Abstract
Background
Breast cancer has become the most common malignant tumour worldwide. Distant metastasis is one of the leading causes of breast cancer-related death. To verify the performance of clinicomics-guided distant metastasis risk prediction for breast cancer via artificial intelligence and to investigate the accuracy of the created prediction models for metachronous distant metastasis, bone metastasis and visceral metastasis.
Methods
We retrospectively enrolled 6703 breast cancer patients from 2011 to 2016 in our hospital. The figures of magnetic resonance imaging scanning and ultrasound were collected, and the figures features of distant metastasis in breast cancer were detected. Clinicomics-guided nomogram was proven to be with significant better ability on distant metastasis prediction than the nomogram constructed by only clinical or radiographic data.
Results
Three clinicomics-guided prediction nomograms on distant metastasis, bone metastasis and visceral metastasis were created and validated. These models can potentially guide metachronous distant metastasis screening and lead to the implementation of individualized prophylactic therapy for breast cancer patients.
Conclusion
Our study is the first study to make cliniomics a reality. Such cliniomics strategy possesses the development potential in artificial intelligence medicine.
Similar content being viewed by others
Introduction
Breast cancer (BC) has become the most common malignant tumour worldwide, its incidence was reported to increase by approximately 0.5% yearly [1]. The 5-year survival of early-stage BC patients is approximately 95%; once distant metastasis (DM) occurs, prognosis significantly deteriorates [2, 3]. Accurate identification of BC patients at high risk for DM risk, prophylactic treatment and close follow-up could improve the prognosis of BC patients. An earlier window for treatment can potentially be created with the identification of BC patients at high metastatic risk.
Prior studies predicted DM in BC patients with established mathematical models [4, 5]. Currently, most models are created based only on clinical or radiographic data. Artificial intelligence (AI) guided models are known to have the potential for wide application [6,7,8]. The clinicomics approach involves multiple disease features that are routinely evaluated [9, 10]. Such features include complete history, epidemiological distribution, physical examination, laboratory testing, imaging evaluation, and histological examination. We hypothesized that the incorporation of multidimensional data into the prediction model could result in a deeper understanding of the disease and a higher prognostic prediction accuracy. No studies have been conducted to validate this hypothesis since it was first described in 2005. Data dimension reduction before the application of AI was the main dilemma for the validation and development of clinicomics methods. Currently, radiomics and deep learning can comprehensively analyse features from imaging and even from videos, making clinicomics a potential reality.
In the present study, we aimed to verify the performance of clinicomics-guided prognostic prediction for breast cancer via artificial intelligence and to investigate the accuracy of the created prediction models for metachronous DM, bone metastasis and visceral metastasis. These models can potentially guide metachronous DM screening and lead to the implementation of individualized prophylactic therapy for BC patients with a high risk for DM.
Materials and methods
Study design and participants
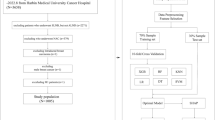
This case–control study protocol was approved by the Ethics Committee of Tianjin Medical University Cancer Institute & Hospital, Tianjin, China (EK2018125). A total of 6,703 consecutive BC patients from the hospital between January 2011 and December 2016 were included. The detailed inclusion and exclusion criteria were as follows: (1) a histopathological diagnosis of invasive BC through surgically resected specimens and/or needle biopsy; (2) availability of diagnostic-quality preoperative magnetic resonance imaging (MRI) scanning and ultrasound (US) images; (3) MRI scanning and US exam conducted before neoadjuvant therapy or surgical resection; (4) no DM present at diagnosis; and (5) follow-up data available for at least five years. Sixty-two patients with DM and 124 randomly selected patients without DM were included in the present study. The flow chart of the present study is shown in Fig. 1. The patients’ demographic and clinicopathological characteristics were collected from their medical records (Table 1). To validate the performance of the prediction model, the included patients were randomly divided into two sets: the training set (70%, N = 131) and the validation set (30%, N = 55).
