Abstract
Background
Papillary thyroid carcinoma (PTC) is globally prevalent and associated with an increased risk of lymph node metastasis (LNM). The role of cancer-associated fibroblasts (CAFs) in PTC remains unclear.
Methods
We collected postoperative pathological hematoxylin–eosin (HE) slides from 984 included patients with PTC to analyze the density of CAF infiltration at the invasive front of the tumor using QuPath software. The relationship between CAF density and LNM was assessed. Single-cell RNA sequencing (scRNA-seq) data from GSE193581 and GSE184362 datasets were integrated to analyze CAF infiltration in PTC. A comprehensive suite of in vitro experiments, encompassing EdU labeling, wound scratch assays, Transwell assays, and flow cytometry, were conducted to elucidate the regulatory role of CD36+CAF in two PTC cell lines, TPC1 and K1.
Results
A significant correlation was observed between high fibrosis density at the invasive front of the tumor and LNM. Analysis of scRNA-seq data revealed metastasis-associated myoCAFs with robust intercellular interactions. A diagnostic model based on metastasis-associated myoCAF genes was established and refined through deep learning methods. CD36 positive expression in CAFs can significantly promote the proliferation, migration, and invasion abilities of PTC cells, while inhibiting the apoptosis of PTC cells.
Conclusion
This study addresses the significant issue of LNM risk in PTC. Analysis of postoperative HE pathological slides from a substantial patient cohort reveals a notable association between high fibrosis density at the invasive front of the tumor and LNM. Integration of scRNA-seq data comprehensively analyzes CAF infiltration in PTC, identifying metastasis-associated myoCAFs with strong intercellular interactions. In vitro experimental results indicate that CD36 positive expression in CAFs plays a promoting role in the progression of PTC. Overall, these findings provide crucial insights into the function of CAF subset in PTC metastasis.
Similar content being viewed by others
Introduction
Thyroid cancer (THCA) stands as the most prevalent malignant tumor within the endocrine system [1]. Among the various pathological types of THCA, papillary thyroid carcinoma (PTC) emerges as the most widespread [2]. Notably, the incidence of PTC has exhibited a rapid increase on a global scale in recent years [3]. The presence of neck lymph node metastasis (LNM) in PTC is acknowledged as a risk factor associated with local recurrence, distant metastasis, and diminished survival rates [4, 5]. Therefore, unraveling the molecular mechanisms governing the invasion and metastasis of PTC assumes paramount significance.
Tumor tissues encompass both tumor parenchyma and stroma. The infiltration and metastasis of tumor cells intricately link to synergistic interactions with stromal components, facilitating mutual nourishment and propelling further cancer progression [6]. Within the tumor microenvironment (TME), stromal cells wield regulatory control over diverse biological behaviors of tumor cells, including proliferation, apoptosis, migration, and invasion. This orchestration facilitates tumor development, while tumor cells reciprocally reshape the TME by modulating stromal cells, ultimately enhancing angiogenesis and metastasis [7, 8]. A pivotal player in this scenario is cancer-associated fibroblasts (CAFs), constituting a major stromal cell population in the TME. They originate from normal fibroblasts at the tumor site or undergo transformation from circulating bone marrow-derived mesenchymal stem cells [9]. CAFs comprise distinct subpopulations with varied functions in tumors [10]. Specific CAF subgroups exert positive influences on diverse facets of tumor growth, encompassing cancer cell survival, proliferation, vascularization, and extracellular matrix (ECM) remodeling, thereby impacting metastasis [9]. CAFs accelerate tumor cell growth and metastasis, directly or indirectly affecting the progression of various cancers, including lung cancer [11], breast cancer [12], colorectal cancer [13], pancreatic cancer [14], and gastric cancer [15]. By secreting growth factors, CAFs stimulate cancer cell proliferation and invasion [16], shape the innate and adaptive immune cell responses to cancer cells via cytokine secretion [17], and transport molecules to cancer cells via extracellular vesicles, promoting their invasiveness [18]. The functionality of CAFs critically affects treatment responses and resistance development [19], positioning CAFs as potential cancer therapy targets.
Despite the established significance of CAFs in various cancers, their precise role in PTC remains incompletely elucidated. Some studies have hinted at a correlation between CAFs and PTC development [20, 21]. However, the specific molecular mechanisms by which CAFs foster the occurrence and progression of PTC remain unexplored. Digital pathology, integral to modern clinical practice [22], has evolved with the advent of whole slide imaging (WSI), enabling the imaging of complete glass slides with high-resolution storage. In this study, we utilized a substantial dataset of pathology WSI from clinical patients to analyze the pathological dimension of the correlation between tumor fibrosis level and LNM. With the advancements of single-cell RNA sequencing (scRNA-seq) technology, in-depth research on the role of CAFs in cancer has been extensively studied using scRNA-seq data analysis [23, 24]. We utilized publicly available scRNA-seq PTC data to characterize the features of CAFs within PTC and describe their interactions with other cell types. The study flowchart is illustrated in Fig. 1A.
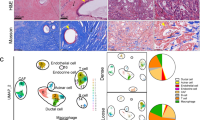
Process flowchart and hematoxylin–eosin slide annotation of tumor invasive front with cancer-associated fibroblasts (CAFs). (A) Process flowchart. (B) Immunofluorescent double staining results for α-SMA and CK19. (C) Smooth muscle actin-alpha (α-SMA) immunohistochemical staining. (D) Schematic representation of Qupath delineation of tumor invasive front and CAF annotation. The orange areas represent fibroblasts and tissues, while the blue areas represent tumor cells
Materials and methods
Data acquisition
Clinical patient data acquisition
Patients with PTC at the First Affiliated Hospital of China Medical University in 2017 were selected. Inclusion criteria comprised an initial PTC diagnosis post-surgery, an absence of prior radiotherapy or chemotherapy, and complete clinical data. Exclusion criteria included non-PTC pathology post-surgery, preoperative treatment, incomplete clinical data, and coexisting malignancies. Following stringent criteria-based selection, a total of 984 patients were enrolled. Postoperative hematoxylin–eosin (HE) slides were systematically collected for panoramic scanning, and patient data, including age, sex, anatomic site, focus type, T grade, N grade, M grade, clinical stage, coexistence with Hashimoto’s disease, and extrathyroidal extension, were meticulously recorded.
scRNA-seq data
Single-cell transcriptome data from GSE193581 and GSE184362 were meticulously downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The analysis included five PTC lesions with LNM and one PTC lesion without metastasis from the GSE193581 dataset, as well as four PTC lesions with LNM and three PTC lesions without metastasis from the GSE184362 dataset. Harmony, a bioinformatics tool, was proficiently employed to seamlessly integrate and harmonize the datasets.
