Abstract
Background
L-2-hydroxyglutaric aciduria (L2HGA) is a progressive neurometabolic disease of brain caused by mutations of in L-2-hydroxyglutarate dehydrogenase (L2HGDH) gene. Cardinal clinical features include cerebellar ataxia, epilepsy, neurodevelopmental delay, intellectual disability, and other clinical neurological deficits.
Case presentation
We describe an index case of the family presented with generalised tonic-clonic seizure, developmental delay, intellectual disability, and ataxia. Initially, the differential diagnosis was difficult to be established and a SNP genome wide scan identified the candidate region on chromosome 14q22.1. DNA sequencing showed a novel homozygous mutation in the candidate gene L2HGDH (NM_024884.2: c.178G > A; p.Gly60Arg). The mutation p.Gly60Arg lies in the highly conserved FAD/NAD(P)-binding domain of this mitochondrial enzyme, predicted to disturb enzymatic function.
Conclusions
The combination of homozygosity mapping and DNA sequencing identified a novel mutation in Pakistani family with variable clinical features. This is second report of a mutation in L2HGDH gene from Pakistan and the largest family with L2HGA reported to date.
Similar content being viewed by others
Background
L-2-hydroxyglutaric aciduria (L2HGA) is a rare autosomal recessive neurodegenerative disorder of metabolism [OMIM #236792] which is due to the accumulation of L-2-hydroxyglutaric acid (LGA) in urine, plasma and cerebrospinal fluid (CSF) [1, 2]. The phenotypic features of this organic aciduria are diverse, including developmental delay, cerebellar ataxia, epilepsy, severe intellectual disability, and macrocephaly [3,4,5]. The onset of disease has been reported to occur at an early age with severe epileptic fits or neurodegenerative symptoms, although it may also appear in adulthood with less severe presentations. There are reports of increased incidence of the development of brain tumours due to progression in L2HGA [6,7,8,9].
The diagnosis of L2HGA comprises of biochemical, radiological and genetic testing. The MRI abnormalities seen in the subcortical cerebral white matter, putamen, caudate nucleus, globus pallidus, and dentate nucleus are unique to L2HGA, and are used as baseline investigation [6, 10,11,12,13,14,15,16]. The disease-causing gene is L-2-hydroxyglutarate dehydrogenase (L2HGDH-NM_024884.2) which is located on chromosome 14q22.1 [MIM 609584] and comprises of 10 coding exons spanning 75 kb. It is expressed in various tissues with the highest expression found in the brain [15, 17]. The gene encodes a protein of 463 amino acids, specifying a mitochondrial targeting sequence (aa 1-50) and a domain for family of FAD-dependent enzymes [15]. L2HGDH is a mitochondrial enzyme which catalyses oxidation of L-2-hydroxyglutarate (L2HG) to α2-ketoglutarate (α2KG); a metabolic product bound to mitochondrial membrane [15, 17]. Several mutations L2HGDH have been reported worldwide in affected individuals belonging to various ethnic groups [6, 8, 13,14,15,16, 18, 19] (http://grenada.lumc.nl/LOVD2/vumc/status.php) [17].
The present case describes the clinical presentation and mutation analysis of L2HGDH in a large Pakistani consanguineous family comprising multiple individuals affected by a metabolic neurological disorder. Homozygosity and sequencing studies revealed a rare missense mutation (NM_024884.2:c.178G > A; p.Gly60Arg), in exon 2 of L2HGDH as the likely cause of disease in this family.
Case presentation
A 16 year old girl (IV-4) presented to hospital with history of seizures since the age of 8 months, intellectual delay, and ataxia. At the age of 13 years she was described by parents as ‘mentally dull’ and generalized tonic-clonic seizures recurred with increased frequency, mostly at night. Her elder sister (IV-6) and brother (IV-1) also showed symptoms of epilepsy and intellectual delay, as did three first cousins (IV-8, IV-9, and IV-10) although no in depth clinical evaluation was performed.