MRI and US technique
All patients underwent MRI and US examinations within 2 weeks before breast surgery. Magnetic resonance images were acquired using scanners manufactured by two companies, a 1.5-T system (Signa Infinity Excite II, GE Healthcare) before 2013, and a 3.0-T MRI system (Discovery MR750, GE Medical Systems) after 2013. The detailed MRI parameters are shown in the Supplementary Materials and Methods. All ultrasound images were acquired using a GE LOGIQ7 or GE LOGIQ E9 ultrasound machine with a 6 ~ 15.0 MHz probe.
Imaging feature detection and radiomics signature construction
MRI and US images were retrieved from picture archiving and communication systems (PACS) for image segmentation and analysis. The lesions were segmented by a radiologist with more than 8 years of experience using ImageJ (https://imagej.nih.gov/ij/). Another experienced radiologist (with 30 years of experience) was consulted when the lesion boundaries in US were not determined clearly.
A total of 2569 radiomics features (855 features from magnetic resonance T2 weighted images (T2WI), 859 features from dynamic-contrast enhanced MRI (DCE-MRI) and 855 from US images) were extracted for each patient. Detailed information about the feature extraction algorithms is provided in Supplementary Table S1. The feature extraction method was performed using in-house software written with MATLAB R2018b (MathWorks, Inc., Natick, Massachusetts).
In order to test the prediction ability of different image types, we built models based on each type and a model based on the integrated features from all types. We followed a three-step procedure to determine reliable radiomic features. First, the Wilson test was used to identify features that were highly correlated to the biomarkers with a significant value (P < 0.05). Pearson correlation matrices were used to evaluate the correlation between the features, and the correlation coefficient greater than 0.8 was considered redundant. One of two features with a lower P-value was excluded. Subsequently, the optimal prognostic combination of features was selected by using the minor absolute contraction and Selection operator (LASSO) regression method. By calculating the radial score (radscore), the linear combination of each patient's selected features weighted by their respective coefficients was calculated to establish the prediction model [11]. A fixed 70%/30% training/rest set split was used, and tenfold cross-validation was performed to assess the true diagnostic potential of the model.
The clinicopathology and the clinicomics-based nomograms
Univariate analysis was used to evaluate the clinicopathological factors in the training set. Variables with P < 0.05 of univariate analysis was included in the Cox proportional hazards regression model, and the clinicopathological nomogram was established to predict DM risk in BC. We evaluated clinicomics-based nomogram to determine whether the model has the best performance in predicting DM risk in BC.
Statistical analysis
Continuous variables are expressed as the mean ± standard deviation (normally distributed) or median with interquartile range (abnormally distributed), while categorical variables are expressed as numbers and percentages. The predictive accuracy of nomogram was evaluated by the area under the receiver operating characteristic (ROC) curve and Harrell’s concordance index (C-index), while the calibration ability was evaluated by calibration curves. The difference in the area under the curve (AUC) between the training and validation datasets was tested by the P-value of Delong’s test. The integrated discrimination improvement (IDI) values were assessed to quantify the incremental prognostic improvement in the radiomic signature. The statistical analyses were conducted using R software (version 6.1, R Foundation for Statistical Computing, Vienna, Austria). A two-tailed difference with P < 0.05 was considered significant. The packages used in the current study included glmnet, time ROC, rms, survival, Hmisc and rmda.
Results
Characteristics of distant metastasis
The clinicopathological characteristics of the training (n = 131) and test (n = 55) sets are summarized in Table 1. The median incubation time of BC patients with DM was 14 months (range, 1–58 months). visceral (N = 42) was the most frequent metastasis site, followed by bone (N = 33) and brain (N = 10) sites.