Bulk RNA-seq data
The THCA expression and clinical data were sourced from The Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/) and the UCSC cancer browser (https://genome-cancer.ucsc.edu), respectively. The TCGA THCA dataset comprised 510 PTC samples and 58 non-cancer samples. As an external validation cohort, the GSE33630 dataset was systematically utilized, downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Following the exclusion of patients with missing clinical information, a comprehensive total of 49 patients with PTC were included in the analysis. Fragments per Kilobase Million was employed as the normalized value for bulk RNA-seq analysis.
Immunohistochemical staining
Immunohistochemical staining was methodically conducted using a specialized immunohistochemical kit and 3,3’-diaminobenzidine substrate kit, meticulously following the manufacturer’s protocol. Tissue sections were incubated overnight at 4 °C with primary antibodies against smooth muscle alpha-actin (α-SMA) (Abcam, Cambridge, MA; dilution 1:100).
Immunofluorescence staining
Double immunofluorescent staining involved deparaffinization, rehydration, and heat-induced epitope retrieval of tissue sections. Subsequent to permeabilization and blocking, sections were incubated overnight at 4 °C with CK19 (Abcam, #ab76539) and α-SMA (Abcam, #ab7817) antibodies. Following thorough washing, fluorochrome-conjugated secondary antibodies were used for the simultaneous visualization of multiple antigens. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI), and sections were mounted with an anti-fade medium.
Evaluation of fibrosis level in HE pathological tissue sections
An open-source software for digital pathology image analysis, QuPath, was used to meticulously assess fibrosis levels in HE pathological tissue sections. Within QuPath, tumor boundaries were delineated on each HE-stained slide, and an area within 1000 μm from this boundary was designated as the invasive front of the tumor. A pixel classifier was developed to identify fibroblasts and fibrotic regions at the invasive front of the tumor using machine learning techniques. The ratio of the identified fibrotic area to the total area of the tumor invasive front yielded the fibrosis density for each HE slide. All HE slides were subjected to independent completion and verification by three pathology experts (Supplementary Table 1).
Preprocessing and annotation of scRNA-seq data
We obtained single-cell count matrices by downloading data from GSE193581 and GSE184362. The Seurat package, a comprehensive toolkit, was used to analyze gene expression matrices for each sample. To eliminate doublets, DoubletFinder was applied. Each cell was subjected to quality control using specific criteria, including a minimum gene count of 500, a maximum gene count of 5000, and a mitochondrial proportion below 10%. We conducted principal component analysis using the most variable genes, and the top 20 principal components were utilized for Louvain clustering. Dimensionality reduction was conducted using UMAP (https://umap-learn.readthedocs.io/en/latest/).
We identified and annotated cell types based on specific gene markers [25]: TG, EPCAM, KRT18, and KRT19 for thyrocytes; CD3D, CD3E, CD3G, and CD247 for T and natural killer (NK) cells; LYZ, S100A8, S100A9, and CD14 for myeloid cells; CD79A, CD79B, IGHM, and IGHD for B cells; COL1A1, COL1A2, COL3A1, and ACTA2 for fibroblasts; PECAM1, CD34, CDH5, and VWF for endothelial cells.
Cell communication analysis
For the comprehensive inference and quantitative characterization of cell communication networks, the “CellChat” R package, encompassing ligand, receptors, and their interactions, was employed. The analysis was conducted using the secreted signaling pathway, and the reference human ligand-receptor database, CellChatDB, was utilized to evaluate human intercellular communication. Interactions between different cell types were evaluated, with pathways containing fewer than 10 cells filtered out.
Pseudo-time trajectory analysis
Pseudo-time analysis was systematically conducted using the Monocle2 package with default parameters. This advanced analysis included machine learning to simulate temporal progression dynamics based on the expression patterns of crucial genes. Genes exhibiting significant variations in intercellular gene expression were selected, and their expression profiles were downsampled to construct a minimum spanning tree (MST) [23]. Subsequently, we employed the MST to delineate the differentiation trajectory of cells sharing similar transcriptional characteristics along the longest path.
High dimensional weighted gene co-expression network analysis (hdWGCNA)
For the comprehensive co-expression analysis of scRNA-seq data, the hdWGCNA R package (v0.1.1.9002) was employed (https://github.com/smorabit/hdWGCNA) [26]. To establish a co-expression gene network with a signed scale-free nature, soft-threshold parameters of β = 4 and a scale-free R2 = 0.9 were chosen to ensure network robustness.
Gene ontology (GO) analysis and protein-protein interaction (PPI) network construction
GO enrichment analysis was conducted in the GO database (http://www.geneontology.org/). The PPI network was analyzed using the STRING protein interaction database (http://string-db.org).
Construction and validation of cancer-associated myofibroblast (myoCAF)-related gene signature
The top 100 highly expressed genes from the brown module were selected. Furthermore, univariable Cox regression analysis was applied to identify myoCAF-related genes associated with metastasis. A gene signature was developed using least absolute shrinkage and selection operator (LASSO) penalized Cox regression analysis with the “glmnet Version 4.1.4” package in R. Thirteen myoCAF-related genes were identified, forming the gene signature.
We constructed receiver operating characteristic (ROC) curves using the R package “ROCR Version 1.0.11” to evaluate the diagnostic performance of the myoCAF-related gene diagnosis signature.
Machine learning analysis
Seven machine learning methods, including weighted k-nearest neighbor classifier (kknn), linear discriminant analysis, logistic regression, Naïve Bayes, Random Forest regression, Decision Tree, and support vector machine (SVM), were employed to search for the most fitting formula. R packages ‘kknn’, ‘MASS’, ‘glmnet’, ‘Ranger’, and ‘rpart’ were used in this process.
