Abstract
Background
Additional accurate non-invasive biomarkers are needed in the clinical setting to improve prostate cancer (PCa) diagnosis. Here we have developed a new and improved multiplex mRNA urine test to detect prostate cancer (PCa). Furthermore, we have validated the PCA3 urinary transcript and some panels of urinary transcripts previously reported as useful diagnostic biomarkers for PCa in our cohort.
Methods
Post-prostatic massage urine samples were prospectively collected from PCa patients and controls. Expression levels of 42 target genes selected from our previous studies and from the literature were studied in 224 post-prostatic massage urine sediments by quantitative PCR. Univariate logistic regression was used to identify individual PCa predictors. A variable selection method was used to develop a multiplex biomarker model. Discrimination was measured by ROC curve AUC for both, our model and the previously published biomarkers.
Results
Seven of the 42 genes evaluated (PCA3, ELF3, HIST1H2BG, MYO6, GALNT3, PHF12 and GDF15) were found to be independent predictors for discriminating patients with PCa from controls. We developed a four-gene expression signature (HIST1H2BG, SPP1, ELF3 and PCA3) with a sensitivity of 77 % and a specificity of 67 % (AUC = 0.763) for discriminating between tumor and control urines. The accuracy of PCA3 and previously reported panels of biomarkers is roughly maintained in our cohort.
Conclusions
Our four-gene expression signature outperforms PCA3 as well as previously reported panels of biomarkers to predict PCa risk. This study suggests that a urinary biomarker panel could improve PCa detection. However, the accuracy of the panels of urinary transcripts developed to date, including our signature, is not high enough to warrant using them routinely in a clinical setting.
Similar content being viewed by others
Background
During the last two decades, prostate-specific antigen (PSA) has been extensively used for prostate cancer (PCa) screening, detection and follow-up. The routine use of PSA has been the subject of continued controversy owing to its limited specificity, which derives from the fact that elevated serum levels of PSA occur in a variety of non-neoplastic conditions such as prostatitis and benign prostate hyperplasia (BPH) [1]. Furthermore, up to 27 % of men with PSA in the normal range (≤ 4 ng/ml) suffer from PCa [2]. The current gold standard method for diagnosis of PCa in patients with elevated serum PSA is non-targeted transrectal ultrasound-guided needle biopsy, which fails to detect PCa in approximately 20–30 % of cases [3]. Therefore, there is a need for additional non-invasive and more specific markers of early PCa that will permit the stratification of patients according to their risk of developing PCa and thus identify men who will require prostate biopsy.
A great improvement in high-throughput gene expression techniques has yielded several promising molecular biomarkers for PCa detection. Prostatic cells can be collected in urine after an intensive prostatic massage. In 2003, Hessels et al. for the first time used the prostate cancer antigen 3 (PCA3) for the identification of PCa in urine sediments obtained after prostatic massage [4]. Since then, several studies have assessed the diagnostic performance of this marker (reviewed in [5, 6]) and other individual transcripts [7, 8]. However, taking into account the heterogeneity of PCa, several authors have searched for a multiplex detection system of biomarkers, which has proved to outperform the diagnostic value of the individual markers [9–12].
We have previously identified new putative mRNA markers for PCa diagnosis that can be extrapolated to post-prostatic massage (PPM) urine samples [13]. In the present study we aim to test several of those previously identified putative biomarkers in a large cohort of PPM-urine samples in order to develop an improved multiplex mRNA biomarker model for PCa diagnosis to be routinely used in the clinical setting. Furthermore, in our cohort we have validated the commercially available test based on urine PCA3 expression as well as the best performing mRNA panels of biomarkers reported in the literature [9–12].
Methods
Patients and urine samples
Under Institutional Review Board approval (Hospital Clinic ethics committee) and patients’ informed consent, we prospectively collected 273 freshly voided urine samples from PCa patients and age matched controls between January 2009 and September 2012 at the Hospital Clínic of Barcelona. All patients underwent radical prostatectomy. The grade and stage of the tumours were determined according to Gleason criteria and TNM classification, respectively [14, 15]. Systematic prostate biopsy was performed to identify PCa patients included in the present study.