Patient clinical characteristics and development of the clinical factor DM model
Among clinicopathologic characteristics, lymph node metastasis (P < 0.001), higher levels of CA153 (P < 0.001), carcinoma embryonic antigen (CEA) (P < 0.001) and CA125 (P = 0.003) were significantly associated with DM risk among BC patients in the training set (Table 1), and these factors were used to establish the clinicopathological model (Supplementary Figure S1). Subgroup analysis of age (younger than 50 years / older than 50 years) to predict DM was showed in Supplementary Table S2. Reproductive history (P = 0.036), lymph node metastasis (P < 0.001), and higher levels of CA153 (P < 0.001), CEA (P < 0.001) and CA125 (P = 0.015) were associated with bone metastasis risk (Table 2). Lymph node metastasis (P = 0.001), oestrogen receptor (ER)-positive status (P < 0.001), progesterone receptor (PR)-positive status (P < 0.001), higher levels of CA153 (P < 0.001), CEA (P = 0.004) and endocrinotherapy (P = 0.041) were associated with viscera metastasis risk (Table 3). ER positivity (P = 0.005) and higher levels of CA153 (P < 0.001), CEA (P < 0.001) and CA125 (P = 0.009) were associated with brain metastasis risk (Table 4).
Development and testing of the radiomics model
Four different feature sets were selected from the T2WI, DCE-MRI, US, and the combination of the imaging models. Through feature selection, 6, 4 and 1 features were selected from the T2WI, DCE-MRI, and US images, respectively. A total of 8 features were selected from the feature sets, including 6 features from T2WI and 2 features from DCE-MRI. These 8 features were used to build the optimal radiomics model based on the imaging models (Supplemental Materials and Methods).
The T2WI model yielded AUCs of 0.838 (95% CI: 0.753–0.923), 0.917 (95% CI: 0.859–0.975), 0.925 (95% CI: 0.869–0.981) in the training set and 0.792 (95% CI: 0.638–0.946), 0.794 (95% CI: 0.667–0.922), and 0.874 (95% CI: 0.777–0.972) in the validation set for 1-, 3-, and 5-year risk, respectively. The AUCs of the DCE-MRI model were 0.888 (95% CI: 0.816–0.960), 0.916 (95% CI: 0.870–0.961), 0.920 (95% CI: 0.876–0.964) in the training set and 0.729 (95% CI: 0.450–0.100), 0.800 (95% CI: 0.641–0.959), and 0.765 (95% CI: 0.619–0.911) in the validation set. The AUCs of the US model were 0.763 (95% CI: 0.655–0.871), 0.749 (95% CI: 0.662–0.835), 0.757 (95% CI: 0.673–0.842) in the training set and 0.567 (95% CI: 0.162–0.972), 0.538 (95% CI: 0.330–0.747), and 0.512 (95% CI: 0.328–0.696) in the validation set for 1-, 3-, and 5-year risk, respectively. The DeLong test showed that there was no significant difference between the AUCs of the training set and validation set in the four radiomics signature models (all P > 0.05). The comparative analysis among different models was shown in Supplementary Figure S2. There was no statistical significance between the combined model and the DCE-MRI model (all P > 0.05), although the AUC of the combined model was higher than that of the DCE-MRI model.
When T2WI, DCE-MRI, and US images were combined, the radiomics signature model with 8 features exhibited the highest AUC and obtained the best diagnostic accuracy. The AUCs were 0.868 (95% CI: 0.795–0.942), 0.945 (95% CI: 0.900–0.989), and 0.950 (95% CI: 0.907–0.993) in the training set and 0.850 (95% CI: 0.720–0.980), 0.798 (95% CI: 0.673–0.922), and 0.867 (95% CI: 0.772–0.962) in the validation set. The sensitivity, specificity, and AUC of each radiomics model are shown in Fig. 2 and Supplementary Table S3.