Identification of PTC subtypes
Using myoCAF-related genes associated with metastasis, non-negative matrix factorization (NMF) clustering was conducted. NMF clustering is commonly used to delineate molecular subtypes in cancer [27]. We used the NMF R package for unsupervised NMF clustering on the metadata set, and the ideal cluster number was determined based on the co-occurrence correlation coefficient K value.
Immune cell infiltrate analysis
The ‘microenvironment cell population count (MCP-counter)’ approach with the R package MCPcounter was employed for immune infiltration assessment [28]. This method quantifies the absolute abundance of 8 immune cell types and 2 stromal cell populations based on transcriptome data.
Cell culture
After rinsing the LNM-PTC specimens with D-Hanks solution, cut them into tissue blocks of 1 × 1 × 1 mm in size. Fetal bovine serum (FBS) is used to soak the bottom of T25 culture flasks, and then the tissue blocks are evenly spread out and placed in an incubator for pre-attachment. After pre-attachment, a specialized culture medium is added to soak the tissue blocks. This specialized culture medium contains DMEM/F12 medium with 5% FBS, 100 units/ml penicillin, 100ug/ml streptomycin, 5ug/ml insulin, 1ug/ml dexamethasone, and 5ng/ml epidermal growth factor. The medium should be changed daily for the first 3 days, then every 3–4 days afterwards, for a total culture period of 26–30 days. Discard the culture medium and tissue blocks, and proceed with the subculturing process according to the cell passage protocol.
TPC1 and K1 cells, acquired from Wuhan Puno Sai Life Technology Co., Ltd. in Wuhan, China, were grown in 1640 medium enriched with 10% FBS.
Establishment of CD36 knockdown CAF
CD36 knockdown CAF were created via lentivirus transfection, while control cells received negative control lentivirus (NC). Verification of successful CD36 depletion was performed through western blotting. The specific CD36 shRNA sequence used was 5′-CCGGCGGATCTGAAATCGACCTTAACTCGAGTTAAGGTCGATTTCAGATCCGTTTTTG-3′. These CD36 shRNA and negative control vectors were introduced into CAF using Lipofectamine 3000 (Invitrogen, USA).
Western blot
Western blot experiments utilized a CD36-specific antibody (ab133625, Abcam, Cambridge, UK) to probe for protein levels. The detected protein bands were visualized using an ECL chemiluminescence solution (BL520A, Biosharp, China) and imaged with a chemiluminescence detection system (Tano 5200Multi).
Co-culture of CAF and PTC cell lines
The co-culture of CAF and TPC1/K1 cells was conducted using a Transwell system (with a membrane pore size of 0.4 μm). shNC CAF cells or shCD36 CAF cells suspensions were added to the Transwell inserts at a density of 2 × 10^5 cells per well in a six-well plate. The Transwell inserts were then placed into six-well plates that had been pre-seeded with TPC1 or K1 cells. The shared culture medium was prepared by mixing CAF culture medium and PTC cell culture medium at a 1:1 ratio. 1 ml of the shared cell culture medium was added to the Transwell inserts, and 2 ml of the shared cell culture medium was added to the wells of the six-well plate. For the control group, no cells were seeded in the upper chamber. The cells were cultured in a 37 °C, 5% CO2 incubator for 48 h. TPC1/K1 cells in the lower chamber were then collected for subsequent experiments following trypsin digestion.
EdU assay
Cell proliferation was assessed utilizing the EdU incorporation method, facilitated by the EdU kit (Cat. No. K1075, ApexBio, USA). TPC1 or K1 cells were plated at a density of 3 × 104 cells per well in 24-well plates and allowed to adhere overnight at 37 °C. Subsequently, cells were treated with 10µM EDU for a 5-hour incubation period. Following this, cells underwent fixation with 4% paraformaldehyde for 15 min and were then treated with 0.3% Triton X-100 for membrane permeabilization for an additional 15 min. The Click Reaction Mixture was then applied to the cells for 30 min in darkness at room temperature, before being stained with Hoechst 33,342 for DNA visualization for 15 min. Fluorescence microscopy at 100× magnification was used to capture images, and ImageJ software facilitated the quantification of proliferating cells.
Wound scratch assay
To assess the migratory capabilities of cells, the scratch wound healing assay was utilized. TPC1 or K1 cells were grown to achieve 100% confluence in a six-well plate. A precise, linear scratch was then made across the layer of cells using the tip of a pipette, followed by the replacement of the culture medium. The gap created by the scratch was documented using a microscope at 200× magnification at both the initial time (0 h) and after 24 h (24 h). The average distance across the scratch was determined at these two time points using ImageJ software, and the extent of cell migration was evaluated by computing the area percentage of the wound that had healed.
Transwell assay
The upper chamber was pre-coated with Matrix gel, which was diluted tenfold in serum-free culture medium. TPC1 or K1 cell concentrations were adjusted to around 90,000 cells/ml using a serum-free medium. The lower chamber received a medium supplemented with 10% FBS, and 300 µl of the cell suspension was then added to the upper chamber. The setup was incubated at 37 °C in a 5% CO2 atmosphere for 24 h to facilitate cell migration. After incubation, each well was fixed with 700 µl of ice-cold methanol at -20 °C for 30 min at room temperature. This was followed by the addition of 700 µl of crystal violet staining solution to each well for a further 30-minute incubation. Images were taken at 200× magnification, and cell quantification was carried out using ImageJ software.
Cell apoptosis
Apoptosis in cells was evaluated using flow cytometry with the aid of the Cell Apoptosis Detection Kit from MultiSciences (Cat. No. AT105, China). Cells from the TPC1 or K1 lines were first suspended in 1× Binding Buffer, followed by the addition of 5 µl Annexin V-APC and 10 µl 7-AAD for staining. These samples were then incubated in the dark at room temperature for 5 min. The analysis was carried out using a FACS C6 flow cytometer.
Statistical analyses
Visualization analyses were performed using R software (version 4.1.1) with a significance level set at p < 0.05. Student’s t-test and analysis of variance were employed to compare quantitative data, whereas the Chi-squared test was employed for categorical variables. Correlation analysis was conducted using Spearman’s correlation test.
