Abstract
Background
Asthma and allergic rhinitis are respiratory diseases with a significant global burden. Forkhead box O3 (FOXO3) is a gene involved in the etiology of a number of respiratory diseases. The objective of this study is to assess the association of rs13217795, an intronic FOXO3 single-nucleotide polymorphism, with asthma and allergic rhinitis.
Methods
In this case–case–control genetic association study, genotyping was conducted using the PCR–RFLP method. Genotype-based associations were investigated under the general, recessive, and dominant models of disease penetrance using binomial logistic regression; and, allele-based associations were tested using Pearson’s chi-squared test.
Results
The final study population consisted of 94 controls, 124 asthmatics, and 110 allergic rhinitis patients. The general and recessive models of disease penetrance were statistically significant for both case–control comparisons. Under the general model, the odds of the asthma phenotype were 1.46 (0.64 to 3.34) and 3.42 (1.37 to 8.57) times higher in heterozygotes and derived allele homozygotes, respectively, compared to ancestral allele homozygotes. The corresponding odds ratios for the allergic rhinitis phenotype were 1.05 (0.46 to 2.40) and 2.35 (0.96 to 5.73), respectively. The dominant model of disease penetrance was not statistically significant. The minor allele in all study groups was the ancestral allele, with a frequency of 0.49 in controls. There was no deviation from Hardy–Weinberg equilibrium in controls. Both case–control allele-based associations were statistically significant.
Conclusions
Herein we present the first report of the association between rs13217795 and allergic rhinitis, and the first independent verification of the association between rs13217795 and asthma. Marker selection in future genetic association studies of asthma and allergic rhinitis should include functional polymorphisms in linkage disequilibrium with rs13217795.
Similar content being viewed by others
Background
Asthma is a chronic respiratory disease characterized by airway inflammation and remodeling [1]. The number of asthmatics worldwide is estimated to be 235 million, and the global burden of this disease is projected to rise [2]. Asthma is a polygenic, multifactorial disorder; more than a hundred susceptibility genes have been identified [3]. The complex risk architecture of asthma is typified by involvement of both genetic and environmental factors [4]. Indeed, asthma is not a singular disease entity. The novel concept of asthma endotypes emerges from this understanding [5]. The clinical delineation of asthma subtypes will further enable benchside-to-bedside translation of genetic studies.
Allergic rhinitis, an IgE-mediated disease of the nasal mucosa, is the most common form of noninfectious rhinitis. The prevalence of allergic rhinitis in the global population is at least twice that of asthma [6]. Allergic rhinitis and asthma are interrelated inflammatory disorders. The former also involves mucosal remodeling, albeit to a lesser extent [7]. It has been postulated that asthma and allergic rhinitis are analogous inflammatory disorders of a continuous airway under the “integrated airway” hypothesis [8].
Forkhead box O3 (FOXO3) is a protein-coding gene on chromosome 6q21. The product, forkhead box protein O3 (FOXO3), is a member of the forkhead family of transcription factors. These transcription factors play a key role in immune homeostasis; as such, loss-of-function variants may be associated with chronic inflammatory processes [9]. FOXO3 variation has been implicated—through a myriad of mechanisms—in the etiology of idiopathic pulmonary fibrosis, non-small cell lung cancer, chronic obstructive pulmonary disease, and bronchiolitis [10,11,12,13,14,15].
A recent human genome assembly by the Genome Reference Consortium (GRCh38.p7) identified 6499 (5372 intron) single-nucleotide variants in FOXO3, 135 of which are common (minor allele frequency greater than or equal to 0.05). Within this subset, three polymorphisms (rs2764264, rs13217795, and rs2802292) were associated with healthy aging and longevity [16]. The findings were independently confirmed in numerous studies [17].
Genetic association studies are also useful in identifying causal polymorphisms associated with the risk of disease. Recently, a novel association between the FOXO3 single-nucleotide polymorphism rs13217795 and asthma susceptibility was described [18]. This finding has yet to be independently verified. To the best of our knowledge, no studies to date have investigated the association between rs13217795, a cytosine-to-thymine substitution, and allergic rhinitis.
The aim of the present study is to assess the association of rs13217795 with asthma and allergic rhinitis.