Differences in the prediction performance between the clinicopathological-feature model and the clinicomics-based model for distant metastasis
As shown in Fig. 3, the clinicomics-based model provided a better performance in the training set (clinical: C-index = 0.725; clinicomics-based model: C-index = 0.882) and the validation set (clinical: C-index = 0.659; clinicomics-based model: C-index = 0.812). The areas under the curve (AUCs) at different follow-up times (1, 2, and 3 years) also confirmed that the clinicomics-based model had good prognostic accuracy in the training and validation sets. The calibration curves for the clinicomics-based model at 1 year, 2 years, and 3 years showed good agreement between the actual and predicted risk in the training and validation sets (Fig. 3). The clinicomics-based model showed a relatively better performance than the clinical model (IDI = 0.302, 95% CI: 0.174–0.431, P < 0.001).
Construction of the clinicomics-based prediction model for non-distant metastasis (DM). A A nomogram was developed in the training data set with clinicopathological characteristics and RadScore. Calibration curves and ROC of the nomogram for the training set (B and D) and validation set (C and E). F Decision curve analysis derived from the validation cohort
The decision curve analysis revealed that the clinicomics-based nomogram had relatively good clinical performance compared with other models. These results suggested that the radiomic signature provided additional value for personalized DM prediction (Fig. 3F).
Construction and validation of the predictive bone metastasis nomogram
A predictive bone metastasis nomogram was constructed, which included reproductive history, lymph node metastasis, CA153, CEA, CA125, and radiomics data (Fig. 4). The model showed good performance in both the primary (C index, 0.931; 95% CI: 0.868, 0.975) and validation cohorts (C index, 0.956; 95% CI: 0.926, 0.986). The ROC and calibration curves are shown in Fig. 4.
Construction and validation of the predictive visceral metastasis nomogram
A predictive visceral metastasis nomogram was constructed, which included lymph node metastasis, CA153, CA153, ER, PR, HER2 and radiomics data (Fig. 5). The model showed good performance in both the primary (C index, 0.895; 95% CI: 0.850–0.941) and validation cohorts (C index, 0.946; 95% CI: 0.918–0.975). The ROC and calibration curves are shown in Fig. 5.
Discussion
Distant metastasis in breast cancer can be divided into two types based on DM diagnosis: synchronous DM at initial and metachronous DM that arises later in the disease course. There is sufficient evidence that the significant difference between synchronous DM and metachronous DM can be found, including clinicopathologic characteristics, treatment responses and survival outcomes [12,13,14]. Compared with patients with synchronous DM, patients with metachronous DM usually show better survival outcomes [15]. Two main potential explanations were summarized: 1. BC patients with synchronous DM usually visit the hospital later than patients with metachronous DM. 2. Treatment response is usually worse in BC patients with synchronous DM due to faster BC progression [16]. Few studies of metachronous DM prediction in BC have been performed, and conducting such studies may guide individualized DM screening during the disease course.
This is the first study to investigate and validate DM risk prediction through cliniomics in BC patients. The concept of “omics” has been widely studied and applied, including genomics, proteomics, radiomics, and metabolomics [17, 18]. “Omics” are developed based on patterns of changes in complex processes. Thus, this concept and approach can also be applied to patients in the clinic using the multidimensional features (complete history, epidemiological distribution, physical examination, laboratory test, imaging evaluation and histological examination) that are routinely investigated in a clinical evaluation of a patient. Before the development of AI techniques, clinicomics remained a hypothesis due to the difficulty of imaging/video data dimension reduction and integration.
Breast US, with a proper balance of specificity and sensitivity, is widely accepted as the first choice for breast lesion evaluation [19,20,21]. Compared with other imaging evaluations, MRI has the highest sensitivity for invasive lesion detection, and such sensitivity is not impaired by fibroglandular tissue, fibrous scarring, radiotherapy, breast implants, or other breast reconstruction [19, 22, 23]. A recent study combined clinical features and MRI features and suggested that this method performs well for brain metastasis prediction before radiosurgery [24]. MRI features were suggested to be of significance for DM prediction in locally advanced rectal cancer [25]. Our previous study verified the significance of MRI for DM prediction in BC [26]. In the latest study, the combination of MRI and US showed satisfactory prediction ability for residual tumour size in early breast cancer [27]. A study evaluating the accuracy of various imaging methods in BC concluded that MRI had advantages for evaluating suspicious breast lumps but had low specificity. US was able to compensate for MRI’s low specificity in image formation [28]. Therefore, we combined the features from MRI and US to develop a clinicomics approach to DM prediction in BC patients.