Results
Correlation between fibrosis density at the tumor invasive front and LNM in PTC
A comprehensive study involving 984 patients meeting the inclusion criteria from the First Affiliated Hospital of China Medical University was undertaken. Postoperative pathological HE-stained sections underwent panoramic scanning, with three independent pathologists delineating the tumor invasive front on all HE sections. CAFs at the invasive front of the tumor were identified using QuPath software (Fig. 1D). Immunohistochemical staining for the CAF marker α-SMA demonstrated a high concordance between the fibrotic areas identified by QuPath and α-SMA-positive expression areas (Fig. 1C). Further validation through α-SMA and CK19 immunofluorescent double staining affirmed CAF distribution delineated by QuPath at the tumor invasive front (Fig. 1B).
The fibrosis density, defined as the ratio of the fibrotic area at the tumor invasive front to the total area at the tumor invasive front, was determined. This density was ranked from high to low across all HE pathological sections, with the median value serving as the cutoff. Sections with higher fibrosis density than the median were classified as high fibrosis infiltration, and those with lower fibrosis density than the median were classified as low fibrosis infiltration. Analyzing the correlation between fibrosis infiltration levels and clinical-pathological indicators revealed a significantly higher proportion of patients with high fibrosis infiltration levels experiencing LNM. Patients with bilateral PTC and those with extrathyroidal extension exhibited a higher proportion of high fibrosis infiltration. However, no significant correlation was observed between fibrosis infiltration levels and patient sex, age, focus type, T grade, M grade, clinical stage, or concurrent Hashimoto’s disease (Table 1).
Differential expression of CAFs between PTC tissues with LNM (LNM-PTC) and PTC tissues without LNM (non-LNM-PTC)
In the combined GSE193581 and GSE184362 datasets, including four non-LMN (non-LNM-PTC) samples and nine PTC lesions with concurrent LNM (LNM-PTC), cell clustering based on cell markers (Figure S1A) resulted in the visualization of six distinct cell clusters through UMAP analysis. These clusters encompassed T/NK cells, myeloid cells, thyrocytes, fibroblasts, and endothelial cells (Fig. 2A). Bar plots illustrated the proportional distribution of these cell types for LNM-PTC and non-LNM-PTC (Fig. 2B). Obviously, the fibroblasts in LNM-PTC are significantly higher than that in non-LNM-PTC.
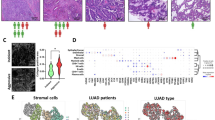
Analysis of cancer-associated fibroblasts (CAFs) in papillary thyroid carcinoma (PTC). (A) UMAP analysis based on cell markers revealed six distinct cell clusters within the combined GSE193581 and GSE184362 datasets. (B) The bar plots depict the proportional distribution of cell types in lymph node metastasis (LNM-PTC) and non-metastasis (non-LNM-PTC) PTC tissues. (C) Subcluster analysis and UMAP visualization of CAFs in LNM-PTC and non-LNM-PTC tissues. (D) Distinct marker characteristics for each CAF subcluster. (E) UMAP plots of myoCAFs and iCAFs in PTC tissues. (F) Cellular communication analysis (interaction strength). (G) Cellular communication analysis (number of interactions). (H-I) Temporal trajectory analysis of CAF clusters. (J) The heat map shows the incoming and outgoing signaling patterns of cell clusters. (K) Network visualization of the pleiotrophin (PTN) signaling pathway
Metastasis-associated myoCAFs strongly interacted with other cells
Subsequently, subcluster analysis and UMAP visualization of CAFs in LNM-PTC and non-LNM-PTC tissues were conducted, resulting in the subdivision of CAFs into 15 clusters (Fig. 2C), with distinct marker characteristics for each cluster highlighted in Fig. 2D. Clusters 0, 1, 3, 6, 7, 12, 14, and 15 exhibited increased MYL9, MYLK, TAGLN, and ACTA2 expression, denoted as inflammatory CAF (iCAF), whereas clusters 2, 4, 5, 8, 9, 10, 11, and 13 exhibited increased expression of S100A10, MCL1, CCDC80, PLA2G2A, and CFD, identified as myoCAF (Fig. 2E).
The subpopulation of myoCAFs that is increased in proportion in LNM-PTC tissues compared to non-LNM-PTC tissues is defined as metastasis-associated myoCAFs. Subsequently, metastasis-associated myoCAFs were extracted and annotated from LNM-PTC tissues. Cellular communication analysis revealed that these myoCAFs exhibited a higher number of communication interactions with other cells (Fig. 2F) and exhibited stronger communication intensities (Fig. 2G).
Temporal trajectory analysis of the 15 CAF clusters revealed a differentiation pattern from cluster 2 toward clusters 6, 7, 14, and 15, indicating a transition from iCAF to myoCAF, potentially associated with late-stage metastasis (Fig. 2H-I).
Signal pathway analysis of intercellular communication revealed that among all cells, metastasis-associated myoCAFs exhibited the highest output signal count. In particular, only metastasis-associated myoCAFs were identified as output signal cells in the pleiotrophin (PTN) pathway, with thyrocytes being the top recipients of this pathway (Fig. 2J). Furthermore, network visualization of the PTN signaling pathway validated similar results (Fig. 2K, Figure S1B). These results reveal that metastasis-associated myoCAFs may influence cancer cells via the PTN signaling pathway.
Identification of the myoCAF module using hdWGCNA
In the hdWGCNA analysis, we selected a threshold at the inflection point β = 4 (Fig. 3A). The application of hdWGCNA for CAF module analysis identified six WGCNA modules—turquoise, blue, green, red, yellow, and brown (Fig. 3B). The brown module, specifically, exhibited high consistency with metastasis-associated myoCAFs, showcasing elevated expression of genes such as TINAGL1, TAGLN, LHFP, CALD1, and ACTA2 (Fig. 3D). Additional details, including mapping and highly expressing genes from other modules, are presented in Figure S1D-E. The bubble chart depicting the correlation between various CAF clusters and WGCNA modules is displayed in Fig. 3E. Notably, the brown module, especially clusters 0, 1, 3, 6, 7, 12, 14, and 15, exhibited higher proportions and expression. Correlation analysis among modules revealed a positive correlation between the brown module and the turquoise and yellow modules (associated with myoCAFs), whereas negative correlations were observed with the blue, green, and red modules (associated with iCAFs) (Fig. 3C). Subsequently, univariate Cox analysis focused on the top 100 highly expressed genes in the brown module, identifying 43 genes associated with metastasis. The results of the GO enrichment analysis for these 43 genes are depicted in Fig. 3F. The PPI network of these genes is demonstrated in Fig. 3G, indicating involvement in processes such as the cell leading edge, protein complex involved in cell adhesion, cell-substrate adhesion, homotypic cell-cell adhesion, and cell adhesion mediator activity. Pseudo-time trajectory analysis of these 43 genes revealed predominantly high expression in later stages (Figure S1F).
