Abstract
Background
Salinomycin is a monocarboxylic polyether antibiotic and is a potential chemotherapy drug. Our previous studies showed that salinomycin inhibited cell growth and targeted CSCs in prostate cancer. However, the precise target of salinomycin action is unclear.
Methods
In this work, we analyzed and identified differentially expressed genes (DEGs) after treatment with or without salinomycin using a gene expression microarray in vitro (PC-3 cells) and in vivo (NOD/SCID mice xenograft model generated from implanted PC-3 cells). Western blotting and immunohistochemical staining were used to analyze the expression of ATP2A3 and endoplasmic reticulum (ER) stress biomarkers. Flow cytometry was used to analyze the cell cycle, apoptosis and intracellular Ca2+ concentration.
Results
A significantly upregulated gene, ATPase sarcoplasmatic/endoplasmatic reticulum Ca2+ transporting 3 (ATP2A3), was successfully identified. In subsequent studies, we found that ATP2A3 overexpression could trigger ER stress and exert anti-cancer effects in PC-3 and DU145 cells. ATP2A3 was slightly expressed, but the ER stress biomarkers showed strong staining in prostate cancer tissues. We also found that salinomycin could trigger ER stress, which might be related to ATP2A3-mediated Ca2+ release in PC-3 cells. Furthermore, we found that salinomycin-triggered ER stress could promote apoptosis and thus exert anti-cancer effects in prostate cancer cells.
Conclusion
This study demonstrates that ATP2A3 might be one of the potential targets for salinomycin, which can inhibit Ca2+ release and trigger ER stress to exert anti-cancer effects.
Similar content being viewed by others
Background
Prostate cancer is the second most common malignant cancer in male patients and has high morbidity and mortality [1]. In the past, hormone therapy, surgery and radiation therapy were requirements for advanced prostate cancer; however, since 2004, chemotherapy (docetaxel) has begun to play a significant role [2]. Consequently, finding and studying more specific chemotherapy drugs has important significance for advanced prostate cancer patients. Salinomycin is a monocarboxylic polyether antibiotic isolated from Streptomyces albus [3]. In 2009, Gupta and colleagues found that salinomycin had nearly 100-fold higher potency against breast cancer stem cells (CSCs) than paclitaxel in a screen of 16,000 compounds [4]. Salinomycin is considered a promising anti-tumor chemotherapy drug, which may reduce the resistance and relapse of cancer by killing cancer cells and CSCs [5].
It has been reported that salinomycin is an ionophore that transports cations (K+, Na+, Ca2+, and Mg2+) through cell membranes [6]. Salinomycin can increase intracellular cation concentrations and disrupt the osmotic balance, resulting in apoptosis [7]. In addition, salinomycin is found to inhibit the Wnt/β-catenin signaling pathway and selectively induces apoptosis [8, 9]; reduce the activity of ABC transporters [10]; induce oxidative stress [11], autophagy [12, 13], and anti-angiogenic and anti-tumorigenic activities [14]; inhibit EMT (Epithelial-mesenchymal transition) [15]; and inhibit cancer cell growth [16, 17]. Despite all of this evidence, the molecular mechanism for salinomycin remains elusive, and the precise target of salinomycin action is unclear.
In our previous studies, we found that salinomycin could kill CSCs in lung cancer and inhibit cell growth and target CSCs in prostate cancer [5, 18]. The cytotoxicity of salinomycin to human prostate cancer PC-3 cells was stronger than to nonmalignant prostate cells RWPE-1. Salinomycin induced apoptosis of PC-3 cells by Wnt/β-catenin signaling pathway. Salinomycin, but not paclitaxel, triggered more apoptosis in aldehyde dehydrogenase- (ALDH-) positive PC-3 cells, which were considered as the prostate cancer stem cells, suggesting that salinomycin may be a promising chemotherapeutic to target CSCs [5]. Furthermore, we found that salinomycin-induced autophagy blocks apoptosis via the ATG3/AKT/mTOR signaling axis in prostate cancer PC-3 cells [19]. Salinomycin induced apoptosis and autophagy in PC-3 cells. Interestingly, autophagy inhibition enhanced salinomycin-induced apoptosis. ATG3 was involved in the blockage of apoptosis by autophagy in salinomycin-treated PC-3 cells. ATG3 regulation might occur through the AKT/mTOR signaling axis [19]. However, our previous studies did not address the precise target of salinomycin action.
To investigate the mechanism of salinomycin, a microarray analysis was used to identify DEGs in vitro (PC-3 cells) and in vivo (NOD/SCID mice xenograft model generated from implanted PC-3 cells). ATPase sarcoplasmatic/ endoplasmatic reticulum Ca2+ transporting 3 (ATP2A3), a significantly upregulated gene, was successfully identified, which encoded one of the Ca2+-ATPases. It is known that ATP2A3 is localized in the ER membrane and involves in Ca2+ transport [20, 21]. Griffin et al. found that the expression of ATP2A3 was downregulated in Jurkat cells, reducing the transport of Ca2+ from the cytoplasm into the ER [22]. Other studies found that upregulation of ATP2A3 caused increases in reticular calcium content in the pheochromocytoma cell line PC12 and ultimately resulted in apoptosis [23].
In this study, we found that ATP2A3 might be a potential targets for salinomycin, which inhibits Ca2+ release and triggers ER stress. This finding could provide new clues for the mechanism of the salinomycin anti-cancer effects.
