16(1):133–59.
Lv Y, et al. Apolipoprotein L3 enhances CD8 + T cell antitumor immunity of colorectal cancer by promoting LDHA-mediated ferroptosis. Int J Biol Sci. 2023;19(4):1284–98.
Liu X, et al. Ferroptosis in osteosarcoma: a promising future. Front Oncol. 2022;12:1031779.
He C et al. Amplified ferroptosis and apoptosis facilitated by differentiation therapy efficiently suppress the progression of Osteosarcoma. Small. 2023. p. e2302575.
Liu Z, et al. Gambogenic acid induces cell death in human osteosarcoma through altering iron metabolism, disturbing the redox balance, and activating the P53 signaling pathway. Chem Biol Interact. 2023;382:110602.
Shoaib Z, Fan TM, Irudayaraj JMK. Osteosarcoma mechanobiology and therapeutic targets. Br J Pharmacol. 2022;179(2):201–17.
Yang B, et al. Circular RNA circ_001422 promotes the progression and metastasis of osteosarcoma via the miR-195-5p/FGF2/PI3K/Akt axis. J Exp Clin Cancer Res. 2021;40(1):235.
Chen Y, et al. MAFB promotes Cancer Stemness and Tumorigenesis in Osteosarcoma through a Sox9-Mediated positive Feedback Loop. Cancer Res. 2020;80(12):2472–83.
Shi Z, et al. Exploring the key genes and pathways of osteosarcoma with pulmonary metastasis using a gene expression microarray. Mol Med Rep. 2017;16(5):7423–31.
Kihm AJ, et al. An abundant erythroid protein that stabilizes free alpha-haemoglobin. Nature. 2002;417(6890):758–63.
Raess PW, et al. α-Hemoglobin-stabilizing protein is a sensitive and specific marker of erythroid precursors. Am J Surg Pathol. 2012;36(10):1538–47.
Rahuel C, Elouet JF, Cartron JP. Post-transcriptional regulation of the cell surface expression of glycophorins a, B, and E. J Biol Chem. 1994;269(52):32752–8.
Wang X, et al. Joint efficacy of the three biomarkers SNCA, GYPB and HBG1 for atrial fibrillation and stroke: analysis via the support vector machine neural network. J Cell Mol Med. 2022;26(7):2010–22.
Liu W, et al. Functional evaluation of KEL as an oncogenic gene in the progression of Acute Erythroleukemia. Oxid Med Cell Longev. 2022;2022:5885342.
Chen CP, et al. Synergistic antitumor activity of artesunate and HDAC inhibitors through elevating heme synthesis via synergistic upregulation of ALAS1 expression. Acta Pharm Sin B. 2019;9(5):937–51.
Dey S, et al. Heme Sequestration effectively suppresses the development and progression of both lung adenocarcinoma and squamous cell carcinoma. Mol Cancer Res. 2022;20(1):139–49.
Shaham L, et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood. 2015;125(8):1292–301.
Whitman SP, et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(18):3622–6.
Namløs HM, et al. Mir-486-5p expression is regulated by DNA methylation in osteosarcoma. BMC Genomics. 2022;23(1):142.
Li S et al. Construction of Osteosarcoma diagnosis model by Random Forest and Artificial neural network. J Pers Med. 2023. 13(3).
Li X, Liu J. FANCD2 inhibits ferroptosis by regulating the JAK2/STAT3 pathway in osteosarcoma. BMC Cancer. 2023;23(1):179.
Wen RJ, et al. Baicalin induces ferroptosis in osteosarcomas through a novel Nrf2/xCT/GPX4 regulatory axis. Phytomedicine. 2023;116:154881.
Lv H, et al. β-Phenethyl isothiocyanate induces cell death in human osteosarcoma through Altering Iron Metabolism, Disturbing the Redox Balance, and activating the MAPK signaling pathway. Oxid Med Cell Longev. 2020;2020:5021983.
Ren T, et al. Zoledronic acid induces ferroptosis by reducing ubiquinone and promoting HMOX1 expression in osteosarcoma cells. Front Pharmacol. 2022;13:1071946.
Funding
This research was funded by Science and Technology Research Project of Jiangxi Provincial Department of Education (No: GJJ201527); Research Project of Gannan Medical University (No: YB201922); Natural Science Foundation of Zhejiang Province (Approval number: LQ20H170001) and Natural Science Foundation of Zhejiang Province Joint Fund Project (Approval number: LBY21H170001).
Author information
Authors and Affiliations
Contributions
Conceptualization, F.Z. and J.M.W.; Data curation, J.M.W. and J.H.Z.; Formal analysis, F.Z. and J.H.Z.; Funding acquisition, F.Z. and J.M.W.; Investigation, J.M.W. and J.H.Z.; Methodology, J.M.W. and J.W.M.; Supervision, J.W.M.; Writing—original draft, F.Z. and J.M.W.; Writing—review and editing, F.Z., J.M.W., J.H.Z. and J.W.M. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were approved by Scientific Research Ethics Committee of the First Affiliated Hospital of Gannan Medical University (No. LLSC-2024-236).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, F., Wan, J., Zhong, J. et al. ANK1 inhibits malignant progression of osteosarcoma by promoting ferroptosis. BMC Cancer 24, 1075 (2024). https://doi.org/10.1186/s12885-024-12836-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12836-z