The most important finding of the present study was that we created and validated DM prediction in BC through clinicomics. This approach can potentially be used in various clinical fields. The features extracted from the high-dimensional images can provide additional information. Factors including complete history, epidemiological distribution, physical examination, and laboratory tests can reflect each patient’s reaction to the specific tumour. Thus, a comprehensive judgement can be reached using AI-guided clinicomics analyses, and such analyses have potential applications.
Three prediction models were created to respectively predict DM risk, bone metastatic risk and visceral metastasis risk. Each model showed a good ability to predict DM in BC, which could be used to stratify BC patients into different groups according to their risk for DM. Among the created models, the DM prediction model can be used for survival evaluation and general DM screening. A bone metastasis prediction model is of significance for the prevention and treatment of bone metastasis and potentially reduces adverse skeletal-related events. A visceral metastasis prediction model can guide DM screening of viscera through imaging examination and reduce unnecessary radiation exposure.
Our study has some limitations. First, the external validation with large population and various human ethnicities will be needed. Second, further studies will be needed to analyse the effect of incorporating other imaging data into the predictive nomogram, such as mammography. Finally, several serological biomarkers indicating metastasis potential of tumors such as EZH2 and PDGF were not analyzed in the study [29].
Conclusion
We validated the importance of clinicomics for predicting the risk for DM and organ-specific DM in BC. Three AI-guided clinicomics prediction models in BC were created: (1) the DM prediction model, (2) the bone metastasis prediction model, and (3) the visceral metastasis prediction model. These models can potentially guide metachronous DM screening and the implementation of individualized therapy in BC. AI-guided clinicomics strategies possess the potential for wide application in the clinic.
Availability of data and materials
The related data and materials are available for sharing upon request to Wenjuan Ma and Chao Zhang.
Abbreviations
- DM:
-
Distant Metastasis
- BC:
-
Breast Cancer
- MRI:
-
Magnetic Resonance Imaging
- ER:
-
Estrogen Receptor
- PR:
-
Progesterone Receptor
- T2WI:
-
T2-Weighted Imaging
- LASSO:
-
Least Absolute Shrink Age and Selection Operator
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area Under the Curve
- IDI:
-
Integrated Discrimination Improvement
- NRI:
-
Net Reclassification Improvement
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Wang R, Zhu Y, Liu X, Liao X, He J, Niu L. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer. 2019;19(1):1091. https://doi.org/10.1186/s12885-019-6311-z.
Riggio AI, Varley KE, Welm AL. The lingering mysteries of metastatic recurrence in breast cancer. Brit J Cancer. 2021;124(1):13–26. https://doi.org/10.1038/s41416-020-01161-4.
Gouri A, Benarba B, Dekaken A, Aoures H, Benharkat S. Prediction of late recurrence and distant metastasis in early-stage breast cancer: overview of current and emerging biomarkers. Curr Drug Targets. 2020;21(10):1008–25. https://doi.org/10.2174/1389450121666200312105908.
Wu Q, Li J, Zhu S, Wu J, Chen C, Liu Q, et al. Breast cancer subtypes predict the preferential site of distant metastases: a SEER based study. Oncotarget. 2017;8(17):27990–6. https://doi.org/10.18632/oncotarget.15856.
Liu X, Zhang D, Liu Z, Li Z, Xie P, Sun K, et al. Deep learning radiomics-based prediction of distant metastasis in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy: a multicentre study. EBioMedicine. 2021;69:103442. https://doi.org/10.1016/j.ebiom.2021.103442.
Tian L, Zhang D, Bao S, Nie P, Hao D, Liu Y, et al. Radiomics-based machine-learning method for prediction of distant metastasis from soft-tissue sarcomas. Clin Radiol. 2021;76(2):119–58. https://doi.org/10.1016/j.crad.2020.08.038.