Clinical evaluation of index case
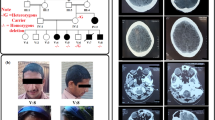
On physical and clinical examination IV-4 was alert, with an ataxic gait. Manual muscular testing did not note any weakness of limbs, but mild finger and nose ataxia was apparent along with a retarded capability of speech. Her deep tender reflexes were +++ and symmetrically preserved, while the plantar responses were bilaterally flexor. Her brain MRI showed abnormal diffuse T2 hyperintense signals in the subcortical white matter and bilateral symmetrical T2 hyperintense signals in bilateral basal ganglia (Fig. 1). Mild cortical cerebellar atrophy was also seen. Electroencephalogram (EEG) examination showed moderate diffuse encephalopathy/moderate diffuse brain dysfunction and observed epileptiform activity arising from the right hemisphere (Fig. 1). Urine testing for L2-hydroxyglutarate was not possible due to unavailability of this test in regional diagnostic laboratories, and remote setting of the family involved. Genetic investigation was recommended and she was advised for tablet Neurobion 1 BD, tablet Folic acid 5 mg OD, capsule Coenzyme Q-10, 50 mg BD, tablet Loprin 75 mg for symptomatic management.
Clinical features of individuals homozygous for L2HGDH c.178G > A. Patient VI:4 at 16 years of age, showed diffuse T2 hyperintense signals abnormality in the subcortical white matter (a), bilateral symmetrical T2 hyperintense signals in bilateral basal ganglia (b) and cerebellar atrophy (c) and epileptiform changes in EEG (d)
Molecular genetic analysis
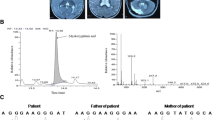
Genomic DNA was extracted from peripheral blood samples. Due to the availability of multiple affected family members and the consanguinity of the parents of the index case (Fig. 1a), autozygosity mapping through genome-wide SNP genotyping was conducted as previously described using Illumina Human CytoSNP-12 v2.1 microarrays [20]. A 24.5 Mb homozygous region on chromosome 14 (chr14:36,976,285-61,626,155 [hg38]) shared among all affected individuals delimited by recombinant SNP markers rs10483479 and rs2244057. The disease locus was predicted to contain 327 genes including L2HGDH [MIM: 236,792], previously implicated in causing overlapping neurological phenotypes. Subsequent di-deoxy sequence analysis of exon 2 (amplicon size 891 bp; primers: 5’-CCAATACATTGCTCTGTCGC-3′; 5’-AAAGTGAGCACAATCCTGGG-3′; Cycling conditions: Denaturation at 95 °C for 2 min; 2 cycles of 30 s at 95 °C, 30 s at 66 °C, 30 s at 72 °C; 2 cycles of 30 s at 95 °C, 30 s at 64 °C, 30 s at 72 °C; 35 cycles of 30 s at 95 °C, 30 s at 62 °C, 30 s at 72 °C and a final extension of 2 min at 72 °C) revealed a rare/novel missense variant with a single heterozygous individual of South Asian origin reported in 60,680 samples by the ExAC Consortium (http://exac.broadinstitute.org/variant/14-50769698-C-T; rs771556952; NM_024884.2:c.178G > A; p.Gly60Arg), which was found to co-segregate in the extended pedigree. The variant was not present in 150 chromosomes of Pakistani ancestry.
Computational analysis
Amino acid sequence alignment using the program ClustalW 2.1 showed high conservation of the Gly60 residue in related vertebrates (Fig. 2). The variant alters a stringently conserved amino acid residue and is predicted to be highly damaging by standard prediction programs using the Emsembl Variant Effect Predictor web interface (FATHMM score = − 6.21 (Damaging); MutationTaster score = 1 (Disease causing); PolyPhen2 score (HumDiv) = 1 (Probably damaging); PolyPhen2 score (HumVar) = 1 (Probably damaging); PROVEAN score = − 7.46 (Damaging); SIFT score = 0 (Deleterious)).