Voided urine samples (20 to 50 ml including the initial portion of the urine,) were collected following prostatic massage in sterile containers containing 2 ml of 0.5 M EDTA, pH 8.0. Urines were immediately stored at 4 °C and processed within the next 8 h. The samples were centrifuged at 1000xg for 10 min, at 4 °C. The cell pellets were re-suspended in 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and frozen at −80 °C until RNA extraction.
RNA extraction, cDNA synthesis and pre-amplification
RNAs from the urinary cell pellets were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and quantified with a NanoDrop (NanoDrop Technologies, Wilmington, DE, USA).
cDNA was synthesized from 100 ng of total RNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA USA; hereafter referred to as AB) following manufacturer’s instructions, except that the final volume of the reaction was 25 μl. A total of 1.25 μl of each cDNA sample, 2.5 μl of TaqMan PreAmp Master Mix kit 2X (AB) and 1.25 μl of pooled assay mix 0.2X containing 46 Gene Expression Assays (AB) were used for the multiplex pre-amplification of the target cDNAs following manufacturer’s instructions (AB). The 46 assays included in the pooled assay mix were selected from previous data from our group [13] and literature [10, 12, 16, 17] and contains 42 target genes and four endogenous controls; B2M, GAPHDH, KLK2 and KLK3 (Additional file 1: Table S1). Of note, 23 of the 42 target genes selected here were previously analyzed in urine samples by our group [13].
Quantitative PCR using BioMark 48.48 Dynamic Arrays
A total of 2.25 μl of each pre-amplified cDNA was loaded into the Dynamic Array along with 0.25 μl of GE Sample Loading Reagent 20X (Fuidigm) and 2.5 μl of TaqMan Universal PCR Master Mix 2X (AB). For the assays, 2.5 μl of TaqMan® Gene Expression Assays 20X (AB) were combined with 2.5 μl of Assay Loading Reagent and were pipetted into the assay inputs. Reaction conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The real-time quantitative PCR (qPCR) experiments were performed on the BioMark instrument.
Quantitative PCR data analysis
The real-time qPCR analysis software was used to obtain cycle quantification (Cq) values. Threshold was manually calculated for each gene. Since experimental errors such as inaccurate pipetting or contamination can result in amplification curves that look significantly different from a typical amplification curve, all amplification plots were checked both computationally and manually. Relative expression levels of target genes within a sample was expressed as ΔCq (ΔCq = Cqendogenous control-Cqtarget gene). We used as endogenous control the mean Cq value of KLK2 and KLK3, which allowed us to normalize the prostate epithelial cell content in the collected urine sample [4]. Most of the studies seeking urinary transcripts for PCa diagnosis have used KLK3 as a prostate-specific endogenous control [4, 18, 19]. In this study, to minimize the possibility of erroneous relative gene expression quantification, we also selected KLK2 as a second prostate-specific endogenous control since its expression level is highly correlated with KLK3 [20].
All 273 urine samples initially included in the study were positive for both housekeeping genes, the B2MG (B2MG mean Cq = 8.79; range 5.07–14.58) and GAPDH (GAPDH mean Cq = 10.85; range 7.6–16.17), indicating that all samples contained cells. Moreover, all samples were also positive for KLK2 (KLK2 mean Cq = 13.12; range 9.87–17.85) and for KLK3 (KLK3 mean Cq = 12.91; range 9.58–17.65) genes, indicating that all samples contained cells of prostate origin. Cq values for all other biomarkers are in the range for those of KLK2 and KLK3 (data not shown). All Cq values (except 2 cases in B2MG gene) fall in the optimal range of quantifiable Cq values in BioMark instrument (Cq = 6 to Cq = 23) [21]. Moreover, to assure the quality of the expression data obtained, low RNA quality samples were identified as outliers according to their average expression by the Mahalanobis Distance Quality Control (MDQC) method [22] and were excluded from the study. Fold change values were generated from the median expression of the genes from the BioMark 48.48 Dynamic Arrays in the groups compared.
Statistical analysis
The association of each variable with final radical prostatectomy pathology results was analyzed by univariate logistic regression. Significance was defined as p values < 0.05.