Liu Z, Meng X, Zhang H, Li Z, Liu J, Sun K, et al. Predicting distant metastasis and chemotherapy benefit in locally advanced rectal cancer. Nat Commun. 2020;11(1):4308. https://doi.org/10.1038/s41467-020-18162-9.
Rennard SI, Stoner JA. Challenges and opportunities for combination therapy in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2005;2(4):391–3, 394-5. https://doi.org/10.1513/pats.200504-046SR.
Maojo V, Crespo J, de la Calle G, Barreiro J, Garcia-Remesal M. Using web services for linking genomic data to medical information systems. Method Inform Med. 2007;46(4):484–92. https://doi.org/10.1160/me9056.
Qi L, Chen D, Li C, Li J, Wang J, Zhang C, et al. Diagnosis of ovarian neoplasms using nomogram in combination with ultrasound image-based radiomics signature and clinical factors. Front Genet. 2021;12:753948. https://doi.org/10.3389/fgene.2021.753948.
Bindea G, Mlecnik B, Galon J. Tumor spread or siege immunity: dissemination to distant metastasis or not. Oncoimmunology. 2021;10(1):1919377. https://doi.org/10.1080/2162402X.2021.1919377.
Liu Q, Zhang R, Michalski CW, Liu B, Liao Q, Kleeff J. Surgery for synchronous and metachronous single-organ metastasis of pancreatic cancer: a SEER database analysis and systematic literature review. Sci Rep-Uk. 2020;10(1):4444. https://doi.org/10.1038/s41598-020-61487-0.
Sabet A, Binse I, Dogan S, Koch A, Rosenbaum-Krumme SJ, Biersack HJ, et al. Distinguishing synchronous from metachronous manifestation of distant metastases: a prognostic feature in differentiated thyroid carcinoma. Eur J Nucl Med Mol I. 2017;44(2):190–5. https://doi.org/10.1007/s00259-016-3485-3.
Miyata M, Ohguri T, Yahara K, Yamaguchi S, Imada H, Korogi Y. Salvage radiotherapy for solitary metachronous bone metastasis in patients with breast cancer. Anticancer Res. 2017;37(5):2575–9. https://doi.org/10.21873/anticanres.11601.
Alkabban FM, Ferguson T. Breast cancer. 2021.
Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics data integration, interpretation, and its application. Bioinform Biol Insights. 2020;14:1215734613. https://doi.org/10.1177/1177932219899051.
Chakraborty S, Hosen MI, Ahmed M, Shekhar HU. Onco-Multi-OMICS approach: a new frontier in cancer research. Biomed Res Int. 2018;2018:9836256. https://doi.org/10.1155/2018/9836256.
Pereira RO, Luz L, Chagas DC, Amorim JR, Nery-Júnior EJ, Alves A, et al. Evaluation of the accuracy of mammography, ultrasound and magnetic resonance imaging in suspect breast lesions. Clinics (Sao Paulo, Brazil). 2020;75:e1805. https://doi.org/10.6061/clinics/2020/e1805.
Guo R, Lu G, Qin B, Fei B. Ultrasound imaging technologies for breast cancer detection and management: a review. Ultrasound Med Biol. 2018;44(1):37–70. https://doi.org/10.1016/j.ultrasmedbio.2017.09.012.
Brem RF, Lenihan MJ, Lieberman J, Torrente J. Screening breast ultrasound: past, present, and future. Am J Roentgenol. 2015;204(2):234–40. https://doi.org/10.2214/AJR.13.12072.
Partovi S, Sin D, Lu Z, Sieck L, Marshall H, Pham R, et al. Fast MRI breast cancer screening - ready for prime time. Clin Imag. 2020;60(2):160–8. https://doi.org/10.1016/j.clinimag.2019.10.013.
Houssami N, Turner RM, Morrow M. Meta-analysis of pre-operative magnetic resonance imaging (MRI) and surgical treatment for breast cancer. Breast Cancer Res Tr. 2017;165(2):273–83. https://doi.org/10.1007/s10549-017-4324-3.