Methods
Cell culture, drugs and cell survival assay
Human prostate cancer PC-3 and DU145 cells (ATCC, Manassas, VA, USA) were cultured as previously described [5]. Salinomycin (Sigma-Aldrich, St Louis, MO, USA), BAPTA-AM (Selleckchem, Houston, TX, USA) were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich, St Louis, MO, USA). Sodium phenylbutyrate (4-PBA) was dissolved in water.
Tumorigenic studies in NOD/SCID mice
For tumorigenic studies, PC-3 cells were subcutaneously inoculated into the flanks of NOD/SCID male mice (5 weeks of age; Beijing HFK BioScience Co., Ltd. Beijing China). Mice were housed in a standard laboratory environment (temperature: 24 ± 2 °C; humidity: 50 ± 5%; 12 h day-night cycle) and treated intraperitoneally (i.p.) daily with either DMSO or salinomycin at a dose of 10 mg/kg/day/200 μL (each group was 5). After 3 weeks, the mice were euthanized by carbon dioxide inhalation followed by cervical dislocation. The xenografts were excised and pulverized in liquid nitrogen. Animal studies have approved by the animal ethics committee from South China university.
Gene expression microarray analysis
Cultured PC-3 cells were treated with 1.0 μM salinomycin or DMSO control for 24 h. Then, total RNA from the abovementioned cells or tumors was extracted with TRIzol reagent (Life Technologies, Inc., Carlsbad, CA, USA) according to the manufacturer’s protocol. Double-stranded cDNA (ds-cDNA) was synthesized from 5 μg of total RNA using a SuperScript ds-cDNA synthesis kit (Life Technologies, Inc., Carlsbad, CA, USA). Human 12 × 135 K Gene Expression Arrays (Roche NimbleGen) were hybridized at 42 °C for 16 to 20 h with 4 μg of Cy3 labeled ds-cDNA in NimbleGen hybridization buffer/hybridization component in a hybridization chamber (Hybridization System-NimbleGen Systems, Inc., Madison, WI, USA).
Microarray data acquisition and analysis
Slides were scanned at 5 μm/pixel resolution using an Axon GenePix 4000B scanner (Molecular Devices Corporation) piloted by GenePix Pro 6.0 software (Axon). Scanned images (TIFF format) were then imported into NimbleScan software (version 2.5) for grid alignment and expression data analysis. The expression data were normalized through quantile normalization and the Robust Multichip Average (RMA) algorithm included in the NimbleScan software. All gene level files were imported into Agilent GeneSpring GX software (version 11.5.1) for further analysis. Differentially expressed genes were identified through fold Change filtering.
Quantitative RT-PCR analysis
Quantitative RT-PCR (reverse transcription-polymerase chain reaction) was performed with FastStart Essential DNA Green Master (Roche, Mannheim, Germany). All reactions were performed on the Roche LightCycler 96 Real-Time PCR system (Roche Diagnostics GmbH, Mannheim, Germany). Individual values were normalized to the GAPDH loading control. Sequences of gene primers are listed in Table 1. The mRNA expression levels were analyzed using delta cycle threshold (ΔCt) values.
Immunohistochemistry staining
Human prostate cancer tissues and para-carcinoma tissues were obtained from the Second Affiliated Hospital, University of South China with institutional review board approval. Some sections were stained with HE. Immunohistochemistry staining was performed as described previously [24, 25] using ATP2A3 (1:500, GeneTex, San Antonio, TX, USA), BIP and ATF4 antibodies (all from CST, Danvers, MA, USA). Images were captured by a CCD camera (Olympus, Center Valley, PA).
Western blot analysis
Protein extraction and Western blotting were performed as previously described [26]. The primary antibodies BIP, PERK, ATF4, eIF2a, CHOP, Caspase 12, p-PERK, p-eIF2a, p-CaMK-II (all from CST, Danvers, MA, USA), CaMK-II (Bioworld Technology, Inc., Minneapolis, MN, USA) and ATP2A3 (GeneTex, San Antonio, TX, USA) were used to detect the ER stress. PARP, cleaved PARP antibodies (all from CST, Danvers, MA, USA) were used to detect apoptosis. The β-actin antibody (CST, Danvers, MA, USA) was used as an internal control.
Transmission electron microscopy
For electron microscopy, PC-3 cells were plated at a density of 106 cells in 100 mm cell culture dishes. The cells were treated with salinomycin for 24 h. Subsequently, the cells were harvested, washed in PBS, and fixed in 2.5% glutaraldehyde/0.2 M phosphate buffer solution (pH 7.4) at 4 °C. Finally, cells were detected by transmission electron microscopy (Servicebio Co., Ltd. Wuhan, China).
Immunofluorescence staining
Immunofluorescence staining was performed after salinomycin treatment, as previously described [27]. BIP (1:200, Abcam, Cambridge, MA, USA) and CHOP (1:200, CST, Danvers, MA, USA) antibodies were used in this experiment. Secondary antibodies conjugated with Alexa Fluor-647 (Abcam, Cambridge, MA, USA) and Alexa Fluor-488 (Abcam, Cambridge, MA, USA) were used. The fluorophore-labeled cells were examined and analyzed by laser scanning confocal microscopy (Olympus, Tokyo, Japan).