Methods
Participants
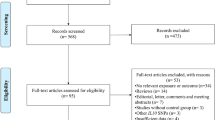
In this case–case–control, the initial study population consisted of 110 controls, 160 asthmatics, and 126 allergic rhinitis patients, for a total of 396 participants. The study population described herein is the aggregate of two previously described samples [19, 20]. Study participants were consecutively recruited from the Jordan University Hospital and the Jordan Hospital during the time period between August 2010 and January 2013. Asthma diagnoses (made by Professor Nathir M. Obeidat) were based on history, physical examination, and pulmonary function tests. Similarly, allergic rhinitis diagnoses (made by Professor Tareq M. Mahafza) were based on history and physical examination. In the latter cases, the skin prick test was used as an ancillary diagnostic tool. Patients with both conditions were excluded from the study. The control group comprised patients’ companions. Controls were unrelated to patients and had no history of respiratory disease, as determined by clinical interview and examination. The chief inclusion criterion for all groups was ethnicity. Participants were exclusively drawn from the same ethnic group (namely Arabs) in order to minimize population stratification. Ethnicity was determined by self-report. Institutional Review Board (Jordan University Hospital, Amman, Jordan) approval was obtained, and all participants or their guardians provided written informed consent.
DNA extraction and genotyping
A sample of 5 mL venous blood was collected in a Vacutainer blood collection tube containing EDTA. Genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega Corporation, USA). Purified DNA was stored at −20 °C, pending analysis. FOXO3 (rs13217795) genotyping was conducted using the PCR–RFLP method. The procedure was adapted from a previous study [18]. Investigators who performed the genotyping were blinded to the participants’ case–control status. A subset of participants was not genotyped due to low DNA yield; these individuals were excluded from all subsequent analyses.
Power analysis
A priori power analysis was conducted using the PGA software package [21]. The following assumptions were made for sample size calculations: 50% minor allele frequency, 10% disease prevalence, complete linkage disequilibrium, 5:4 case–control ratio, 5% Type I error rate, 20% Type II error rate, and one effective degree of freedom. Under the general model of disease penetrance, the anticipated odds ratios of heterozygotes and derived allele homozygotes in reference to ancestral allele homozygotes were 2.5 and 4, respectively. An odds ratio of 3 was anticipated under both the recessive and dominant models of disease penetrance. Minimum sample size estimates for the general, recessive, and dominant models of disease penetrance were 89, 57, and 97 cases, respectively. The assumptions were informed by a previous study, published guidelines, and data from the 1000 Genomes Project [18, 22].
Data analysis
Data were manually entered into the IBM SPSS Statistics Data Editor. All data analyses were conducted using this software package, unless otherwise indicated. Descriptive statistics were generated. The Mann–Whitney U Test was used to compare the median age of each case group with that of controls. Pearson’s chi-squared test was used to test for an association between gender and case–control status. The phi coefficient (φ) was used as a measure of the strength of association.
Binomial logistic regression was used to relate case–control status, the response variable, to its predictors (namely, genotype, age, and gender) under the general, recessive, and dominant models of disease penetrance [23]. The linearity assumption was assessed using the Box–Tidwell test; multicollinearity was assessed using variance inflation factors; and, studentized residuals were used to detect outliers. Genotype data of controls were tested for deviation from Hardy–Weinberg equilibrium using an exact test implemented in PLINK [24]. Allele-based associations with case–control status were tested using Pearson’s chi-squared test.
All underlying assumptions were met. A P value <0.05 was considered to indicate statistical significance for all but one test; deviation from Hardy–Weinberg equilibrium was assessed at a threshold of 0.001 [25]. Numerical data are presented according to the recommendations of T. J. Cole [26]. Odds ratios are presented as: odds ratio (95% confidence interval). Frequencies are presented as: absolute frequency (relative frequency).
Results
The final study population consisted of 94 controls, 124 asthmatics, and 110 allergic rhinitis patients (see Additional file 1). The demographic characteristics of the study population—namely age and gender—are shown in Table 1. The median age (range) of controls, asthmatics, and allergic rhinitis patients was 37.5 (17 to 73) years, 47.5 (17 to 84) years, and 30 (11 to 61) years, respectively. The median age of asthmatics and allergic rhinitis patients was statistically significant different from the median age of controls (P = 0.005 and P < 0.001, respectively). The proportion of female controls, asthmatics, and allergic rhinitis patients was 47%, 73%, and 56%, respectively. There was a statistically significant, weak association between gender and case–control status in the asthma arm of the study (P < 0.001, φ = 0.3). No such association was found between controls and allergic rhinitis patients (P = 0.2).
