Abstract
Background
Data on the dynamics and persistence of humoral immunity against SARS-CoV-2 after primary vaccination with two-dose inactivated vaccine (CoronaVac) are limited. This study evaluated the sequential effects of prior infection, heterologous boosting with mRNA-1273 (Moderna), and the occurrence of Omicron vaccine-breakthrough infection (VBI) thereafter.
Methods
We evaluated anti-spike IgG (Abbott) and neutralising (cPASS/GenScript) antibody (nAb) titers up to one year after mRNA-1273 boost in two-dose-CoronaVac-primed Indonesian healthcare workers (August 2021-August 2022). We used linear mixed modeling to estimate the rate of change in antibody levels, and logistic regression to examine associations between antibody levels and VBI.
Results
Of 138 participants, 52 (37.7%) had a prior infection and 78 (56.5%) received an mRNA-1273 booster. After two-dose CoronaVac, antibody titers had significantly declined within 180 days, irrespective of prior infection. After mRNA-1273 booster, anti-spike IgG (1.47% decline/day) and Omicron B.1.1.529/BA.2 nAbs declined between day 28–90, and IgG titers plateaued between day 90–360. During the BA.1/BA.2 wave (February–March 2022), 34.6% (27/78) of individuals experienced a VBI (median 181 days after mRNA-1273), although none developed severe illness. VBI was associated with low pre-VBI anti-spike IgG and B.1.1.529/BA.2 nAbs, which were restored post-VBI.
Conclusions
mRNA-1273 booster after two-dose CoronaVac did not prevent BA.1/BA.2 VBI. Periodic vaccine boosters may be warranted against emerging SARS-CoV-2 variants.
Key points
An ancestral-strain mRNA-1273 (Moderna) vaccine boost after two-dose inactivated virus vaccine (CoronaVac) did not prevent Omicron BA.1/BA.2 breakthrough infections in Indonesian healthcare workers, and were associated with declined anti-spike and neutralizing antibodies.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has significantly impacted global economies, societies, and public health. By February 1st, 2024, Indonesia, the world’s fourth most populous country with 275 million people, reported 6.8 million cases and 161.000 deaths. Indonesian frontline healthcare workers (HCWs) were disproportionately impacted during the first 18 months of the pandemic, with an estimated mortality rate of approximately 1.7 deaths per 1000, which is five times higher than that of the general population [1].
Vaccination is a crucial global strategy to control the COVID-19 pandemic, with several vaccines proving to be safe and effective in preventing COVID-19-related hospitalization and death. Data primarily from studies on viral vector and mRNA vaccines indicate that humoral responses wane within 6 months after the second dose [2, 3] and there is a time-progressive increase in vaccine-breakthrough infection (VBI), particularly with the Omicron variant (B.1.1.529) and its subvariants [4]. However, a third vaccine dose significantly restores antibody levels [5].
CoronaVac (CV) (SinoVac Life Sciences Co. Ltd., Beijing, China), an inactivated whole-virus vaccine, has been the predominant vaccine used in Indonesia’s mass vaccination campaign. Despite reports of high effectiveness [6], data suggest that CV has lower immunogenicity compared to viral vector and mRNA vaccines [7], with a marked decline in neutralizing antibody titers within a few months [8,9,10].
To date, most of the global population has developed heterogenous immune histories from various exposures to infection with different viral variants, and diverse vaccination types and regimens, referred to as “hybrid immunity” [11, 12]. Both infection and vaccination can induce strong, but short-lived, immune protection against reinfection [13]. While many countries have implemented third or even successive vaccine doses, there is limited data on the durability and dynamics of neutralizing antibodies for primary regimens based on inactivated vaccines, including the effects of prior infection, heterologous vaccine boosters, and VBI.