Detection of [Ca2+]i by Fluo-3/AM
Intracellular Ca2+ release was detected using the fluorescent probe Fluo-3/AM (Beyotime Biotechnology Co., Haimen, China). For salinomycin treatment, PC-3 and DU145 cells were incubated with Fluo-3/AM stock solution (0.5–5 μM) at 37 °C in the dark for 10 min following three washes with PBS. Samples were analyzed by a FACS-Calibur (BD Biosciences, San Jose, CA, USA), and fluo-3 was detected using a 530/30 nm filter. The arithmetic mean of fluo-3 fluorescence intensity was expressed as [Ca2+]i. Images of these cells were also captured using an inverted fluorescence microscope (Carl Zeiss, Jena, Germany) to observe the green fluorescence. The results were averaged from three independent experiments.
Plasmid construction and cell transfection
To construct recombinant ATP2A3 plasmids, the ORF of ATP2A3 was amplified by PCR and subcloned into the pCDH cDNA cloning lentivector (Cat#CD513B-1; SBI, Mountain View, CA) using the following primers:5′-CCCAAGCTTATGGAGGCGGCGCATCTG-3′ (forward, Hind III included) and 5′-CCGCTCGAGCTTCTGGCTCATTTCGTGC-3′ (reverse, Xho I included). All constructs were verified by sequencing (Sanggon Biotech Co., Ltd., Shanghai, China). The pDsRed2-ER plasmid was purchased from Clontech (Palo Alto, CA, USA). Cell transfection was performed with TurboFect™ in vitro Transfection Reagent (Fermentas, Glen Burnie, MD, USA) according to the manufacturer’s instructions.
Small interfering RNA knockdown
Small interfering RNA (siRNA) was used to knockdown ATP2A3 expression in PC-3 cells. ATP2A3-siRNAs and negative control sequences were synthesized by RiboBio Co., Ltd. (Guangzhou, China). The siRNAs were transfected by Lipofectamin 2000 (Invitrogen, Life Technologies, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell cycle and apoptosis analysis
PC-3 cells were seeded in six-well plates and then transfected with pCDH cDNA-ATP2A3 or empty vector for 24–48 h. For cell cycle analysis, cells were fixed and stained as previously described [26]. For apoptosis analysis, apoptosis and necrosis were evaluated by annexin V-FITC/PI staining as previously described [26]. The samples were analyzed by a FACS-Calibur (BD Biosciences, San Jose, CA, USA).
Statistical analysis
The results were representative of at least three replicates except where specified and are shown as the mean ± SD. To compare the rates of cell apoptosis, a chi-square test was used. For WB of protein expression, ImageJ was used to quantify the density of each band, and then the significance of the fold change was determined with one-way analysis of variance (ANOVA); the significance of the difference of mean fluorescence between groups was also tested with ANOVA. The LSD t-test was used to compare the groups. All statistical analyses were performed using the SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) for Windows. A P value < 0.05 was considered statistically significant.
Results
Identification of DEGs induced by salinomycin in vitro and in vivo
Previously, we found that salinomycin inhibited PC-3 cell proliferation and decreased xenograft tumor size [5]. To investigate the mechanism of salinomycin, a microarray analysis was used to identify differentially expressed genes (DEGs) in vitro (PC-3 cells) and in vivo (NOD/SCID mice xenograft model generated from implanted PC-3 cells) (Fig. 1a). Based on the threshold of P-value ≤0.001 and fold change ≥2.0, Venn diagrams showed that 150 DEGs were obtained (Fig. 1b). All target values were log-2 base transformed, and then a heatmap was generated for the 150 DEGs (Fig. 1c). The heatmap results showed distinguishable gene expression profiling between samples.
Gene expression changes induced by salinomycin in vitro and in vivo. a. Log–log scatter-plot of the microarray results. Plots in region Y represent upregulated genes, while plots in region X represent downregulated genes. b. Venn diagram displaying the number of DEGs. c. Heatmap representation of genes whose expression is differently regulated by salinomycin in vitro and in vivo. d. Chart showing the enriched gene ontology (GO) terms in the category of biological processes for DEGs. e. Chart showing the enriched gene ontology (GO) terms in the category of molecular functions for DEGs. f. Validation of the expression of DEGs by quantitative RT-PCR. The expression levels of mRNA were analyzed using delta cycle threshold (ΔCt) values
For the identified DEGs, we performed enrichment analysis in GO categories. The results showed that secondary metabolic process (GO:0019748, false discovery rate (FDR) =1E0) and plasma membrane organization (GO:0007009, FDR =1E0) were significantly enriched in the biological processes (Fig. 1d), and G-protein coupled receptor activity (GO:0004930, FDR =1.77E-1) was significantly enriched in molecular functions (Fig. 1e).
The P-value ≤0.001 and fold change ≥3.0 thresholds were set to select genes from the 150 DEGs. This search identified 10 genes that fit these criteria (Table. 2). Then, the 10 identified genes were validated by quantitative RT-PCR. Seven gene expression profiles were similar to those revealed by the microarray data. A significantly upregulated gene, ATP2A3, which is known to be involved in Ca2+ transport [20], was successfully identified (Fig. 1f). Thus, we hypothesized that ATP2A3 may be a potential target for salinomycin in PC-3 cells.