Family pedigree showing L2HGDH c.178G > A genotype data and images of affected individuals. a Simplified pedigree of the extended Pakistani family investigated, with pictorial representation of genotypes across ∼24 Mb of chromosome 14 encompassing the disease locus (dashed blue boxed region, red boxed region). All affected individuals were subsequently shown to be homozygous for the L2HGDH variant NM_024884.2:c.178G > A (indicated). Parental samples were heterozygous, and unaffected siblings were either WT or heterozygous carriers. b-d Electropherograms showing the DNA sequence at the position of L2HGDH c.178G > A in a homozygous affected (b), heterozygous father (c) and WT control (d) and amino acid alignment using ClustalW showing high conservation of the G60 residue across vertebrates (e)
On structural analysis, the Gly60 residue of L2HGDH resides in the helix region. This short non-polar glycine residue is replaced in the mutant protein by the larger, more positively charged and hydrophilic arginine residue. The Gly60Arg mutation is predicted to produce a minor local conformational change due to the difference in the observed contacts and surface area. The native glycine residue is only involved in intermolecular interactions with threonine at position 90 while the replaced basic arginine introduces an electrically charged, basic guanidium group which, unlike glycine, has more hydrogen bonding capabilities leading to the formation of an a inter molecular hydrogen bond with Thr90 as well as with Arg196 and Thr195. This leads to a slight local perturbation of the helix conformation for mutated protein (Fig. 3).
a Representation of predicted structure for human L2HGDH by means of Molecular Operating Environment (MOE v2013) software package, computationally predicted mutation is highlighted by circle (b) Representation of wild type protein interactions (c) Mutant type protein interactions (d) Secondary structure pattern of predicted wild and (e) Mutant type protein of human L2HGDH
Discussion and conclusions
In this study, we describe a large pedigree from Pakistan showing multiple neurological symptoms. Homozygosity mapping and Sanger sequencing revealed a novel missense mutation in L2HGDH gene. This is the second report of L2HGDH mutation in a Pakistani family; previously a nonsense mutation (p.Arg335Ter) was reported in a family showing a neuro-degenerative disorder of metabolism with two affected individuals [8]. Clinical and radiologic examinations of affected individuals identified presence of a slowly progressive neurodegenerative disease with cerebellar ataxia, seizures, delay in growth and abnormal subcortical white matter. MRI showed the persistent changes in the subcortical white matter characteristic in L2HGA leukoencephalopathy while the brain stem involvement in other leukoencephalopathy [11, 15]. Although additional phenotypic characteristics are described in the literature, including macrocephaly, pyramidal and extra-pyramidal features, these were not present in our patients, [15, 21].
L2HGDH encodes L-2 hydroxyglutare dehydrogenase which is the key contributor for this neurodegenerative disease. A large number of families and cases are reported with more than 100 pathogenic mutations in this gene. These mutations are mostly repeated in different ethnic populations. Interestingly, the disease is mostly reported in families from Mediterranean origin with numerous families reported from Turkey, Tunisia, Italy and Lebanon [6, 8, 15, 17]. Currently, there are 162 families with 283 cases which have been investigated for mutations in L2HGDH gene comprising a total of 112 mutations, 36 of which are found repeatedly found in different ethnic groups (http://grenada.lumc.nl/LOVD2/vumc/status.php).
The possible impact of our mutation on protein function was investigated using in silico bioinformatics tools. The mutation was predicted to affect the hydrogen bonding, and thus alter the stereochemistry of the protein (Fig. 3) [22].
To conclude, this case report provides the molecular diagnosis of a large consanguineous Pakistani family with six individuals. We identified a novel L2HGDH mutation predicted to cause in a loss of stability of L2HGDH protein.
Abbreviations
- aa:
-
Amino acid
- Arg:
-
(amino acid symbol) Arginine
- CSF:
-
Cerebrospinal fluid
- Glu:
-
(amino acid symbol) Glutamic Acid
- Gly:
-
(amino acid symbol) Glycine
- L2HG:
-
L-2-hydroxyglutarate
- L2HGA:
-
L-2-hydroxyglutaric aciduria
- L2HGDH :
-
(gene symbol) L-2-Hydroxyglutarate Dehydrogenase
- L2HGDH:
-
(protein symbol)L-2-Hydroxyglutarate Dehydrogenase
- Leu:
-
(amino acid symbol) Leucine
- LGA:
-
L-2-hydroxyglutaric acid
- Ser:
-
(amino acid symbol) Serine
- Thr:
-
(amino acid symbol) Threonine
- α2KG:
-
α2-ketoglutarate
References
Duran M, Kamerling JP, Bakker HD, van Gennip AH, Wadman SK. L-2-Hydroxyglutaric aciduria: an inborn error of metabolism? J Inherit Metab Dis. 1980;3:109–12.