Genotype-based associations were investigated under the general, recessive, and dominant models of disease penetrance. The results of these analyses are presented in Table 2. The control group comprised 19 (20%) ancestral allele homozygotes (CC), 54 (57%) heterozygotes (CT), and 21 (22%) derived allele homozygotes (TT). The proportion of ancestral allele homozygotes and heterozygotes was lower in both case groups compared to the control group. By reciprocity, the proportion of derived allele homozygotes in both case groups was higher; 49 (40%) asthmatics and 47 (43%) allergic rhinitis patients were homozygous for the derived allele.
Indeed, the general model of disease penetrance was statistically significant for both arms of the study (asthma, P = 0.01; allergic rhinitis, P = 0.046). The odds of the asthma phenotype was 1.46 (0.64 to 3.34) and 3.42 (1.37 to 8.57) times higher in heterozygotes and derived allele homozygotes, respectively, compared to ancestral allele homozygotes. The corresponding odds ratios for the allergic rhinitis phenotype were 1.05 (0.46 to 2.40) and 2.35 (0.96 to 5.73), respectively.
In addition, the recessive model of disease penetrance was statistically significant (asthma, P = 0.005; allergic rhinitis, P = 0.01). The odds of asthma and allergic rhinitis phenotypes given derived allele homozygosity were 2.54 (1.33 to 4.85) and 2.26 (1.19 to 4.30) times higher, respectively, in reference to the pooled count of heterozygotes and ancestral allele homozygotes. The dominant model of disease penetrance was not statistically significant (asthma, P = 0.1; allergic rhinitis, P = 0.4).
There was no deviation from Hardy–Weinberg equilibrium in controls (P = 0.2). Thus, allele frequencies were computed, and allele-based associations were tested. The results are presented in Table 3. The relative frequency of the ancestral allele was 49%, 36%, and 35% in controls, asthmatics, and allergic rhinitis patients, respectively. Both case–control allele-based associations were statistically significant (asthma, P = 0.006; allergic rhinitis, P = 0.006).
Discussion
In this study, we explored the association between an intronic single-nucleotide variant in FOXO3 (rs13217795) and two disease phenotypes—asthma and allergic rhinitis. The two independent case groups were compared with ethnically matched controls in age- and gender-adjusted analyses. We found a significant association between rs13217795 and both diseases under multiple standard models of disease penetrance.
For both arms of the study, general model analytics suggest that more than one specific model—namely the recessive and multiplicative models—is highly plausible. Indeed, both the recessive and multiplicative models were statistically significant. The dominant model of disease penetrance did not provide significant evidence for association in either study arm.
The single-nucleotide variant under study (rs13217795) was polymorphic in our study population. The minor allele in all study groups was the ancestral allele, with a frequency of 0.49 in controls. Coincidentally, the minor (ancestral) allele frequency reported by the 1000 Genomes Project, based on 26 populations, was 0.49. Frequency data across these populations were highly variable. However, South Asian population data at the rs13217795 locus were a very close fit to the control data in this study.
The association between rs13217795 and asthma has only been explored in one previous study [18]. Incidentally, the study population in question comprised Indians of South Asia. The reported odds of the asthma phenotype were 5.54 (2.48 to 12.62) and 21.45 (8.78 to 53.84) higher in heterozygotes and derived allele homozygotes compared to ancestral allele homozygotes. The independent verification of this association in the present study, albeit to a modest extent, is of marked importance in a landscape of studies wherein the reproducibility of results is a matter of great concern [27].
The case–case–control design implemented in this study enabled a direct comparison of asthma and allergic rhinitis in terms of the risk conferred by rs13217795. Curiously, the genotypic and allelic distributions of the two diseases were strikingly similar. In turn, the significance pattern of all associations tested were identical for both case–control analyses. Odds ratios for specific alleles influencing complex diseases are expected to be in the range of 1.1 to 3.0 [28]. The odds ratios reported herein—for both case–control comparisons—fall within this range.