ATP2A3 overexpression triggered ER stress in prostate cancer cells
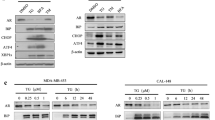
We validated the microarray and quantitative PCR results by Western blotting in prostate cancer cells. PC-3 or DU145 cells were treated with 0 (vehicle, DMSO), 0.5, 1.0 and 2.0 μM salinomycin for 24 h, and the expression of ATP2A3 was measured by WB. The results showed that ATP2A3 was upregulated by salinomycin treatment, and higher concentrations resulted in higher expression levels. To investigate the time-course of salinomycin-induced ATP2A3 expression, PC-3 or DU145 cells were also treated with 1.0 μM salinomycin for 0, 6, 12 or 24 h. The WB showed that ATP2A3 was progressively induced by salinomycin (Fig. 2a). The data therefore showed that salinomycin upregulates ATP2A3 expression in dose- and time-dependent manners.
ATP2A3 overexpression triggered ER stress. a. Salinomycin induced ATP2A3 upregulation in PC-3 and DU145 cells. b. Flow cytometry showed that ATP2A3 overexpression induced cell-cycle arrest in PC-3 and DU145 cells. *P < 0.05, vs. empty vector. c. Western blotting showed that ATP2A3 overexpression changed cell-cycle regulatory protein expression levels. d. Annexin V-FITC/PI staining showed that ATP2A3 overexpression induced PC-3 and DU145 cells apoptosis. *P < 0.05, vs. empty vector. e. Western blotting showed that ATP2A3 overexpression induced cleavage of PARP. f. ATP2A3 overexpression triggered ER stress in PC-3 and DU145 cells. Western blotting analysis of BIP, CHOP, Caspase 12, ATF4 expression and PERK, eIF2a phosphorylation level. g. Immumohistochemical staining showed that ATP2A3 and ER stress biomarkers (BIP and ATF4) expression in prostate cancer and para-carcinoma tissues. Scale bars, 10 μm
To further confirm the role of ATP2A3, we overexpressed ATP2A3 in prostate cancer cells. The data showed that ATP2A3 overexpression arrested the progression of the cell cycle (Fig. 2b) and decreased the protein levels of cyclin D1 and cyclin E1 (Fig. 2c). Furthermore, ATP2A3 overexpression induced apoptosis (Fig. 2d), and triggered PARP cleavage (Fig. 2e). These results suggest that ATP2A3 overexpression can cause cell cycle arrest and induce apoptosis in prostate cancer cells.
It is known that ATP2A3 controls Ca2+ transport and that its aberrant expression can cause ER stress [28, 29]. Thus we analyzed the effect of ATP2A3 overexpression on ER stress in prostate cancer. As expected, ATP2A3 overexpression caused the upregulation of ER stress biomarkers (BIP, CHOP, Caspase 12 and ATF4). Furthermore, the phosphorylation levels of PERK and eIF2a also increased significantly (Fig. 2f). These results suggest that ATP2A3 overexpression can trigger ER stress.
Immunohistochemistry staining was used to evaluate the expression of ATP2A3 and ER stress biomarkers (BIP and ATF4) in human prostate cancer and para-carcinoma tissues. The data showed weak ATP2A3 staining, but the ER stress biomarkers were strongly stained in cancer tissues (Fig. 2g). In conclusion, our results suggest that salinomycin can upregulate ATP2A3 expression, which triggers ER stress to exert anti-cancer effects in prostate cancer cells.
Salinomycin-triggers ER stress in PC-3 cells
We analyzed the effect of salinomycin on ER stress in prostate cancer PC-3 cells. As expected, salinomycin upregulated the expression of BIP, ATF4, CHOP and caspase-12. The phosphorylation levels of PERK and eIF2a also increased significantly (Fig. 3a and b). Furthermore, we measured the protein levels of BIP and CHOP by immunofluorescence staining. Consistent with the Western blotting data, the BIP and CHOP antibody staining signals were stronger after salinomycin treatment than before treatment (Fig. 3c).
Salinomycin triggers ER stress. a. Western blotting analyzed BIP, CHOP, Caspase 12 and ATF4 expression, and PERK and eIF2a phosphorylation in PC-3 cells treated with indicated concentration of salinomycin or DMSO as control (CON). b. The histogram shows fold protein expression normalized to control (ImageJ quantification of blots density from A. was performed, with three independent replicates.*P < 0.05 vs. control). c. The expression of BIP and CHOP in PC-3 cells treated as A. were measured by immunofluorescence. Scale bars, 10 μm. d. Representative immunofluorescence images of PC-3 cells transfected with pDsRed-ER and then treated with salinomycin. Scale bars, 10 μm. e. Transmission electron microscopy to show salinomycin-treated PC-3 cells. Scale bars, 1 μm
To further confirm that salinomycin-triggers ER stress, we transfected PC-3 cells with the pDsRed2-ER plasmid, which was designed for fluorescent labeling of the endoplasmic reticulum and then treated cells with salinomycin. The results showed that the fluorescent labeling of the endoplasmic reticulum became dim (Fig. 3d). Next, we observed the endoplasmic reticulum by transmission electron microscopy. The results showed that the ER membrane of salinomycin-treated PC-3 cells was dilated (Fig. 3e). Our data suggest that salinomycin can trigger ER stress in PC-3 cells.
