Abstract
Background
The KMT2A gene encoded lysine methyltransferase plays an essential role in regulating gene expression during early development and hematopoiesis. To date, 92 different mutations of KMT2A have been curated in the human gene mutation database (HGMD), resulting in Wiedemann-Steiner syndrome (WDSTS) and intellectual disability (ID)/developmental delay (DD).
Case presentation
In this report, we present a de novo heterozygous deletion mutation [c.74delG; p. (Gly26Alafs*2)] in the KMT2A gene, which was identified by trio-based whole exome sequencing from a 5.5-year-old boy with ID/DD, stereotypic hand movements and blood eosinophilia. Many deleterious germline mutations of KMT2A have been documented to affect development of central nervous system, oral and craniofacial tissues, but not blood eosinophils.
Conclusions
This is the first report of a rare case with ID/DD as well as eosinophilia, resulting from a previously undescribed null mutation of KMT2A. Our findings expand the phenotypical spectrum in affected individuals with KMT2A mutations, and may shed some insight into the role of KMT2A in eosinophil metabolism.
Similar content being viewed by others
Background
Neurodevelopmental disorders are a wide range of developmental brain dysfunctions, such as intellectual disability (ID), developmental delay (DD), autism spectrum disorder (ASD), epilepsy, and attention deficit hyperactivity disorder (ADHD). It has been estimated approximately 2% of children with ID/DD [1]; many of them are resulted from genetical alterations. Recent analysis showed that 42% neurodevelopmental disorders in children are caused by de novo mutations [2]. With the development of high-throughput sequencing technology, such as the whole-exome sequencing (WES), thousands of genes have been identified to be associated with neurodevelopmental disorders [3,4,5,6].
Germline mutations in the KMT2A gene (OMIM: 159555) were first identified in patients with Wiedemann-Steiner syndrome [7], which is characterized by hypertrichosis cubiti associated with short stature, consistent facial features, mild to moderate ID, behavioral difficulties, and hypertrichosis on the back (WDSTS, OMIM: 605130). On the other hand, many different somatic mutations or chromosomal rearrangements involving the KMT2A gene have been identified in affected individuals or families with leukemia, myeloid/lymphoid or mixed-lineage (OMIM: 159555).
In our previous studies, we utilized trio-based whole-exome sequencing (trio-WES) on hundreds of affected individuals with neurodevelopmental diseases, and identified multiple de novo or inherited genetic mutations in different causal genes, such as ANK3, PLA2G6, BCL11A, and PAK2 [8,9,10,11]. In the present study, a de novo deletion mutation in KMT2A, which is predicted to be a null allele, is identified in a boy with ID/DD, stereotypic hand movements, and blood eosinophilia.
Case presentation
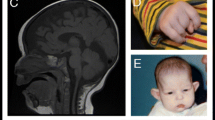
The proband was from a nonconsanguineous family; his parents were phenotypically normal. He was first referred to the hospital when he was 3.5-year-old (Fig. 1A, left panel) with a language developmental delay, a middle level of intellectual disability (ID), and stereotypic hand movements. He was born at 40 weeks in an uneventful spontaneous delivery. His height, body weight, and head circumference at birth were 50.0 cm, 3450 g, and 33 cm respectively, which implies no significant prenatal growth retardation. There were no feeding difficulties and epilepsy. He was able to hold his head up at around 5 months old, sit unaided at 7 months, and walk at 17 months. He started babbling words like “baba”, “mama” around 6 months. At age of 3.5 years, he could only speak a single word, like “yi”, but not a complete sentence. Also, he could not point at an object. He could not communicate with others, and avoided eye-to-eye contact with others. He made repetitive and purposeless movements, like hand waving, tapping and scratching. According to the standard of growth and development of children (0~7-year-old Chinese boy), his growth in height (90 cm, − 3 SD), weight (12 kg, − 2.5 SD) and head circumference (47.5 cm, − 2 SD) was significantly delayed. His score in the infant-junior middle school students social living ability scale (S-M Scale) was low. Physical examination in neurological system and cranial MRI found no obvious abnormalities. At the age of 5.5 years (Fig. 1A, right panel), his height was 102 cm (− 3 SD), weight 15 kg (− 2.5 SD), and head circumference 48.5 cm (− 2 SD).