All transcripts analyzed were subjected to variable selection using the lars function with method LASSO in the lars R statistical package (http://CRAN.R-project.org/package=lars) [23]. As all the samples were used for the model generation, the performance of the model may be over-optimized. To correct this bias, we further performed a leave-one-out cross-validation (LOOCV) and 100 randomisations with 5- fold cross-validation (5fCV) (http://CRAN.R-project.org/package=rms).
The optimal probability cutoff for the univariate study variables and logistic regression models (our model and those previously described in the literature [9–12]) was computed through a ROC analysis. To evaluate the performance of the models, we computed sensitivity (SN), specificity (SP), negative predictive value (NPV), positive predictive value (PPV) and overall error rates (ER) for the mRNA expression signature. Analysis of variance (ANOVA) of the Risk score probability versus three groups of PSA was done. Pairwise comparisons were made with Tukey’s HSD procedure. R-software was used for all calculations.
Results
Study population and informative rate
Among the 273 urine samples initially collected from 180 PCa patients and 93 control individuals, we excluded 29 urines from PCa patients (16 %) and 20 from controls (22 %) because they were flagged as low-quality samples when tested using MDQC method [22]. Thus, in total, the urine samples of 224 men, 151 with PCa and 73 controls were successfully analyzed (82 %). Table 1 shows characteristics and clinicopathological information for the 224 evaluable subjects. Only 10 patients with PSA levels > 4 were included as controls. Pathological reports from these patients confirmed the absence of malignity at the time of sample collection and they have not presented PCa during a mean follow-up of 45.6 months (range 19.5 to 78.9).
Development of a new multiplex mRNA model
All 42 selected genes were first tested by univariate logistic regression analysis, with 7 genes (PCA3, ELF3, HIST1H2BG, MYO6, GALNT3, PHF12 and GDF15) showing significant association for discriminating PCa patients from control individuals (Table 2 and Additional file 2: Table S2). Notably, no significant differences in TMPRSS2-ERG status between tumor (mean Cq = 13.54; range 10.28–18.21) and control (mean Cq = 13.88; range 10.28–18.71) urine samples were found. Differences in Cq values for TMPRSS2-ERG across the different Gleason stages (mean Cq = 13.54 for Gleason ≤ 6; mean Cq = 13.64 for Gleason = 7; mean Cq = 13.27 for Gleason ≥ 8) were not found either.
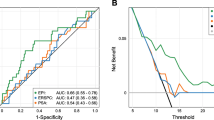
To evaluate the performance of individual markers for diagnosing PCa, we performed a ROC analysis (Table 2). Then, individual biomarkers were subjected to variable selection to develop a multiplex model that could improve performance over single biomarkers. This analysis resulted in a final selection of a four-gene model that contains HIST1H2BG, SPP1, ELF3 and PCA3. The four gene model outperformed single genes and previously reported models in the literature in detecting PCa in urinary sediments (SN = 77 %; SP = 67 %; PPV = 83 %; NPV = 58 %; ER = 26 %; AUC = 0.763). After applying LOOCV analysis to the four-gene model, we obtained a SN of 79 % for discriminating between tumor and control urines with a SP of 60 % (PPV = 80 %; NPP = 58 %; ER = 27 %; AUC 0.735). By using 5fCV analysis, we found a SN of 72.52 % for discriminating between tumor and control urines with a SP of 64.83 % (PPV = 80.86 %; NPV = 53.5 %; ER = 30 %; AUC 0.732) (Fig. 1a). To note, the four-gene model also performs well in the diagnostic PSA gray-zone (PSA 3–10 ng/ml) yielding a SN of 79 % for discriminating between tumor urines from patients with PSA serum values between 3 and 10 ng/ml and control urines, with a SP of 59 % (PPV = 72 %; NPP = 68 %; ER = 29 %; p < 0.001) (Fig. 1b).