Salinomycin-triggered ER stress might be related to ATP2A3-mediated Ca2+ release
Because ATP2A3 re-sequesters cytoplasmic Ca2+ to the sarcoplasmatic/ endoplasmatic reticulum store, we determined whether salinomycin influences Ca2+ release. PC-3 or DU145 cells were treated with the indicated concentration of salinomycin for 24 h, stained with the calcium indicator Fluo-3 AM and analyzed with flow cytometry. As illustrated in Fig. 4a and b, salinomycin significantly inhibited Ca2+ release in prostate cancer cells. Next, we transfected PC-3 cells with siRNA to inhibit ATP2A3 expression. Of the three synthetic siRNAs, ATP2A3#2 and ATP2A3#3 greatly knocked down ATP2A3 expression (Fig. 4c). PC-3 cells transfected with siRNA ATP2A3#2 or ATP2A3#3 for 48 h were further treated with 1.0 μM salinomycin for 24 h; afterward, WB was performed to detect ER stress markers (Fig. 4d), or staining with Fluo-3 AM was performed to detect Ca2+ concentration changes (Fig. 4e). The results showed that ATP2A3 silencing weakened salinomycin-triggered ER stress, as salinomycin induced upregulation of BIP and CHOP was constrained in cells transfected with siRNA ATP2A3#2 and ATP2A3#3. Similarly, reduced salinomycin-inhibited Ca2+ release was also observed. Our results suggest that salinomycin inhibition of Ca2+ release might be related to ATP2A3 in PC-3 cells.
Salinomycin-triggered ER stress might be related to ATP2A3-mediated Ca2+ release. a. Flow cytometry showed that salinomycin inhibited Ca2+ release in PC-3 and DU145 cells. b. Images were captured using an inverted fluorescence microscope after salinomycin treatment. Scale bars, 10 μm. c. ATP2A3 siRNAs (#1, #2 and #3) were respectively transfected in PC-3 cells for 48 h. Western blotting analyzed ATP2A3 protein expression. d. ATP2A3 silencing weakened salinomycin-triggered ER stress in PC-3 cells. e. ATP2A3 silencing reduced salinomycin-inhibited Ca2+ release in PC-3 cells. f. Salinomycin changed CaMK activity. The CaMK-II protein expression level and phosphorylation level (Thr 286) were analyzed by Western blotting. g. Western blotting showed that BAPTA-AM (10 μM, an intracellular calcium chelator) weakened salinomycin-triggered ER stress
The calcium/calmodulin kinases (CaMKs) are important mediators of intracellular Ca2 + [30]. Thus, we measured the CaMK-II protein expression level and phosphorylation level (Thr 286) to indirectly ascertain CaMK activity changes [31, 32]. The results showed that salinomycin treatment reduced the protein level of CaMK-II, but the phosphorylation level did not change significantly (Fig. 4f). These data suggest that salinomycin-mediated inhibition of Ca2+ release might be related to CaMKs via ubiquitin mediated protein degradation [33], which will be one focus of our future studies.
It has been reported that alterations in Ca2+ homeostasis are implicated in ER stress. Thus, we pretreated PC-3 cells with BAPTA-AM (10 μM, an intracellular calcium chelator) to assess whether salinomycin triggers ER stress by releasing Ca2+. As expected, the data showed that BAPTA-AM could weaken salinomycin-triggered ER stress (Fig. 4g). Our results suggest that salinomycin-triggered ER stress might be related to ATP2A3-mediated Ca2+ release in PC-3 cells.
Salinomycin triggers ER stress leading to apoptosis
Since ER stress has been shown to play a crucial role in apoptosis [34], we co-treated PC-3 cells with salinomycin and 4-phenylbutyrate (4-PBA, 1 mM, an ER stress antagonist) for 24 h. WB was performed to detect the expression of BIP, CHOP, cleaved caspase3 and cleaved PARP. The data showed that 4-PBA significantly reduced salinomycin-triggered ER stress (Fig. 5a) and hampered caspase3 and PARP cleavage (Fig. 5b). The treated cells were also stained with Annexin V-FITC to detect apoptosis. Flow cytometry showed that 4-PBA also significantly decreased salinomycin-induced apoptosis (Fig. 5c). These results suggest that ER stress can promote salinomycin-triggered apoptosis in PC-3 cells.
Salinomycin triggers ER stress leading to apoptosis. a. Western blotting showed that 4-PBA weakened salinomycin-triggered ER stress. b. Western blotting showed that 4-PBA weakened salinomycin-induced apoptosis. c. Flow cytometry showed that 4-PBA weakened salinomycin-induced apoptosis. d. Flow cytometry showed that ATP2A3 silencing weakened salinomycin-induced apoptosis
We also transfected PC-3 cells with siRNA to inhibit ATP2A3 expression. Annexin V-FITC staining illustrated that ATP2A3 silencing could inhibit salinomycin-induced apoptosis (Fig. 5d). In conclusion, our data suggest that ER stress promotes salinomycin-induced apoptosis, which might be related to ATP2A3 in PC-3 cells.
Discussion
Salinomycin has been shown to exert anti-cancer effects in human prostate cancer as well as in other cancers [35]. However, the molecular mechanism of salinomycin is not completely known. Therefore, a microarray-based approach was used and a significantly upregulated gene, ATP2A3, was successfully identified in this study. ATP2A3 is involved in Ca2+ transport [20]. Coincidentally, a recent study showed that salinomycin’s action is comparable to that of nigericin (K+/H+ exchanger) [36]. Therefore, we hypothesized that the anti-cancer effects of salinomycin might be associated with cation transport in prostate cancer cells.