Photographs of the affected boy and the pedigree of his family. (a) Facial appearance of the child at 3.5 (left panel) and 5.5 (right panel) years old, respectively. (b) The filled square represents the affected proband, empty symbols represent unaffected individuals, square represents male; the circle, female
First peripheral venous blood analysis on 3.5-year-old showed increased absolute count of eosinophils (1.847 × 10^9/L, compared to normal level < 0.5*109/L) as well as increased eosinophil ratio (17.1%) [12]. After eight months, his eosinophils numbers and eosinophil ratio still remained at higher levels (1.112 × 10^9/L and 10.8%, respectively). Furthermore, bone marrow puncture analysis showed granulocyte hyperplasia (Fig. 2) and increased eosinophils ratio (5.5%). In addition, blood lymphocyte subset analysis showed no alteration. Virus detection was negative. C-reactive protein and erythrocyte sedimentation rate were normal. Allergy testing was negative. Other clinical data, such as EKG, echocardiogram, chest X-ray, and ultrasonography of abdomen, were all normal.
A trio-based whole exome sequencing (WES) was performed on the affected individual and his parents. After removing low quality readings and adapter or contaminant sequences, 60.8 Mb cleaning readings per person were produced. Further variant analysis was based on Genome Analysis Toolkit (GATK) annotation, which was described in more details in previous studies [9, 13, 14]. Approximately 91.54% of target sequences were covered for more than 10 times. After relatively common variants (MAF ≥ 0.1%) were removed based upon several commonly used databases (i.e., dbSNP142, ExAC, 1000 Genomes and 2000 Chinese Han Exome Sequence database, ChES). Potential inherited and de novo mutations (DNM) related to brain developmental disorders [8, 9] and blood cell development were screened using a recently developed program mirTrios [15]. Deleterious variants were predicted utilizing several commonly used online programs, such as SIFT (http://sift.jcvi.org), Polyphen2 (http://genetics.bwh.harvard.edu/pph2/), MutationTaster (http://www.mutationtaster.org) and PROVEAN (http://provean.jcvi.org/index.php), etc. Sequence variants were interpreted based on ACM standards and guidelines [16].
Finally, bioinformatic analysis identified a de novo mutation of c.74delG in exon 1 of the KMT2A gene (GenBank acc. no., NM_001197104.1), which was predicted to result in a premature stop-gain mutation p.(Gly26Alafs*2) in the encoded protein (i.e., histone-lysine N-methyltransferase 2A, GenBank acc. no., NP_001184033.1). Since the mutation is located at the beginning of the N-terminal region of the gene product, it is assumed to be a null allele, thereby predicting to be a very strong pathogenic mutation.
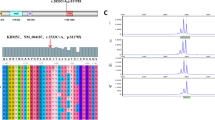
In HGMD database, there are 67 different mutations responsible for Wiedemann-Steiner syndrome (WDSTS), compared to additional 25 mutations for ID/DD (Table 1). Further analysis revealed more nonsense and loss-of-function (LoF) mutations in the affected individuals with WDSTS compared to the ID/DD group, suggesting LoF alleles are implicated in more severe clinical manifestations in WDSTS group. Since the present case mainly showed mild ID and short stature, but not distinctive facial appearance, we compared the genotype of the boy with additional 25 mutations detected in affected individuals with ID/DD in HGMD (Fig. 3). Approximately 68% of the mutations, including non-sense, indel, and splicing alleles, are predicted to be LoF alleles, while eight of them (32%) are hypomorphic missense alleles. However, none of these previously genotyped cases, to the best of our knowledge, were reported with blood eosinophilia.
Locations of ID/DD-associated KMT2A mutations. 22 mutations in affected individuals with ID/DD disorders are mapped. Colored boxes depict specific functional domains of the encoded protein using SMART program (http://smart.embl-heidelberg.de/). The mutation in red represents the current case. Four additional mutations with no predicted protein positions are not shown in the diagram
Discussion and conclusions
The KMT2A gene-encoded H3K4 methyltransferase contains multiple conserved functional domains, such as DNA-binding AT-hook motifs, a cysteine-rich CXXC domain, and a C-terminal SET domain (Fig. 3). KMT2A is abundantly expressed in brain, blood and bone marrow and interacts with multiple target genes for transcriptional regulation [17], thereby playing an important role in the development of central nervous system and hematopoietic stem cell differentiation [18, 19].
Different from the effect of germline mutations in KMT2A on brain development, somatic truncations or fusion mutations in KMT2A are frequently found to be associated with lymphoid leukemia and myeloid leukemia [20,21,22,23]. And yet, a germline missense mutation in KMT2A also is identified by whole-exome sequencing to segregate with four patients in a family with B-cell lymphoma [24], suggesting a role of KMT2A-encoded protein in hematopoietic stem cell development.