hdWGCNA analysis. (A) The inflection point threshold β = 4 was considered for hdWGCNA analysis. (B) hdWGCNA was used to analyze CAF modules, followed by the identification of six WGCNA modules. (C) Correlation analysis among the modules. (D) Mapping and high-expressing genes of brown module. (E) Bubble chart of the correlation between various CAF subclusters and WGCNA modules. (F) Gene ontology enrichment analysis of 43 genes associated with metastasis. (G) Protein–protein interaction network of the 43 genes associated with metastasis
Development and validation of a metastasis diagnostic model based on LNM-associated myoCAF genes
We constructed a diagnostic model using LASSO regression for the 43 LNM-associated myoCAF-related genes (Fig. 4A-B). A subset of 13 genes was identified for diagnosing LNM in PTC, and their relationship with stromal cells is depicted in Fig. 4C. Notably, besides COX8C and LDHB, which were significantly negatively correlated with CAFs, the other genes demonstrated significant positive correlations. To refine the diagnostic model, we used seven deep learning methods. Ultimately, we selected the SVM method to construct the final diagnostic model, which exhibited an area under the curve (AUC) of 0.706 in the external validation dataset GSE33630 (Fig. 4E).
Establishment and refinement of the diagnostic model. (A) Least absolute shrinkage and selection operator (LASSO) coefficient profiles. (B) The tuning parameter (lambda) in the LASSO model was selected by performing a 10-fold cross-validation using the minimum criteria approach. (C) Correlation analysis of 13 metastasis-associated myoCAF-related genes with immune cells. (D) Area under the curve of seven deep learning models in the training set of The Cancer Genome Atlas. (E) Receiver operating characteristic curve of the prognostic model optimized using the support vector machine algorithm applied to the external validation set GSE33630
Identification of cancer subtypes and immune infiltration characteristics in patients with PTC
NMF analysis categorized patients with PTC in TCGA into three subgroups (Fig. 5A, Figure S1G), and consensus clustering results indicated distinct boundaries among these subgroups (Fig. 5B). Correlation analysis of metastasis-associated CAF infiltration levels across these three subgroups revealed that subgroup 1 exhibited the highest level of CAF infiltration, whereas subgroup 3 displayed the lowest CAF infiltration (Fig. 5C). Analysis of immune cell infiltration levels suggested that the expression of NK cells, monocytic lineage, neutrophils, endothelial cells, and fibroblasts among the three subgroups were significantly different (Fig. 5D). We generated heatmaps demonstrating the correlation of 13 metastasis-associated myoCAF-related genes and stromal cells in each patient. Deep learning was then performed for the heatmaps for all patient heatmaps (Fig. 5E). Deep learning significantly enhanced the diagnostic efficacy of the model to 0.951 (Fig. 5F-I).
Subgroup analysis and immune infiltration characteristics in patients with papillary thyroid carcinoma (PTC). (A) Non-negative matrix factorization analysis stratified patients with PTC from The Cancer Genome Atlas into three distinct subgroups. (B) Consensus clustering outcomes showed well-defined boundaries among these subgroups. (C) Correlation analysis of metastasis-associated CAF infiltration across the three subgroups. (D) Immune cell infiltration analysis of the three subgroups. (E) The heatmaps show the correlation between 13 metastasis-associated myoCAF-related genes and stromal cells for each patient, followed by machine learning applied to heatmaps of all patients. (F-I) The utilization of deep learning significantly boosted the diagnostic accuracy of the model to 0.951
In vitro experiments confirm the pro-oncogenic role of CD36+CAF in PTC cell line
Previous studies have shown that the expression of CD36 in CAFs varies across different types of cancer. Our analysis of single-cell data revealed a significant increase in CD36 expression in CAFs within PTC (Figure S1C). Primary CAF cells expressing α-SMA were isolated from PTC tissues (Fig. 6A). Subsequently, CD36 knockdown CAFs (shCD36 CAF) were successfully established through lentivirus transfection (Fig. 6B-C). Further, a Transwell co-culture system was established to investigate the effects of the control group (Transwell inserts without cell seeding), shNC CAF (Transwell inserts seeded with CAF transfected with control lentivirus vector), and shCD36 CAF (Transwell inserts seeded with CAF transfected with shCD36 lentivirus) on PTC cell proliferation, migration, invasion, and apoptosis. The results showed that the proliferation (Fig. 6D, H), migration (Fig. 6F, J), and invasion abilities (Fig. 6G, K) of TPC1 and K1 cells co-cultured with shNC CAF were significantly higher than those co-cultured with shCD36 CAF and the control group cells, and the apoptosis rate (Fig. 6E, I) was significantly lower than that in TPC1 co-cultured with shCD36 CAF and the control group cells. The proliferation (Fig. 6D, H), migration (Fig. 6F, J), and invasion abilities (Fig. 6G, K) of TPC1 and K1 cells co-cultured with shCD36 CAF were significantly higher than those of the control group cells, and the apoptosis rate (Fig. 6E, I) was significantly lower than that in the control group cells.