Evaluation of previously reported diagnostic biomarkers of urinary transcripts in our cohort
First, we evaluated the PCA3 marker (TaqMan PCR test for PCA3) as a single marker. Univariate logistic regression analysis showed that expression of PCA3 was a significant discriminator of PCa from control individuals (p < 0.01). PCA3 alone achieved an overall SN of 49 % and a SP of 85 % (AUC = 0.708) to discriminate controls from PCa urines (Table 2 and Additional file 2: Table S2). Then, we evaluated in our cohort some of the most potentially promising PCa diagnostic panels of urinary transcripts reported in the literature, to validate their performance in an independent set. Table 3 summarizes the diagnostic performance of the biomarkers panels in our case-control setting in comparison to the results obtained in the original studies. As shown, all the biomarker combinations roughly maintain their performance when tested in an independent set, the combination described by Laxman et al. (2008) having the best performance [10].
Discussion
Currently, PSA is considered the most valuable tool in the early detection, staging and monitoring of PCa. However, as mentioned in the introduction, PSA has several limitations as a PCa diagnostic biomarker, especially in deciding the necessity of a prostate biopsy. Actually, PCa is detected in only about a third of patients with elevated serum PSA who undergo random prostate biopsy. Repeated biopsies reveal the presence of PCa in another 10–35 % of the cases [24]. Not only economic aspects but also anxiety, discomfort, and sometimes severe complications are associated with prostate biopsies. Therefore, the development of a non-invasive diagnostic tool for the early detection and screening of PCa as well as to increase the probability of detecting PCa at repeat biopsy, reducing the number of unnecessary biopsies, is needed in urological practice. Detection of aberrantly expressed transcripts in PCa cells shed into the urine after prostatic massage are promising biomarkers for the development of a reliable non-invasive PCa diagnostic method. In fact, several promising RNA-based urine PCa biomarkers are described in the literature, but only the PCA3 assay (Progensa) is approved by the FDA and currently is the only molecular diagnostic assay for PCa commercially available. However, PCA3 is not routinely used in the clinical setting mainly because clinicians feel that the increase in accuracy over serum PSA testing is not significant enough to warrant a biopsy. Furthermore, since PCa is a heterogeneous disease, it is reasonable that a combination of markers outperforms single marker detection. In this regard, several authors have described combinations of RNA-markers in urine samples but to our knowledge, none of them, except one [25], has been externally validated nor is currently used in the clinical setting. In the present work, we have developed a four-gene panel that outperforms those previously described in the literature. In addition, in our cohort we have validated PCA3 as well as the most promising panels of biomarkers described.
From our analysis, we have been able to identify six new candidates that independently predict PCa in PPM-urine samples, besides PCA3. This has been possible since we have explored target genes selected from previous PCa microarray data [13, 17] instead of analyzing only previously described prostate related biomarkers. Actually, all target genes explored were used to develop the four-gene set model that contains the previously described PCA3 gene and three new biomarkers: HIST1H2BG, SPP1 and ELF3. This model outperforms individual biomarkers and previously reported models in the literature. Although LOOCV indicates a certain degree of overfitting, all data obtained after cross validation corroborate the SN and SP for the final model. Moreover, the model performs well in the diagnostic PSA gray-zone (PSA 3–10 ng/ml) where a reduction in the number of unnecessary biopsies is necessary.
Notably, the three new biomarkers of the model had been previously associated with PCa. Alterations in expression of histone HIST1H2BG were associated with biochemical recurrence in PCa patients after radical prostatectomy [26]. The transcription factor ELF3 (E74-like factor 3), that acts as a negative modulator of androgen receptor transcriptional activity, was found underexpressed in PCa [27], according to our results. On the other hand, SPP1 (secreted phosphoprotein 1) encodes the protein osteopontin (OPN). Both, OPN RNA and protein have been found overexpressed in a number of human tumor types, including PCa [28]. In some cases, OPN overexpression has been shown to be associated directly with poor patient prognosis or with other indicators of poor prognosis. Thus, OPN has a dual interest, as a biomarker of malignancy as well as a candidate for testing as a poor prognostic factor. Even though in the present study we did not achieve statistical significance for SPP1, the addition of this gene to the model improved the AUC from 0.740 (HIST1H2BG, PCA3 and ELF3) to 0.763 (SPP1, HIST1H2BG, PCA3 and ELF3), indicating that effectively its expression adds information to the model.