Aberrant expression of FOXO3 is associated with various pulmonary pathologies [10,11,12,13,14,15]. In addition, the gene product is a transcription factor involved in immune homeostasis [9]. Thus, the association of FOXO3 (rs13217795) with asthma and allergic rhinitis is highly plausible from a biologic standpoint. The robustness of this finding remains to be established.
Therapeutic interventions targeting the FOXO3 gene or its products are of great interest, particularly in relation to longevity. In a recent study of healthy rats, long-term consumption of epigallocatechin gallate was associated with increased lifespan through a FOXO3-driven mechanism [29]. Additionally, curcumin has been shown to increase FOXO3-mediated gene expression by twofold [30]. Following further biologic elucidation of the role of FOXO3 in asthma and allergic rhinitis, these phytochemicals may represent attractive therapeutic alternatives.
The main limitation of the present study is the likely causal irrelevance of rs13217795. While intronic single-nucleotide variants may very well influence gene expression, a significant association is more likely to be indirect. In that case, the marker locus may be in linkage disequilibrium with the causal variant. Indeed, rs13217795 has not been detected as an expression quantitative trait locus of FOXO3 by the Genotype-Tissue Expression (GTEx) project [31]. Therefore, future genetic association studies of asthma and allergic rhinitis should include marker loci with putative causal relevance. In addition, anticipated odds ratios may have been overestimated in the power analysis. Therefore, the study was underpowered to detect low odds ratios despite a larger operative sample size than computed estimates. However, anticipated odds ratios were reasonably overestimated based on the results of a previous study [18]. Ultimately, the small operative sample size does not appear to compromise the study’s conclusions. The null hypothesis was rejected for both case–control comparisons under the general, recessive, and multiplicative models of disease penetrance. The pattern of significance is in support of the apparent differential distribution of genotype frequencies between cases and controls.
Conclusions
We present the first independent verification of the association between rs13217795 and asthma. In addition, we report, for the first time, an association between rs13217795 and allergic rhinitis. By extension, this is a novel characterization of rs13217795 as a shared genetic marker for both asthma and allergic rhinitis. Marker selection in future genetic association studies of asthma and allergic rhinitis should include functional polymorphisms in linkage disequilibrium with rs13217795.
Abbreviations
- COPD:
-
Chronic obstructive pulmonary disease
- FOXO3 :
-
Forkhead box O3
- FOXO3:
-
Forkhead box protein O3
- PCR–RFLP:
-
Polymerase chain reaction–restriction fragment length polymorphism
References
Bousquet J, Chanez P, Lacoste JY, White R, Vic P, Godard P, Michel FB. Asthma: a disease remodeling the airways. Allergy. 1992;47:3–11.
Ferkol T, Schraufnagel D. The global burden of respiratory disease. Ann Am Thorac Soc. 2014;11:404–6.
Thomsen SF. Genetics of asthma: an introduction for the clinician. Eur Clin Respir J. 2015;2:24643.
Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse north American populations. Nat Genet. 2011;43:887–92.
Lötvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, Lemanske RF Jr, Wardlaw AJ, Wenzel SE, Greenberger PA. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–60.
Bousquet J, Khaltaev N, Cruz AA, Denburg J, Fokkens WJ, Togias A, Zuberbier T, Baena-Cagnani CE, Canonica GW, van Weel C, et al. Allergic rhinitis and its impact on asthma (ARIA) 2008. Allergy. 2008;63:8–160.
Bousquet J, Jacquot W, Vignola AM, Bachert C, Van Cauwenberge P. Allergic rhinitis: a disease remodeling the upper airways? J Allergy Clin Immunol. 2004;113:43–9.
Meltzer EO, Szwarcberg J, Pill MW. Allergic rhinitis, asthma, and Rhinosinusitis: diseases of the integrated airway. J Manag Care Spec Pharm. 2004;10:310–7.
Peng SL. Forkhead transcription factors in chronic inflammation. Int J Biochem Cell Biol. 2010;42:482–5.
Im J, Hergert P, Nho RS. Reduced FoxO3a expression causes low autophagy in idiopathic pulmonary fibrosis fibroblasts on collagen matrices. Am J Physiol Lung Cell Mol Physiol. 2015;309:L552–61.
Mikse OR, Blake DC Jr, Jones NR, Sun YW, Amin S, Gallagher CJ, Lazarus P, Weisz J, Herzog CR. FOXO3 encodes a carcinogen-activated transcription factor frequently deleted in early-stage lung Adenocarcinoma. Cancer Res. 2010;70:6205–15.