Our results showed that salinomycin upregulated the expression of ATP2A3 in PC-3 and DU145 prostate cancer cells. The ATP2A3 gene encodes Ca2+-ATPase3 from the sarco/endoplasmic reticulum (SERCA3), which is expressed in endothelial and epithelial tissues [37, 38]. We found that ATP2A3 was slightly expressed in human prostate cancer tissues, which suggested that the expression of ATP2A3 might be correlated with prostate tumorigenesis. Thus, we investigated the effects of ATP2A3 in PC-3 and DU145 prostate cancer cells. The results showed that ATP2A3 overexpression inhibited cell cycle progression, induced apoptosis, and markedly triggered ER stress. Consistent with previous reports [12, 39], salinomycin triggered ER stress in prostate cancer PC-3 cells.
ER stress is a physiological response that is caused by internal or external stimuli such as oxidative stress [40], ischemia [41], and Ca2+ disorders [20]. ER stress is orchestrated by the unfolded protein response (UPR), and failure to adapt to ER stress results in apoptosis [42]. Transient ER stress could activate UPR and promote cell survival [43]. When ER stress is not mitigated, the UPR triggers apoptosis. The UPR is mediated by at least three major stress sensors: IRE1, ATF6 and PERK [42]. Each stress sensor uses a unique mechanism to promote the activation of a specific transcription factor and the upregulation of a subset of UPR target genes. In this study, we found that ATP2A3 overexpression and salinomycin treatment triggered ER stress by the PERK sensor in PC-3 cells. However, ATP2A3 silencing reduced salinomycin-triggered ER stress. Based on these findings, we postulated that salinomycin-triggered ER stress might be related to ATP2A3 upregulation in PC-3 cells.
ATP2A3 is a fundamental for maintaining intracellular [Ca2+] homeostasis by pumping Ca2+ into the ER of eukaryotic cells [44]. [Ca2+]i is a ubiquitous second messenger that operates with versatility in the regulation of several physiological events, such as apoptosis [45]. Alterations to the Ca2+ transport or homeostasis machinery have detrimental effects on survival and health [46]. We found that salinomycin inhibited Ca2+ release, thereby resulting in ER stress in PC-3 cells. As expected, ATP2A3 silencing reduced the salinomycin-mediated inhibition of Ca2+ release. These results suggest ATP2A3 might be a potential target for salinomycin, which inhibits Ca2+ release and triggers ER stress to exert anti-cancer effects. Furthermore, our data suggest that salinomycin-mediated inhibition of Ca2+ release might be related to CaMKs via ubiquitin mediated protein degradation [33], which will be one focus of our future studies.
Conclusions
In this study, we identified DEGs) after salinomycin treatment or no-treatment in vitro and in vivo. We further identified ATP2A3 as a potential target for the molecular mechanism of salinomycin, which might inhibit Ca2+ release and trigger ER stress to exert its anti-cancer effects.
Abbreviations
- [Ca2+]i:
-
Intracellular Ca2+
- ATP2A3:
-
ATPase sarcoplasmatic/ endoplasmatic reticulum Ca2+ transporting 3
- CSCs:
-
Cancer stem cells
- DGEs:
-
Differentially expressed genes
- EMT:
-
Epithelial-mesenchymal transition
- ER stress:
-
Endoplasmic reticulum stress
- FDR:
-
False discovery rate
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- siRNA:
-
small interfering RNA
- UPR:
-
Unfolded protein response
References
Dai X, Liu L, Liang Z, Guo K, Xu S, Wang H. Silencing of lncRNA MALAT1 inhibits cell cycle progression via androgen receptor signaling in prostate cancer cells. Pathol Res Pract. 2019.
Sartor O. Advanced prostate cancer update 2018. Asia Pac J Clin Oncol. 2018;14 Suppl 5:9–12.
Naujokat C, Fuchs D, Opelz G. Salinomycin in cancer: a new mission for an old agent. Mol Med Rep. 2010;3(4):555–9.
Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–59.
Zhang Y, Liu L, Li F, Wu T, Jiang H, Jiang X, Du X, Wang Y. Salinomycin exerts anticancer effects on PC-3 cells and PC-3-derived Cancer stem cells in vitro and in vivo. Biomed Res Int. 2017;2017:13.
Mitani M, Yamanishi T, Miyazaki Y. Salinomycin: a new monovalent cation ionophore. Biochem Biophys Res Commun. 1975;66(4):1231–6.
Zhou J, Li P, Xue X, He S, Kuang Y, Zhao H, Chen S, Zhi Q, Guo X: Salinomycin induces apoptosis in cisplatin-resistant colorectal cancer cells by accumulation of reactive oxygen species. Toxicol Lett 2013, 222(2):139–145.
Lu D, Choi MY, Yu J, Castro JE, Kipps TJ, Carson DA. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc Natl Acad Sci U S A. 2011;108(32):13253–7.
Mao J, Fan S, Ma W, Fan P, Wang B, Zhang J, Wang H, Tang B, Zhang Q, Yu X, et al. Roles of Wnt/beta-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis. 2014;5:e1039.
Meng S, Wu H, Wang L, Zhang B, Bai L. Enhancement of antibiotic productions by engineered nitrate utilization in actinomycetes. Appl Microbiol Biotechnol. 2017;101(13):5341–52.