In vitro experiments confirm the pro-oncogenic role of CD36+CAF in PTC cell line. (A) Immunofluorescence staining of α-SMA in CAFs. (B) WB analysis of CD36 protein expression in CAF after transfecting shCD36 or control lentivirus vector and (C) Statistical graphs for each group. WB images are cropped. (D) EdU staining results and (H) statistical graphs for each group. (F) Scratch assays results and (J) statistical graphs for each group. (G) Transwell assays results and (K) statistical graphs for each group. (E) Flow cytometry analysis of apoptosis levels in each group and (I) statistical graphs. Asterisks indicate statistical comparison with the control group unless indicated otherwise on the graphs. * p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001
Discussion
Patients with PTC and concurrent LNM face elevated mortality and recurrence risks compared to those without LNM [29]. Timely detection of LNM is crucial for reducing recurrence rates and improving the prognosis of PTC, ultimately enhancing cancer survival rates. Although nearly 80% of patients with PTC experience microscopic LNM, its identification typically relies on postoperative histopathological diagnosis [30]. Current methods, especially preoperative ultrasound examination, exhibit only 48.3% accuracy in diagnosing lymph node metastasis [31]. Consequently, establishing a reliable predictive model for diagnosing LNM in PTC, applicable in preoperative biopsies or intraoperative pathological examinations, holds paramount significance for early tumor diagnosis and preventing rapid PTC progression.
This study delves into the pivotal role of CAFs in PTC with LNM. Utilizing postoperative pathological HE slides from an extensive patient cohort, we meticulously delineated the tumor invasive front, a critical region within 1000 μm inside the tumor edge. Our findings revealed a significant correlation between high fibrosis density at the tumor invasive front and LNM. We employed a diverse range of methodologies, leading to the development of a robust diagnostic model predicting LNM in PTC based on metastasis-associated myoCAF genes. Our initial analysis, comparing CAF expression between PTC tissues with and without LNM (LNM-PTC and non-LNM-PTC, respectively), indicated a higher proportion of CAFs in LNM-PTC tissues, suggesting their potential involvement in metastatic processes. Subsequent subcluster analysis and UMAP visualization stratified CAFs into 15 clusters, revealing distinct marker profiles for myoCAFs and iCAFs. Notably, myoCAFs exhibited intense interactions with other cells, implying their significant contribution in shaping the TME. Temporal trajectory analysis unveiled a differentiation pattern from iCAFs to myoCAFs, possibly correlating with late-stage metastasis. Applying the hdWGCNA approach, we identified the brown module highly correlated with metastasis-associated myoCAFs, highlighting genes such as TINAGL1, TAGLN, LHFP, CALD1, and ACTA2. This module exhibited strong associations with metastasis-related processes, shedding light on the molecular underlying mechanisms of myoCAFs in LNM-PTC. Leveraging these findings, we constructed a diagnostic model using machine learning methods for the 43 LNM-associated myoCAF-related genes. This model exhibited remarkable accuracy, showcasing its potential in identifying PTC patients at risk of LNM. Additionally, our study included cancer subtype identification and immune infiltration analysis. NMF analysis classified TCGA patients with PTC into three subgroups, each displaying distinct immune infiltration patterns. The refined diagnostic model further enhanced its efficacy to an impressive 0.951, emphasizing its valuable clinical application potential. In summary, our study demonstrates that in PTC, CAFs infiltrate at high levels at the tumor invasive front, particularly the myoCAF subtype, which promotes PTC progression and increases the likelihood of lymph node metastasis. The diagnostic model based on myoCAF-related genes, developed using deep learning methods, can efficiently predict lymph node metastasis in PTC.
Research on CAFs in PTC is currently limited. A study of 125 PTC samples reported the use of immunohistochemistry to analyze four CAF marker proteins (FAP, α-SMA, vimentin, and PDGFR-α) and correlated them with clinicopathological features [32]. Elevated FAP and α-SMA immunoreactivity scores were associated with unfavorable tumor features, such as BRAF mutation, extrathyroidal invasion, and LNM. Dadafarin et al. proposed that MEG3 expression in tumor CAFs might drive PTC invasiveness and LNM, suggesting a potential therapeutic target [33]. Research including single-cell sequencing [34] revealed high fibroblast infiltration in PTC and their interactions with various cell types, uncovering differentially expressed fibroblast-related genes in THCA tissues. The fibrosis score model emerged as an independent prognostic factor for patients with THCA, with low fibrosis scores correlating with improved overall survival. High CAF scores were linked to aggressive phenotypes, genetic mutations, oncogenic signaling pathways, and alterations in the immune landscape alterations [34, 35]. We identified 43 metastasis-associated myoCAF-related genes, suggesting a close association with cell adhesion-related signaling pathways. We selected 13 genes (ACTB, C1QTNF1, CALD1, CD36, COX6C, CSRP2, FABP4, LDHB, MYL12A, MYO1B, NES, SUCNR1, and TBX2) to establish an LNM prediction model, achieving an optimized AUC of 0.951 via deep learning. However, further exploration and validation are warranted to understand the underlying molecular mechanisms and potential therapeutic targets.
myoCAFs are typically induced by TGF-β1 or SMAD signaling, leading to changes in cellular cytoskeleton and contributing to the formation of the ECM that promotes metastasis [36, 37]. MyoCAFs express α-SMA and secrete collagen-rich ECM [38, 39]. In various solid tumors, including esophageal [40], breast [41], colorectal [42], gastric [43], and prostate [44] cancers, myoCAFs govern malignancy-associated tumor features and are linked to poor prognosis. This study extensively explored the predictive role and underlying mechanisms of CAFs in PTC LNM, with a particular focus on myoCAFs and their strong correlation with PTC metastasis.
CD36 is a scavenger receptor expressed in various cell types, mediating lipid metabolism, immune recognition, inflammation, molecular adhesion, and apoptosis [45]. The lipid metabolism reprogramming driven by CD36 and the functional suppression of tumor-associated immune cells lead to tumor immune tolerance and cancer progression [46]. The uptake of palmitic acid (PA) by CD36 has been shown to induce phosphorylation of AKT in gastric cancer cells, inhibit the degradation of GSK3β/β-catenin, and promote gastric cancer metastasis [47]. Studies have found that in hepatocellular carcinoma (HCC), CD36+ CAFs exhibit high levels of lipid metabolism and expression of macrophage migration inhibitory factors [48]. In this study, CD36 positive expression in CAFs can significantly promote the proliferation, migration, and invasion abilities of PTC cells, while inhibiting the apoptosis of PTC cells. These results imply that CD36+CAF plays a promoting role in PTC, aligning with conclusions drawn in HCC research, thus underscoring its potential as a therapeutic target. CD36 plays a crucial role in lipid uptake, immune recognition, inflammation, molecular adhesion, and apoptosis, impacting the initiation, development, and progression of cancer. Currently, several anti-tumor drugs targeting CD36 have entered clinical trials [49]. Nonetheless, the precise mechanisms underlying CD36+CAF promoting role in PTC warrant further investigation.