The present study confirms that PCA3 can successfully discriminate PCa from controls in randomly selected patients with variable PSA levels (PSA = 0.94–365 ng/ml) [29, 30]. A limitation of most studies based on urinary biomarkers is that the negative PCa patient group consists of patients who have undergone prostate biopsy for suspected PCa with a negative result, but in fact, 20–30 % of such patients will be diagnosed with PCa at a later date [3]. To overcome this limitation, our control group consisted of patients without suspected PCa (PSA < 4.0 ng/ml), thus minimizing the risk of including subjects with PCa in the control group. Moreover, there is no uniform methodological protocol for urinary transcript quantification in the reported studies. For instance, some studies use a multiplex cDNA preamplification step before qPCR transcript quantification [16, 31], while others use a Whole Transcriptome Amplification [10, 32] or even in some studies cDNA is not preamplifed [11]. Also different gene expression normalization methods are used [4, 11, 16, 18, 31]. Thus, it is notable that despite this methodological heterogeneity and the inherent limitations of the sample source (PPM-urine contains different cell types, including renal tubular cells, urothelial cells, prostate cells, etc.… and the proportion of prostate tumor cells in each subject is different), we and the vast majority of the groups identify PCA3 as an independent predictor for PCa diagnosis, making it the most reliable individual biomarker to date.
However, combining urinary biomarkers in a panel has shown higher diagnostic accuracy than PCA3 alone. Regarding this, we have been able to validate some of the previously reported panels of biomarkers [9–12] in our cohort and to develop a new urinary panel of biomarkers that improves serum PSA and previously reported panels of biomarkers. On the contrary, we could not validate differences between control and cancer population for the TMPRSS2-ERG status. This is in all probability due to the methodological approach used here, since others using the same methodology as us (RT-qPCR using the same gene expression assay as us; Hs03063375_ft ) to evaluate TMPRSS2-ERG status also did not find differences between cancer and control urines [33] while other authors using Southern blot [9] or transcription-mediated amplification [32] were able to find such differences.
Of concern, neither the FDA approved PCA3 test alone, or in combination with other biomarkers, is being routinely used in the clinical setting. This is most likely because the addition of urine biomarkers to the current clinical diagnostic tools only shows a limited improvement in the PCa diagnosis accuracy and does not provide sufficient value to affect biopsy decision making. In fact, recently the Evaluation of Genomic Applications in Practice and Prevention Working Group (EWG) has found insufficient evidence to recommend PCA3 testing not only for deciding to conduct initial biopsies for PCa at risk men (e.g. previously elevated PSA test or suspicious digital rectal examination) but also for deciding when to rebiopsy previously biopsy-negative patients for PCa. Furthermore, the EWG did not find convincing evidence to recommend PCA3 testing in men with PCa positive-biopsies to determine whether the disease is indolent or aggressive, in order to develop an optimal treatment plan [34]. Thus, even though many efforts have been made in the last decade to identify urine biomarkers that determine men at high risk of PCa and whether the disease is indolent or aggressive in men with PCa, the results do not seem convincing for clinicians.
We acknowledge that our study has several limitations. First it resides in the relatively low sample size of the studied cohort. This was because 18 % of urine samples collected could not be evaluated (informative specimen rate of 82 %). Although some improvements in the methodological process would be desirable to decrease the percentage of fails, this percentage is in the range of those described by other authors who quantify gene expression in PPM urine samples (informative specimen rates 56 to 92 %) [10–12, 16, 30, 31]. However, sample collection can be repeated if necessary. It could also be argued that we arbitrarily selected the 42 target genes, while the list of differentially expressed genes in PCa is much larger. In this regard, we have tried to include the biomarkers according to previous studies, as being either detectable in urine or appropriate for combined models, and genes highly differentially expressed in PCa tissue samples. We are also aware that we should test the performance of our four-gene expression signature in a real clinical scenario by analyzing patients who undergo prostate biopsy for suspected PCa, even though this study will have the limitation of false negative biopsies, which account for 20–30 % of men at risk of PCa [3]. Lastly, future validation studies are needed to further improve the performance of this test by examination of larger and independent cohorts.