Chen E, Nyhan WL, Jakobs C, Greco CM, Barkovich AJ, Cox VA, Packman S. L-2-Hydroxyglutaric aciduria: neuropathological correlations and first report of severe neurodegenerative disease and neonatal death. J Inherit Metab Dis. 1996;19:335–43.
Barth PG, Hoffmann GF, Jaeken J, Lehnert W, Hanefeld F, van Gennip AH, Duran M, Valk J, Schutgens RB, Trefz FK, et al. L-2-hydroxyglutaric acidemia: a novel inherited neurometabolic disease. Ann Neurol. 1992;32:66–71.
Barth PG, Wanders RJ, Scholte HR, Abeling N, Jakobs C, Schutgens RB, Vreken P. L-2-hydroxyglutaric aciduria and lactic acidosis. J Inherit Metab Dis. 1998;21:251–4.
Hanefeld F, Kruse B, Bruhn H, Frahm J. In vivo proton magnetic resonance spectroscopy of the brain in a patient with L-2-hydroxyglutaric acidemia. Pediatr Res. 1994;35:614–6.
Steenweg ME, Jakobs C, Errami A, van Dooren SJ, Adeva Bartolome MT, Aerssens P, Augoustides-Savvapoulou P, Baric I, Baumann M, Bonafe L, et al. An overview of L-2-hydroxyglutarate dehydrogenase gene (L2HGDH) variants: a genotype-phenotype study. Hum Mutat. 2010;31:380–90.
Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J Neuro-Oncol. 2009;91:233–6.
Haliloglu G, Jobard F, Oguz KK, Anlar B, Akalan N, Coskun T, Sass JO, Fischer J, Topcu M. L-2-hydroxyglutaric aciduria and brain tumors in children with mutations in the L2HGDH gene: neuroimaging findings. Neuropediatrics. 2008;39:119–22.
Moroni I, Bugiani M, D'Incerti L, Maccagnano C, Rimoldi M, Bissola L, Pollo B, Finocchiaro G, Uziel G. L-2-hydroxyglutaric aciduria and brain malignant tumors: a predisposing condition? Neurology. 2004;62:1882–4.
Seijo-Martinez M, Navarro C, Castro del Rio M, Vila O, Puig M, Ribes A, Butron M. L-2-hydroxyglutaric aciduria: clinical, neuroimaging, and neuropathological findings. Arch Neurol. 2005;62:666–70.
Topcu M, Aydin OF, Yalcinkaya C, Haliloglu G, Aysun S, Anlar B, Topaloglu H, Turanli G, Yalnizoglu D, Kesimer M, Coskun T. L-2-hydroxyglutaric aciduria: a report of 29 patients. Turk J Pediatr. 2005;47:1–7.
Steenweg ME, Salomons GS, Yapici Z, Uziel G, Scalais E, Zafeiriou DI, Ruiz-Falco ML, Mejaski-Bosnjak V, Augoustides-Savvopoulou P, Wajner M, et al. L-2-Hydroxyglutaric aciduria: pattern of MR imaging abnormalities in 56 patients. Radiology. 2009;251:856–65.
Vilarinho L, Cardoso ML, Gaspar P, Barbot C, Azevedo L, Diogo L, Santos M, Carrilho I, Fineza I, Kok F, et al. Novel L2HGDH mutations in 21 patients with L-2-hydroxyglutaric aciduria of Portuguese origin. Hum Mutat. 2005;26:395–6.
Goffette SM, Duprez TP, Nassogne MC, Vincent MF, Jakobs C, Sindic CJ. L-2-Hydroxyglutaric aciduria: clinical, genetic, and brain MRI characteristics in two adult sisters. Eur J Neurol. 2006;13:499–504.