The present is a single-center study. This is the limitation of this study. To mitigate the limitations of a single-center study, we expanded our sample size. Furthermore, the First Hospital of China Medical University is a leading hospital in Northeast China, attracting patients from across the region. This somewhat compensates for the single-center limitation by including a diverse patient population from a broad geographic area.
This study has certain limitations. The data utilized in this study originates from a singular institution, potentially constraining the generalizability of the findings. Despite efforts to increase the sample size and incorporate a varied patient demographic, future research endeavors will involve multi-center studies to improve the transferability of the results to diverse populations. Furthermore, the molecular mechanisms of CD36+CAFs in the progression of PTC have not been thoroughly investigated. The present study predominantly confirms their function through in vitro experiments. Subsequently, we aim to pursue comprehensive mechanistic investigations and in vivo experiments to establish a more robust scientific foundation.
Conclusion
In conclusion, in the present study, we addressed the critical issue of the risk of LNM in patients with PTC. The analysis of postoperative HE-stained pathological slides from several patients revealed that high fibrosis density at the tumor invasive front was significantly correlated to LNM. Further, we comprehensively analyzed CAF infiltration in PTC by integrating scRNA-seq data from GSE193581 and GSE184362 datasets. Notably, we identified metastasis-associated myoCAFs exhibiting strong intercellular interactions and established a diagnostic model validated using deep learning. Next, NMF clustering revealed distinct PTC subtypes and immune infiltrate variations. In vitro experimental results indicate that CD36 positive expression in CAFs plays a promoting role in the progression of PTC. Overall, these findings provide crucial insights into the function of CAF subset in PTC metastasis.
Data availability
To protect study participant privacy, the data utilized to substantiate the conclusions of this research can be obtained by reaching out to the corresponding author upon inquiry.
Abbreviations
- THCA:
-
Thyroid cancer
- PTC:
-
Papillary thyroid carcinoma
- LNM:
-
Lymph node metastasis
- CAFs:
-
Cancer-associated fibroblasts
- HE:
-
Hematoxylin–eosin
- scRNA:
-
seq Single-cell RNA sequencing
- HdWGCNA:
-
High dimensional weighted gene co-expression network analysis
- NMF:
-
Non-negative matrix factorization
- TME:
-
Tumor microenvironment
- WSI:
-
Whole slide imaging
- TCGA:
-
The Cancer Genome Atlas
- α-SMA:
-
Smooth muscle alpha-actin
- LASSO:
-
Least absolute shrinkage and selection operator
- ROC:
-
Receiver operating characteristic
- NMF:
-
Non-negative matrix factorization
- MCP:
-
Counter microenvironment cell population count
References
Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7(10):569–80.
Rossi ED, Pantanowitz L, Hornick JL. A worldwide journey of thyroid cancer incidence centred on tumour histology. Lancet Diabetes Endocrinol. 2021;9(4):193–4.
Kim J, Gosnell JE, Roman SA. Geographic influences in the global rise of thyroid cancer. Nat Rev Endocrinol. 2020;16(1):17–29.
Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM, Schlumberger M, et al. 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid Cancer: the American Thyroid Association Guidelines Task Force on thyroid nodules and differentiated thyroid Cancer. Thyroid. 2016;26(1):1–133.
Hall CM, Snyder SK, Lairmore TC. Central Lymph Node Dissection improves lymph node clearance in papillary thyroid Cancer patients with lateral Neck metastases, even after prior total thyroidectomy. Am Surg. 2018;84(4):531–6.
Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–52.
Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1(1):46–54.
De Wever O, Mareel M. Role of tissue stroma in cancer cell invasion. J Pathol. 2003;200(4):429–47.
Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–98.
Ohlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211(8):1503–23.
Zhu Y, Zhang L, Zha H, Yang F, Hu C, Chen L, Guo B, Zhu B. Stroma-derived fibrinogen-like protein 2 activates Cancer-associated fibroblasts to promote Tumor Growth in Lung Cancer. Int J Biol Sci. 2017;13(6):804–14.
Luo H, Tu G, Liu Z, Liu M. Cancer-associated fibroblasts: a multifaceted driver of breast cancer progression. Cancer Lett. 2015;361(2):155–63.
Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–84.
Pereira BA, Vennin C, Papanicolaou M, Chambers CR, Herrmann D, Morton JP, Cox TR, Timpson P. CAF subpopulations: a New Reservoir of stromal targets in pancreatic Cancer. Trends Cancer. 2019;5(11):724–41.
Yang Y, Ma Y, Yan S, Wang P, Hu J, Chen S, Zhu J, Wang J, Chen G, Liu Y. CAF promotes chemoresistance through NRP2 in gastric cancer. Gastric Cancer. 2022;25(3):503–14.
Mishra P, Banerjee D, Ben-Baruch A. Chemokines at the crossroads of tumor-fibroblast interactions that promote malignancy. J Leukoc Biol. 2011;89(1):31–9.
Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18(2):99–115.
Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770–8.
Kadel D, Zhang Y, Sun HR, Zhao Y, Dong QZ, Qin LX. Current perspectives of cancer-associated fibroblast in therapeutic resistance: potential mechanism and future strategy. Cell Biol Toxicol. 2019;35(5):407–21.
Cho JG, Byeon HK, Oh KH, Baek SK, Kwon SY, Jung KY, Woo JS. Clinicopathological significance of cancer-associated fibroblasts in papillary thyroid carcinoma: a predictive marker of cervical lymph node metastasis. Eur Arch Otorhinolaryngol. 2018;275(9):2355–61.
Sun WY, Jung WH, Koo JS. Expression of cancer-associated fibroblast-related proteins in thyroid papillary carcinoma. Tumour Biol. 2016;37(6):8197–207.
Jahn SW, Plass M, Moinfar F. Digital Pathology: advantages, limitations and emerging perspectives. J Clin Med 2020, 9(11).
Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, Teinor JA, Belleau P, Biffi G, Lucito MS, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals Antigen-Presenting Cancer-Associated fibroblasts. Cancer Discov. 2019;9(8):1102–23.