Conclusions
We report a four-gene expression signature with higher diagnostic accuracy than PCA3, the only non-invasive commercially available urinary biomarker, to predict individuals at risk of PCa. Moreover, our four-gene expression signature outperforms previously reported panels of biomarkers for PCa detection. Taken together, these results suggest that new biomarkers can be successfully combined with PCA3, resulting in improvements in PCa detection. However, further sources of new non-invasive biomarkers that enable physicians to accurately predict any PCa at initial prostate biopsy and aggressive PCa should be explored.
Abbreviations
- 5fCV:
-
5-fold cross-validation
- AUC:
-
area under curve
- BPH:
-
benign prostate hyperplasia
- CI:
-
confidence interval
- Cq:
-
cycle quantification
- ER:
-
error rate
- LOOCV:
-
leave-one-out cross-validation
- NPV:
-
negative predictive value
- OD:
-
odds ratio
- PCa:
-
prostate cancer
- PPM:
-
post-prostatic massage
- PPV:
-
positive predictive value
- PSA:
-
Prostate-specific antigen
- qPCR:
-
quantitative PCR
- ROC:
-
receiver operator characteristic
- SD:
-
standard deviation
- SN:
-
sensitivity
- SP:
-
specificity
References
Stamey TA, Caldwell M, McNeal JE, Nolley R, Hemenez M, Downs J. The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol. 2004;172:1297–301.
Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, Mason MD, Matveev V, Mottet N, van der kwast TH, Wiegel T, Zattoni F. Guidelines on Prostate Cancer. In: European Association of Urology Guidelines. edition presented at the 28th EAU Annual Congress edition. members of the European Association of Urology (EAU) Guidelines Office, editor. Milan: 2013. https://uroweb.org/guidelines/.
Taira AV, Merrick GS, Galbreath RW, Andreini H, Taubenslag W, Curtis R, et al. Performance of transperineal template-guided mapping biopsy in detecting prostate cancer in the initial and repeat biopsy setting. Prostate Cancer Prostatic Dis. 2010;13:71–7.
Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15.
Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6:255–61.
Dijkstra S, Mulders PF, Schalken JA. Clinical use of novel urine and blood based prostate cancer biomarkers: a review. Clin Biochem. 2014;47:889–96.
Zielie PJ, Mobley JA, Ebb RG, Jiang Z, Blute RD, Ho SM. A novel diagnostic test for prostate cancer emerges from the determination of alpha-methylacyl-coenzyme a racemase in prostatic secretions. J Urol. 2004;172:1130–3.
Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, Helgeson BE, et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8:885–8.
Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–8.
Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–9.
Ouyang B, Bracken B, Burke B, Chung E, Liang J, Ho SM. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol. 2009;181:2508–13.
Rigau M, Morote J, Mir MC, Ballesteros C, Ortega I, Sanchez A, et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate. 2010;70:1760–7.
Mengual L, Ars E, Lozano JJ, Burset M, Izquierdo L, Ingelmo-Torres M, Gaya JM, Algaba F, Villavicencio H, Ribal MJ, Alcaraz A. Gene expression profiles in prostate cancer: Identification of candidate non-invasive diagnostic markers. Actas Urol Esp. 2014;38:143–9.
Sobin LH, Gospodariwicz M, Wittekind CH. TNM Classification of Malignant Tumours. UICC International Union Against Cancer. New York: Wiley; 2009.
Epstein JI, Allsbrook Jr WC, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–42.
Rigau M, Ortega I, Mir MC, Ballesteros C, Garcia M, Llaurado M, et al. A three-gene panel on urine increases PSA specificity in the detection of prostate cancer. Prostate. 2011;71:1736–45.
Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406.
Rice KR, Chen Y, Ali A, Whitman EJ, Blase A, Ibrahim M, et al. Evaluation of the ETS-related gene mRNA in urine for the detection of prostate cancer. Clin Cancer Res. 2010;16:1572–6.
Ouyang B, Leung YK, Wang V, Chung E, Levin L, Bracken B, et al. alpha-Methylacyl-CoA racemase spliced variants and their expression in normal and malignant prostate tissues. Urology. 2011;77:249. e1-7.
Shaw JL, Diamandis EP. Distribution of 15 human kallikreins in tissues and biological fluids. Clin Chem. 2007;53:1423–32.