KMT2A (a.k.a., MLL) is abundantly expressed in lymphoid-derived T cells and myeloid granulocytes like the eosinophils (BioGPS and Human Protein Atlas). Eosinophils are a kind of white blood cell, which is originated from bone marrow hematopoietic stem cells. Blood eosinophilia (> 0.5*109/L) [25] results from overproduction of eosinophils from abnormal myeloid progenitor cells, thereby serving as an early sign of hematological malignancy [26]. Persistent eosinophilia may be caused by chromosomal rearrangements and gene mutations [27]. Somatic mutations in KMT2A in patients with leukemia or myeloid/lymphoid or mixed-lineage (OMIM: 159555) may accompany with hypereosinophilia. Interestingly, macrophages with a striated eosinophilic cytoplasm were frequently noted in Mll-AF4 fusion gene knock-in mice [28], an animal model of lymphoid and myeloid deregulation and hematologic malignancy. In addition, mutations in six additional genes are found to be associated with the phenotype “blood eosinophilia” (Additional file 1: Table S1). However, no deleterious mutations in any of these genes are found in the trio-whole-exome analysis of the present case.
In summary, we identified a previously undescribed de novo stop-gain mutation in KMT2A in a boy with ID/DD as well as persistent blood eosinophilia. Whether the eosinophilia is directly resulted from the KMT2A null mutation needs to be validated in further studies, preferably in additional cases with loss-of function mutations in the KMT2A gene.
Abbreviations
- ADHD:
-
Attention deficit hyperactivity disorder
- AML-M5:
-
Acute monoblastic leukemia
- AMML-M4:
-
Myelomonocytic leukemia
- ASD:
-
Autism spectrum disorder
- DD:
-
Developmental delay
- H3K4:
-
Histone H3 lysine 4
- HGMD:
-
Human gene mutation database
- ID:
-
Intellectual disability
- KMT2A:
-
Lysine methyltransferase 2A
- LOF:
-
Loss of Function
- MLL:
-
Mixed-lineage leukemia
- NDDs:
-
Neurodevelopmental disorders
- WDSTS:
-
Wiedemann-Steiner syndrome
References
Tarlungeanu DC, Novarino G. Genomics in neurodevelopmental disorders: an avenue to personalized medicine. Exp Mol Med. 2018;50(8):100.
Study DDD. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–8.
Boivin MJ, Kakooza AM, Warf BC, Davidson LL, Grigorenko EL. Reducing neurodevelopmental disorders and disability through research and interventions. Nature. 2015;527(7578):S155–60.
Baer S, Afenjar A, Smol T, Piton A, Gerard B, Alembik Y, Bienvenu T, Boursier G, Boute O, Colson C, et al. Wiedemann-Steiner syndrome as a major cause of syndromic intellectual disability: a study of 33 French cases. Clin Genet. 2018;94(1):141–52.
Min Ko J, Cho JS, Yoo Y, Seo J, Choi M, Chae JH, Lee HR, Cho TJ. Wiedemann-Steiner syndrome with 2 novel KMT2A mutations. J Child Neurol. 2017;32(2):237–42.
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, de Vries BB, Willemsen MH, Kleefstra T, Lohner K, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19(9):1194–6.
Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, Irving M, Saggar AK, Smithson S, Trembath RC, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91(2):358–64.
Cai T, Chen X, Li J, Xiang B, Yang L, Liu Y, Chen Q, He Z, Sun K, Liu PP. Identification of novel mutations in the HbF repressor gene BCL11A in patients with autism and intelligence disabilities. Am J Hematol. 2017;92(12):E653–6.
Guo S, Yang L, Liu H, Chen W, Li J, Yu P, Sun ZS, Chen X, Du J, Cai T. Identification of novel compound mutations in PLA2G6-associated neurodegeneration patient with characteristic MRI imaging. Mol Neurobiol. 2017;54(6):4636–43.
Bi C, Wu J, Jiang T, Liu Q, Cai W, Yu P, Cai T, Zhao M, Jiang YH, Sun ZS. Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum Mutat. 2012;33(12):1635–8.
Wang Y, Zeng C, Li J, Zhou Z, Ju X, Xia S, Li Y, Liu A, Teng H, Zhang K, et al. PAK2 Haploinsufficiency results in synaptic cytoskeleton impairment and autism-related behavior. Cell Rep. 2018;24(8):2029–41.
Brito-Babapulle F. The eosinophilias, including the idiopathic hypereosinophilic syndrome. Br J Haematol. 2003;121(2):203–23.
Li J, Cai T, Jiang Y, Chen H, He X, Chen C, Li X, Shao Q, Ran X, Li Z, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2016;21(2):290–7.