Yu L, Shen N, Shi Y, Shi X, Fu X, Li S, Zhu B, Yu W, Zhang Y. Characterization of cancer-related fibroblasts (CAF) in hepatocellular carcinoma and construction of CAF-based risk signature based on single-cell RNA-seq and bulk RNA-seq data. Front Immunol. 2022;13:1009789.
Pu W, Shi X, Yu P, Zhang M, Liu Z, Tan L, Han P, Wang Y, Ji D, Gan H, et al. Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma. Nat Commun. 2021;12(1):6058.
Morabito S, Miyoshi E, Michael N, Shahin S, Martini AC, Head E, Silva J, Leavy K, Perez-Rosendahl M, Swarup V. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat Genet. 2021;53(8):1143–55.
Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401(6755):788–91.
Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman C, Fridman WH, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218.
Pelizzo MR, Boschin IM, Toniato A, Piotto A, Pagetta C, Gross MD, Al-Nahhas A, Rubello D. Papillary thyroid carcinoma: 35-year outcome and prognostic factors in 1858 patients. Clin Nucl Med. 2007;32(6):440–4.
Lee YM, Sung TY, Kim WB, Chung KW, Yoon JH, Hong SJ. Risk factors for recurrence in patients with papillary thyroid carcinoma undergoing modified radical neck dissection. Br J Surg. 2016;103(8):1020–5.
Xue S, Wang P, Hurst ZA, Chang YS, Chen G. Active surveillance for papillary thyroid Microcarcinoma: challenges and prospects. Front Endocrinol (Lausanne). 2018;9:736.
Zhu L, Zhang X, Zhang S, Zhang Q, Cao L, Zhang Y, Wang D, Liang X, Wu W, Wu S et al. Cancer-associated fibroblasts in papillary thyroid carcinoma. Clin Exp Med 2023.
Dadafarin S, Rodriguez TC, Carnazza MA, Tiwari RK, Moscatello A, Geliebter J. MEG3 Expression Indicates Lymph Node Metastasis and Presence of Cancer-Associated Fibroblasts in Papillary Thyroid Cancer. Cells 2022, 11(19).
Li W, Liu Z, Cen X, Xu J, Zhao S, Wang B, Zhang W, Qiu M. Integrated analysis of fibroblasts molecular features in papillary thyroid cancer combining single-cell and bulk RNA sequencing technology. Front Endocrinol (Lausanne). 2022;13:1019072.
Wen S, Qu N, Ma B, Wang X, Luo Y, Xu W, Jiang H, Zhang Y, Wang Y, Ji Q. Cancer-Associated fibroblasts positively correlate with dedifferentiation and aggressiveness of thyroid Cancer. Onco Targets Ther. 2021;14:1205–17.
Chakravarthy A, Khan L, Bensler NP, Bose P, De Carvalho DD. TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun. 2018;9(1):4692.
Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70(23):9621–30.
Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170(6):1807–16.
Henderson NC, Rieder F, Wynn TA. Fibrosis: from mechanisms to medicines. Nature. 2020;587(7835):555–66.
Ha SY, Yeo SY, Xuan YH, Kim SH. The prognostic significance of cancer-associated fibroblasts in esophageal squamous cell carcinoma. PLoS ONE. 2014;9(6):e99955.
Cook KL, Metheny-Barlow LJ, Tallant EA, Gallagher PE. Angiotensin-(1–7) reduces fibrosis in orthotopic breast tumors. Cancer Res. 2010;70(21):8319–28.
Tsujino T, Seshimo I, Yamamoto H, Ngan CY, Ezumi K, Takemasa I, Ikeda M, Sekimoto M, Matsuura N, Monden M. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin Cancer Res. 2007;13(7):2082–90.
Fuyuhiro Y, Yashiro M, Noda S, Matsuoka J, Hasegawa T, Kato Y, Sawada T, Hirakawa K. Cancer-associated orthotopic myofibroblasts stimulates the motility of gastric carcinoma cells. Cancer Sci. 2012;103(4):797–805.
Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M, Spitler J, Rowley DR. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003;9(13):4792–801.
Li Y, Huang X, Yang G, et al. CD36 favours fat sensing and transport to govern lipid metabolism. Prog Lipid Res. 2022;88:101193.
Yang P, Qin H, Li Y, et al. CD36-mediated metabolic crosstalk between tumor cells and macrophages affects liver metastasis. Nat Commun. 2022;13(1):5782.
Ma X, Xiao L, Liu L, et al. CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell Metab. 2021;33(5):1001–12. e5.
Pan J, Fan Z, Wang Z, et al. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3beta/beta-catenin pathway. J Exp Clin Cancer Res. 2019;38(1):52.
Wang J, Li Y. CD36 tango in cancer: signaling pathways and functions. Theranostics. 2019;9(17):4893–908.
Acknowledgements
We wish to extend our heartfelt gratitude for the invaluable contribution made by the TCGA and GEO public database to the field of human medicine and for the linguistic editing by Bullet Edits Limited.
Funding
The National Natural Science Foundation of China (82170507) provided funding for this study.
Author information
Authors and Affiliations
Contributions
Author contributionYixian Wang: Conceptualization, Writing-Original Draft, Writing-Review & Editing, Formal analysis, Visualization. Xin Li: Methodology, Validation, Data Curation. Qingwei Gang: Methodology, Validation, Data Curation.Yinde Huang : Data Curation, Software, Validation. Han Zhang: Data Curation, Software, Validation. Mingyu Liu: Data Curation, Software, Validation. Shikai Shen: Data Curation, Software, Validation. Yao Qi: Data Curation, Software, Formal analysis. Jian Zhang: Conceptualization, Supervision, Writing-Review & Editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The experiment of the study was approved by the Medical Research Ethics Committee of the First Hospital of China Medical University (Ethics approval number: 2023 − 497).Written informed consent was obtained from all patients. All experiments were carried out in accordance with the Declaration of Helsinki.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, Y., Li, X., Gang, Q. et al. Pathomics and single-cell analysis of papillary thyroid carcinoma reveal the pro-metastatic influence of cancer-associated fibroblasts. BMC Cancer 24, 710 (2024). https://doi.org/10.1186/s12885-024-12459-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12459-4