Topcu M, Jobard F, Halliez S, Coskun T, Yalcinkayal C, Gerceker FO, Wanders RJ, Prud'homme JF, Lathrop M, Ozguc M, Fischer J. L-2-Hydroxyglutaric aciduria: identification of a mutant gene C14orf160, localized on chromosome 14q22.1. Hum Mol Genet. 2004;13:2803–11.
Vilarinho L, Tafulo S, Sibilio M, Kok F, Fontana F, Diogo L, Venancio M, Ferreira M, Nogueira C, Valongo C, et al. Identification of novel L2HGDH gene mutations and update of the pathological spectrum. J Hum Genet. 2010;55:55–8.
Jellouli NK, Hadj Salem I, Ellouz E, Kamoun Z, kamoun F, tlili A, Kaabachi N, Triki C, Fakhfakh F, Tunisian Network on Mental Retardation s. Founder effect confirmation of c.241A>G mutation in the L2HGDH gene and characterization of oxidative stress parameters in six Tunisian families with L-2-hydroxyglutaric aciduria. J Hum Genet. 2014;59:216–22.
Larnaout A, Amouri R, Kefi M, Hentati F. L-2-hydroxyglutaric aciduria: clinical and molecular study in three Tunisian families. Identification of a new mutation and inter-familial phenotype variability. J Inherit Metab Dis. 2008;31(Suppl 2):S375–9.
O'Connor G, King M, Salomons G, Jakobs C, Hardiman O. A novel mutation as a cause of L-2-hydroxyglutaric aciduria. J Neurol. 2009;256:672–3.
Puffenberger EG, Hu-Lince D, Parod JM, Craig DW, Dobrin SE, Conway AR, Donarum EA, Strauss KA, Dunckley T, Cardenas JF, et al. Mapping of sudden infant death with dysgenesis of the testes syndrome (SIDDT) by a SNP genome scan and identification of TSPYL loss of function. Proc Natl Acad Sci U S A. 2004;101:11689–94.
Faiyaz-Ul-Haque M, Al-Sayed MD, Faqeih E, Jamil M, Saeed A, Amoudi MS, Kaya N, Abalkhail H, Al-Abdullatif A, Rashed M, et al. Clinical, neuroimaging, and genetic features of L-2-hydroxyglutaric aciduria in Arab kindreds. Ann Saudi Med. 2014;34:107–14.
Ramachandran G, Kumar M, Selvi Rani D, Annanthapur V, Calambur N, Nallari P, Kaur P. An in silico analysis of troponin I mutations in hypertrophic cardiomyopathy of Indian origin. PLoS One. 2013;8:e70704.
Acknowledgements
We are grateful to the family members who participated in this study. We also acknowledge the Higher Education Commission (HEC) of Pakistan for providing IRSIP fellowship to MIU.
Funding
This research was funded by the Medical Research Council UK (MRC) – grant G1002279 (AHC).
Availability of data and materials
All data supporting our findings are included in the manuscript.
Author information
Authors and Affiliations
Contributions
MIU, AN, AA, WA and MJH carried out family recruitment, blood sampling and clinical analysis. AHC and BAC conceived and designed the experiments. BAC, GVH and MIU performed the experiments. AHC, BAC and MIU analyzed the data. AHC contributed reagents/materials/analysis tools. MIU, BAC, AHC and ELB wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Blood samples were collected from the proband and her family members. The study was approved by the Institutional Review Boards of Quaid-i-Azam University, Shifa International Hospital, Shifa Tameer e Millat University, Islamabad, Pakistan and University of Exeter, UK. Written informed consent was obtained from all family members who participated in the study.
Consent for publication
The participants included in this study signed a written informed consent to publish their data (the parents signed on the behalf of the children).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ullah, M.I., Nasir, A., Ahmad, A. et al. Identification of novel L2HGDH mutation in a large consanguineous Pakistani family- a case report. BMC Med Genet 19, 25 (2018). https://doi.org/10.1186/s12881-018-0532-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-018-0532-x