Ma D, Wang X, Guo J, Zhang J, Cai T. Identification of a novel mutation of RUNX2 in a family with supernumerary teeth and craniofacial dysplasia by whole-exome sequencing: a case report and literature review. Medicine (Baltimore). 2018;97(32):e11328.
Li J, Jiang Y, Wang T, Chen H, Xie Q, Shao Q, Ran X, Xia K, Sun ZS, Wu J. mirTrios: an integrated pipeline for detection of de novo and rare inherited mutations from trios-based next-generation sequencing. J Med Genet. 2015;52(4):275–81.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Lebrun N, Giurgea I, Goldenberg A, Dieux A, Afenjar A, Ghoumid J, Diebold B, Mietton L, Briand-Suleau A, Billuart P, et al. Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. Eur J Hum Genet. 2018;26(1):107–16.
Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–59.
Mishra BP, Zaffuto KM, Artinger EL, Org T, Mikkola HK, Cheng C, Djabali M, Ernst P. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep. 2014;7(4):1239–47.
Ayton P, Sneddon SF, Palmer DB, Rosewell IR, Owen MJ, Young B, Presley R, Subramanian V. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis. 2001;30(4):201–12.
Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10(10):721–8.
Urtishak KA, Robinson BW, Rappaport EF, Sarezky MD, Biegel JA, Nichols KE, Wilmoth DM, Wang LS, Stern JW, Felix CA. Unique familial MLL (KMT2A)-rearranged precursor B-cell infant acute lymphoblastic leukemia in non-twin siblings. Pediatr Blood Cancer. 2016;63(7):1175–80.
Peterson JF, Baughn LB, Pearce KE, Williamson CM, Benevides Demasi JC, Olson RM, Goble TA, Meyer RG, Greipp PT, Ketterling RP. KMT2A (MLL) rearrangements observed in pediatric/young adult T-lymphoblastic leukemia/lymphoma: a 10-year review from a single cytogenetic laboratory. Genes Chromosomes Cancer. 2018;57(11):541–6.
Saarinen S, Kaasinen E, Karjalainen-Lindsberg ML, Vesanen K, Aavikko M, Katainen R, Taskinen M, Kytola S, Leppa S, Hietala M, et al. Primary mediastinal large B-cell lymphoma segregating in a family: exome sequencing identifies MLL as a candidate predisposition gene. Blood. 2013;121(17):3428–30.
Gotlib J. World Health Organization-defined eosinophilic disorders: 2017 update on diagnosis, risk stratification, and management. Am J Hematol. 2017;92(11):1243–59.
Andersen CL, Siersma VD, Hasselbalch HC, Vestergaard H, Mesa R, Felding P, Olivarius ND, Bjerrum OW. Association of the blood eosinophil count with hematological malignancies and mortality. Am J Hematol. 2015;90(3):225–9.
Anastasi J. The myeloproliferative neoplasms including the eosinophilia-related myeloproliferations associated with tyrosine kinase mutations: changes and issues in classification and diagnosis criteria. Semin Diagn Pathol. 2011;28(4):304–13.
Chen W, Li Q, Hudson WA, Kumar A, Kirchhof N, Kersey JH. A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood. 2006;108(2):669–77.
Acknowledgements
The authors thank the family for supporting and participating in this research.
Funding
This study was supported by the research funding from the Chinese Ministry of Health Project (No: 201302002 to XC) and Natural Science Foundation of Hunan Province (No: 2017JJ3461 to HXZ). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The causal mutation data generated in this study are included in this report. The raw sequence data related to this study are available on request from the corresponding author [TC].
Author information
Authors and Affiliations
Contributions
Recruited and phenotyped the participants: BWX, HC, XC, HXZ, TC. Sequencing data analysis: TC. Wrote the manuscript: HXZ, TC. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed consent for participation of this study was obtained from the proband’s parents. This research was approved by the ethics committee of Wenzhou Medical University. As a disclaimer, T.C. represented his own perspective in the article, not that of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
Consent for publication
Written informed consent for publication of the case and any accompanying images were obtained from the patient’s parents.
Competing interests
The authors declare that they have no competing interest. As author Tao Cai is a member of the editorial board (Associate Editor) of this journal.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Human Blood Eosinophilia-associated Mutated Genes (DOCX 26 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, H., Xiang, B., Chen, H. et al. A novel deletion mutation in KMT2A identified in a child with ID/DD and blood eosinophilia. BMC Med Genet 20, 38 (2019). https://doi.org/10.1186/s12881-019-0776-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-019-0776-0