Hsu CY, Xiao F, Liu KL, Chen TL, Lee YC, Wang W. Radiomic analysis of magnetic resonance imaging predicts brain metastases velocity and clinical outcome after upfront radiosurgery. Neuro-oncol Adv. 2020;2(1):a100. https://doi.org/10.1093/noajnl/vdaa100.
Chiloiro G, Rodriguez-Carnero P, Lenkowicz J, Casà C, Masciocchi C, Boldrini L, et al. Delta radiomics can predict distant metastasis in locally advanced rectal cancer: the challenge to personalize the cure. Front Oncol. 2020;10:595012. https://doi.org/10.3389/fonc.2020.595012.
Ma W, Wang X, Xu G, Liu Z, Yin Z, Xu Y, et al. Distant metastasis prediction via a multi-feature fusion model in breast cancer. Aging. 2020;12(18):18151–62. https://doi.org/10.18632/aging.103630.
Graeser M, Schrading S, Gluz O, Strobel K, Herzog C, Umutlu L, et al. Magnetic resonance imaging and ultrasound for prediction of residual tumor size in early breast cancer within the ADAPT subtrials. Breast Cancer Res. 2021;23(1):36. https://doi.org/10.1186/s13058-021-01413-y.
An Y. Comments on “Evaluation of the accuracy of mammography, ultrasound and magnetic resonance imaging in suspect breast lesions.” Clinics (Sao Paulo, Brazil). 2020;75:e2338. https://doi.org/10.6061/clinics/2020/e2338.
Jafari SH, Saadatpour Z, Salmaninejad A, Momeni F, Mokhtari M, Nahand JS, et al. Breast cancer diagnosis: Imaging techniques and biochemical markers. J Cell Physiol. 2018;233(7):5200–13. https://doi.org/10.1002/jcp.26379.
Acknowledgements
Not applicable.
Funding
The present study was sponsored by Natural Science Foundation of China (81801781, 82072004, 81903398, 82011530050). The present study was funded by Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-009A).
Author information
Authors and Affiliations
Contributions
Conceptualization, W.M., X.W., C.Z.; methodology, W.M., X.W., L.Q. and, J.C.; software, H.W., Y.L., Z.L.; validation, W.J., V.C., K.P.; investigation, W.M., X.W., L.Q. and C.Z.; resources, W.M., Q.L., C.Z.; data curation, X.W.; formal analysis, W.M. and X.W.; visualization, H.W., Y.L. and C.Z.; writing—original draft preparation, W.M. and C.Z.; writing—review and editing, C.Z., X.W., V.C.; supervision, V.C., C.Z., K.P.; funding acquisition, W.M., C.Z. and X.W., project administration, W.M., C.Z. and X.W.. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Tianjin Medical University Cancer Institute & Hospital. Informed consent was obtained from all participants and their parents or legal guardians before inclusion into the study.
Consent for publication
All authors expressed great pleasure to publish in this journal.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Supplemental Materials and Methods. Supplementary Table S1. Texture features used in this study. Supplementary Table S2. Clinicopathologic characteristics between breast cancer patients with /without distant metastasis in different age groups (Training set). Supplementary Table S3. Performance comparison among radiomics model in the training and validation set of different image types. Supplementary Figure S1. Construction of the clinicopathological model for predicting distant metastasis (DM). (A) A nomogram was developed in the training data set with clinicopathological characteristics. Calibration curves and ROC of the nomogram for the training set (B and D) and validation set (C and E). Supplementary Figure S2. The difference analysis among different radiomics signature models with DeLong test in training set (A-C) and validation set (D-F).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, C., Qi, L., Cai, J. et al. Clinicomics-guided distant metastasis prediction in breast cancer via artificial intelligence. BMC Cancer 23, 239 (2023). https://doi.org/10.1186/s12885-023-10704-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-023-10704-w