Hwang J, Rajendrasozhan S, Yao H, Chung S, Sundar IK, Huyck HL, Pryhuber GS, Kinnula VL, Rahman I. FOXO3 deficiency leads to increased susceptibility to cigarette smoke-induced inflammation, airspace enlargement, and chronic obstructive pulmonary disease. J Immunol. 2011;187:987–98.
Yao H, Chung S, Hwang JW, Rajendrasozhan S, Sundar IK, Dean DA, McBurney MW, Guarente L, Gu W, Rönty M, Kinnula VL, Rahman I. SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J Clin Invest. 2012;122:2032–45.
Ganesan S, Unger BL, Comstock AT, Angel KA, Mancuso P, Martinez FJ, Sajjan US. Aberrantly activated EGFR contributes to enhanced IL-8 expression in COPD airways epithelial cells via regulation of nuclear FoxO3A. Thorax. 2013;68:131–41.
Lunghi B, De Cunto G, Cavarra E, Fineschi S, Bartalesi B, Lungarella G, Lucattelli M. Smoking p66Shc knocked out mice develop respiratory Bronchiolitis with fibrosis but not emphysema. PLoS One. 2015;10:e0119797.
Donlon TA, Curb JD, He Q, Grove JS, Masaki KH, Rodriguez B, Elliott A, Willcox DC, Willcox BJ. FOXO3 gene variants and human aging: coding variants may not be key players. J Gerontol A Biol Sci Med Sci. 2012;67:1132–9.
Bao JM, Song XL, Hong YQ, Zhu HL, Li C, Zhang T, Chen W, Zhao SC, Chen Q. Association between FOXO3A gene polymorphisms and human longevity: a meta-analysis. Asian J Androl. 2014;16:446–52.
Barkund S, Shah T, Ambatkar N, Gadgil M, Joshi K. FOXO3a gene polymorphism associated with asthma in Indian population. Mol Biol Int. 2015;2015:638515.
Zihlif M, Zihlif N, Obeidat NM, Mahafza T, Froukh T, Ghanim MT, Al-Akhras FM, Naffa R. Association between ADAM33 polymorphisms and susceptibility with adult and childhood asthma among Jordanians. Genet Test Mol Biomarkers. 2014;18:767–74.
Zihlif M, Mahafza T, Obeidat NM, Froukh T, Shaban M, Al-Akhras FM, Zihlif N, Naffa R. Frequency of genetic polymorphisms of ADAM33 and their association with allergic rhinitis among Jordanians. Gene. 2013;531:462–6.
Menashe I, Rosenberg PS, Chen PE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36.
Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 2012;10:117–22.
Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6:121–33.
Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93.
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–73.
Cole TJ. Too many digits: the presentation of numerical data. Arch Dis Child. 2015;100:608–9.
Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61.
Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2:2492–501.
Niu Y, Na L, Feng R, Gong L, Zhao Y, Li Q, Li Y, Sun C. The phytochemical, EGCG, extends lifespan by reducing liver and kidney function damage and improving age-associated inflammation and oxidative stress in healthy rats. Aging Cell. 2013;12:1041–9.
Zingg J, Hasan ST, Cowan D, Ricciarelli R, Azzi A, Meydani M. Regulatory effects of curcumin on lipid accumulation in monocytes/macrophages. J Cell Biochem. 2012;113:833–40.
The GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–5.
Acknowledgments
Not applicable.
Funding
This work was supported by the Deanship of Academic Research, The University of Jordan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information file.
Author information
Authors and Affiliations
Contributions
MAZ, RGN, JZA, and HHS conceived and designed the study. NMO and TMM recruited study participants and conducted clinical investigations. RGN, JZA, HHS, and YMA genotyped participants. RGN and JZA curated the data. JZA conducted formal analysis and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Institutional Review Board approval was obtained from the Jordan University Hospital. All participants or their guardians provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Raw data. Demographic and genotype data for all participants. (XLS 56 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Amarin, J.Z., Naffa, R.G., Suradi, H.H. et al. An intronic single-nucleotide polymorphism (rs13217795) in FOXO3 is associated with asthma and allergic rhinitis: a case–case–control study. BMC Med Genet 18, 132 (2017). https://doi.org/10.1186/s12881-017-0494-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-017-0494-4