Ketola K, Hilvo M, Hyotylainen T, Vuoristo A, Ruskeepaa AL, Oresic M, Kallioniemi O, Iljin K. Salinomycin inhibits prostate cancer growth and migration via induction of oxidative stress. Br J Cancer. 2012;106(1):99–106.
Li T, Su L, Zhong N, Hao X, Zhong D, Singhal S, Liu X. Salinomycin induces cell death with autophagy through activation of endoplasmic reticulum stress in human cancer cells. Autophagy. 2013;9(7):1057–68.
Jangamreddy JR, Jain MV, Hallbeck AL, Roberg K, Lotfi K, Los MJ. Glucose starvation-mediated inhibition of salinomycin induced autophagy amplifies cancer cell specific cell death. Oncotarget. 2015;6(12):10134–45.
Li T, Liu X, Shen Q, Yang W, Huo Z, Liu Q, Jiao H, Chen J. Salinomycin exerts anti-angiogenic and anti-tumorigenic activities by inhibiting vascular endothelial growth factor receptor 2-mediated angiogenesis. Oncotarget. 2016;7(18):26580–92.
Zhang C, Lu Y, Li Q, Mao J, Hou Z, Yu X, Fan S, Li J, Gao T, Yan B, et al. Salinomycin suppresses TGF-beta1-induced epithelial-to-mesenchymal transition in MCF-7 human breast cancer cells. Chem Biol Interact. 2016;248:74–81.
He L, Wang F, Dai WQ, Wu D, Lin CL, Wu SM, Cheng P, Zhang Y, Shen M, Wang CF, et al. Mechanism of action of salinomycin on growth and migration in pancreatic cancer cell lines. Pancreatology. 2013;13(1):72–8.
Wu D, Zhang Y, Huang J, Fan Z, Shi F, Wang S. Salinomycin inhibits proliferation and induces apoptosis of human nasopharyngeal carcinoma cell in vitro and suppresses tumor growth in vivo. Biochem Biophys Res Commun. 2014;443(2):712–7.
Wang Y. Effects of salinomycin on cancer stem cell in human lung adenocarcinoma A549 cells. Med Chem. 2011;7(2):106–11.
Zhang Y, Li F, Liu L, Jiang H, Jiang X, Ge X, Cao J, Wang Z, Zhang L, Wang Y. Salinomycin-induced autophagy blocks apoptosis via the ATG3/AKT/mTOR signaling axis in PC-3 cells. Life Sci. 2018;207:451–60.
Ravier MA, Daro D, Roma LP, Jonas JC, Cheng-Xue R, Schuit FC, Gilon P. Mechanisms of control of the free Ca2+ concentration in the endoplasmic reticulum of mouse pancreatic beta-cells: interplay with cell metabolism and [Ca2+]c and role of SERCA2b and SERCA3. Diabetes. 2011;60(10):2533–45.
Bublitz M, Musgaard M, Poulsen H, Thogersen L, Olesen C, Schiott B, Morth JP, Moller JV, Nissen P. Ion pathways in the sarcoplasmic reticulum Ca2+−ATPase. J Biol Chem. 2013;288(15):10759–65.
Griffin JB, Rodriguez-Melendez R, Dode L, Wuytack F, Zempleni J. Biotin supplementation decreases the expression of the SERCA3 gene (ATP2A3) in Jurkat cells, thus, triggering unfolded protein response. J Nutr Biochem. 2006;17(4):272–81.
Krizanova O, Markova J, Pacak K, Skultety L, Soltysova A, Hudecova S. Triptolide induces apoptosis through the SERCA 3 upregulation in PC12 cells. Gen Physiol Biophys. 2014;33(1):137–44.
Teng Y, Ren MQ, Cheney R, Sharma S, Cowell JK. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103(7):1066–75.
Davis JE, Xie X, Guo J, Huang W, Chu WM, Huang S, Teng Y, Wu G. ARF1 promotes prostate tumorigenesis via targeting oncogenic MAPK signaling. Oncotarget. 2016;7(26):39834–45.
Zhang Y, Yang L, Lin Y, Rong Z, Liu X, Li D. SPATA12 and its possible role in DNA damage induced by ultraviolet-C. PLoS One. 2013;8(10):e78201.
Zhang Y, Wu X, Li Y, Zhang H, Li Z, Zhang L, Ju J, Liu X, Chen X, Glybochko PV, et al. Endothelial to mesenchymal transition contributes to arsenic-trioxide-induced cardiac fibrosis. Sci Rep. 2016;6:33787.
Cardozo AK, Ortis F, Storling J, Feng YM, Rasschaert J, Tonnesen M, Van Eylen F, Mandrup-Poulsen T, Herchuelz A, Eizirik DL. Cytokines downregulate the sarcoendoplasmic reticulum pump Ca2+ ATPase 2b and deplete endoplasmic reticulum Ca2+, leading to induction of endoplasmic reticulum stress in pancreatic beta-cells. Diabetes. 2005;54(2):452–61.
Guo W, Diao Z, Liu W. Asymmetric dimethylarginine downregulates sarco/endoplasmic reticulum calciumATPase 3 and induces endoplasmic reticulum stress in human umbilical vein endothelial cells. Mol Med Rep. 2017;16(5):7541–7.