Sorg D, Danowski K, Korenkova V, Rusnakova V, Kuffner R, Zimmer R, et al. Microfluidic high-throughput RT-qPCR measurements of the immune response of primary bovine mammary epithelial cells cultured from milk to mastitis pathogens. Animal. 2013;7:799–805.
Cohen Freue GV, Hollander Z, Shen E, Zamar RH, Balshaw R, Scherer A, et al. MDQC: a new quality assessment method for microarrays based on quality control reports. Bioinformatics. 2007;23:3162–9.
Efron B, Johnstone I, Hastie T, Tibshirani R. Least angle regression (with discussion). Ann Stat. 2004;32:407–99.
Djavan B, Remzi M, Schulman CC, Marberger M, Zlotta AR. Repeat prostate biopsy: who, how and when? A review. Eur Urol. 2002;42:93–103.
Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB, de Reijke TM, Vergunst H, Kil P, Knipscheer BC, van Oort IM, Mulders PF, Hulsbergen-van de Kaa CA, Schalken JA. Prospective Multicentre Evaluation of PCA3 and TMPRSS2-ERG Gene Fusions as Diagnostic and Prognostic Urinary Biomarkers for Prostate Cancer. Eur Urol. 2014;65:534–42.
Chen X, Xu S, McClelland M, Rahmatpanah F, Sawyers A, Jia Z, et al. An accurate prostate cancer prognosticator using a seven-gene signature plus Gleason score and taking cell type heterogeneity into account. PLoS One. 2012;7:e45178.
Shatnawi A, Norris JD, Chaveroux C, Jasper JS, Sherk AB, McDonnell DP, Giguere V. ELF3 is a repressor of androgen receptor action in prostate cancer cells. Oncogene. 2014;33:862–71.
Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, et al. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145:610–23.
Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, Mulders PF, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081–8.
van Gils MP, Hessels D, van Hooij O, Jannink SA, Peelen WP, Hanssen SL, et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin Cancer Res. 2007;13:939–43.
Jamaspishvili T, Kral M, Khomeriki I, Vyhnankova V, Mgebrishvili G, Student V, et al. Quadriplex model enhances urine-based detection of prostate cancer. Prostate Cancer Prostatic Dis. 2011;14:354–60.
Salami SS, Schmidt F, Laxman B, Regan MM, Rickman DS, Scherr D, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol. 2013;31:566–71.
Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masia E, Casanova J, et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol. 2014;192:252–9.
Recommendations from the EGAPP Working Group: does PCA3 testing for the diagnosis and management of prostate cancer improve patient health outcomes? Genet Med. 2014; 16:338–46.
Acknowledgements
We thank the patients for their collaboration and all the staff from the Urology Departments and nurses from Hospital Clínic for collaborating in the sample collection. This work was supported by grants from Laboratorios FINA BIOTECH, Ministerio de Economia y Competividad (IPT-2012-1311-300000) and Fundación para la Investigación en Urología (FIU 2010).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LM participated in study concept and design, acquisition and analysis of data, drafting of the manuscript and supervision of the study conduct. JJL participated in study concept and design, analysis of data, critical revision of the manuscript and statistical analysis. MIT participated in acquisition and analysis of data, critical revision of the manuscript and supervision of the study conduct. LI and MM participated in acquisition of data and critical revision of the manuscript. MJR participated in study concept and design, analysis of data, critical revision of the manuscript and supervision of the study conduct. AA participated in study concept and design, analysis of data, critical revision of the manuscript and supervision of the study conduct. All authors read and approved the final manuscript.
Additional files
Additional file 1: Table S1.
Commercial Gene Expression Assays from Life Technologies used in this study. Target exons for transcript detection as well as amplicon length for each trasncrip are shown. (DOCX 18 kb)
Additional file 2: Table S2.
Univariate logistic regression and ROC analyses of the biomarkers. (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mengual, L., Lozano, J.J., Ingelmo-Torres, M. et al. Using gene expression from urine sediment to diagnose prostate cancer: development of a new multiplex mRNA urine test and validation of current biomarkers. BMC Cancer 16, 76 (2016). https://doi.org/10.1186/s12885-016-2127-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-016-2127-2