Monaco S, Rusciano M, Maione A, Soprano M, Gomathinayagam R, Todd L, Campiglia P, Salzano S, Pastore L, Leggiero E, et al. A novel crosstalk between calcium/calmodulin kinases II and IV regulates cell proliferation in myeloid leukemia cells. Cell Signal. 2015;27(2):204–14.
Nakanishi A, Hatano N, Fujiwara Y, Bin Shari A, Takabatake S, Akano H, Kanayama N, Magari M, Nozaki N, Tokumitsu H. AMP-activated protein kinase-mediated feedback phosphorylation controls the Ca2+/calmodulin (CaM) dependence of Ca2+/CaM-dependent protein kinase kinase beta. J Biol Chem. 2017.
Wei XH, Yu SD, Ren L, Huang SH, Yang QM, Wang P, Chu YP, Yang W, Ding YS, Huo Y, et al. Inhibition of late sodium current suppresses calcium-related ventricular arrhythmias by reducing the phosphorylation of CaMK-II and sodium channel expressions. Sci Rep. 2017;7(1):981.
Menconi MJ, Wei W, Yang H, Wray CJ, Hasselgren PO. Treatment of cultured myotubes with the calcium ionophore A23187 increases proteasome activity via a CaMK II-caspase-calpain-dependent mechanism. Surgery. 2004;136(2):135–42.
Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012.
Zhou S, Wang F, Wong ET, Fonkem E, Hsieh TC, Wu JM, Wu E. Salinomycin: a novel anti-cancer agent with known anti-coccidial activities. Curr Med Chem. 2013;20(33):4095–101.
Manago A, Leanza L, Carraretto L, Sassi N, Grancara S, Quintana-Cabrera R, Trimarco V, Toninello A, Scorrano L, Trentin L, et al. Early effects of the antineoplastic agent salinomycin on mitochondrial function. Cell Death Dis. 2015;6:e1930.
Kovacs T, Felfoldi F, Papp B, Paszty K, Bredoux R, Enyedi A, Enouf J. All three splice variants of the human sarco/endoplasmic reticulum Ca2+−ATPase 3 gene are translated to proteins: a study of their co-expression in platelets and lymphoid cells. Biochem J. 2001;358(Pt 3:559–68.
Martin V, Bredoux R, Corvazier E, Van Gorp R, Kovacs T, Gelebart P, Enouf J. Three novel sarco/endoplasmic reticulum Ca2+−ATPase (SERCA) 3 isoforms. Expression, regulation, and function of the membranes of the SERCA3 family. J Biol Chem. 2002;277(27):24442–52.
Yu SN, Kim SH, Kim KY, Ji JH, Seo YK, Yu HS, Ahn SC. Salinomycin induces endoplasmic reticulum stressmediated autophagy and apoptosis through generation of reactive oxygen species in human glioma U87MG cells. Oncol Rep. 2017;37(6):3321–8.
Guo ZY, Zhang YH, Xie GQ, Liu CX, Zhou R, Shi W. Down-regulation of Homer1 attenuates t-BHP-induced oxidative stress through regulating calcium homeostasis and ER stress in brain endothelial cells. Biochem Biophys Res Commun. 2016.
Bai X, Liu S, Yuan L, Xie Y, Li T, Wang L, Wang X, Zhang T, Qin S, Song G, et al. Hydrogen-rich saline mediates neuroprotection through the regulation of endoplasmic reticulum stress and autophagy under hypoxia-ischemia neonatal brain injury in mice. Brain Res. 2016.
Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102.
Gotoh T, Mori M. Nitric oxide and endoplasmic reticulum stress. Arterioscler Thromb Vasc Biol. 2006;26(7):1439–46.
Izquierdo-Torres E, Rodriguez G, Meneses-Morales I, Zarain-Herzberg A. ATP2A3 gene as an important player for resveratrol anticancer activity in breast cancer cells. Mol Carcinog. 2017;56(7):1703–11.
Izquierdo-Torres E, Rodríguez G, Meneses-Morales I, Zarain-Herzberg A. ATP2A3 gene as an important player for resveratrol anticancer activity in breast cancer cells. Mol Carcinog. 2017.
Brini M, Carafoli E. Calcium pumps in health and disease. Physiol Rev. 2009;89(4):1341–78.
Acknowledgments
We are grateful to Professor Dan Li from the College of Biology at Hunan University and Zhiqiang Liu from the Department of Pathophysiology at Tianjin Medical University for helpful scientific discussion. We are also grateful to Research assistant Lanhua Zhao from the Medical College at University of South China for flow cytometric analysis.
Funding
This work was supported by the Zhengxiang Scholar Program of the University of South China, the National Natural Science Foundation of China (nos.81241090), the Natural Science Foundation of Hunan Province (nos.2018JJ3462 and nos.2016WK2013), the Scientific Research Project of Education Department of Hunan Province (nos.18B278) and the Scientific Research Project of Hunan Provincial Health Commission (nos. B2019107).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent to publish
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: YZ and YW. Performed the experiments: YZ, FL, LL, HJ, HH, XD, XG and JC. Analyzed the data: YZ, FL, LL and YW. Wrote the manuscript: YZ. All authors read and approved the final manuscript to be published.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committeeof the University of South China.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, Y., Li, F., Liu, L. et al. Salinomycin triggers endoplasmic reticulum stress through ATP2A3 upregulation in PC-3 cells. BMC Cancer 19, 381 (2019). https://doi.org/10.1186/s12885-019-5590-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-019-5590-8