In a longitudinal adult cohort in Jakarta, Indonesia, we described the dynamics and persistence of spike-specific IgG and neutralizing antibodies against SARS-CoV-2 variants in adults who had received two-dose CoronaVac primary vaccination, and examined the sequential effects of prior infection, a third heterologous vaccine dose (booster) of mRNA-1273 (Moderna), and the occurrence of Omicron VBI thereafter. For comparison, the cohort also included a sub-group of participants who had only received two-dose CoronaVac primary vaccination.
Methods
Design and population
The Indonesia Vaccine Immunity & Infection Evaluation (INVITE) study is a longitudinal observational cohort, as described elsewhere [12]. Briefly, we consecutively enrolled HCWs (aged ≥ 18 years) on the day they received their 100-μg mRNA-1273 third dose (between August 6th and October 4th, 2021), after having completed the two-dose CoronaVac primary regimen at least 6 months prior (denoted as CV-CV-mRNA vaccinees). We also enrolled individuals from the general population (aged ≥ 12 years) on the day they received the first dose of their two-dose CoronaVac primary regimen (between November 16th, 2021 and June 10th, 2022) (denoted as CV-CV vaccinees). The present analysis included all participants for whom at least two serial stored serum samples were available, including the pre-vaccine (day 0) and at least one post-vaccine dose (timepoints day 28, 90, 180 for all; and day 360 and at time of VBI for CV-CV-mRNA vaccinees) (Figure S1, Table S1).
The outcomes of interest were:
-
1)
Titers and the rate of change over time of anti-Spike Receptor Binding Domain IgG, and neutralizing antibody inhibition (nAbs), comparing CV-CV-mRNA and CV-CV vaccinees with or without history of prior infection (PI versus noPI); PI was defined as, for CV-CV vaccinees, presence of anti-SARS-CoV-2 nucleocapsid protein, or a history of rapid antigen test or PCR-confirmed SARS-CoV-2 before first vaccine dose, and, for CV-CV-mRNA vaccinees, a history of rapid antigen test or PCR-confirmed SARS-CoV-2 infection before third dose vaccine.
-
2)
Occurrence of VBI after third (mRNA-1273) vaccine dose, defined as a PCR-confirmed SARS-CoV-2 infection at least 14 days post-vaccination. For this analysis, we compared participants with VBI (n = 27) with randomly selected participants without a history of VBI (noVBI, n = 50), matched for sex, age group and calendar month.
This study is reported as per Strengthening the Reporting of Observational Studies in Epidemiology guidelines.
Procedures and assays
Study data were collected and managed using a REDCap electronic data capture tool. To detect SARS-CoV-2 infection, we adopted the following test strategies: 1) all participants were asked to undergo either RT-PCR or a rapid antigen test in case of exposure to an infected individual or COVID-19-like symptoms); 2) all participants who reported any COVID-19-like symptoms at a study visit were RT-PCR tested; 3) During the successive SARS-CoV-2 surges, HCW participants were routinely tested every month using either RT-PCR (January to April 2022) or rapid antigen testing (May to December 2022).
RT-PCR-positive specimens were submitted for SARS-CoV-2 whole genome sequencing at Genomik Solidaritas Laboratory (GSI) in Jakarta. Anti-nucleocapsid protein antibody seropositivity was measured using the SARS-CoV-2 NP IgG ELISA Kit (My Biosource, MBS398004) following the kit manual instructions; sera were incubated in a recombinant nucleocapsid protein pre-coated well followed by HRP-conjugated solution at room temperature for an hour. Absorbance at 450 nm was measured with a ratio above 1.1 considered positive. Titers of anti-S IgG were determined using the chemiluminescent microparticle immunoassay (CMIA) SARS-CoV-2 IgG II Quant assay (Abbott Laboratories, Abbott Park, IL, US) on the Architect i2000sr platform, in accordance with the manufacturers’ instructions, and expressed in WHO International Standard binding antibody units (BAU)/mL using the manufacturer’s conversion factor (1 BAU/mL = 0.142 × arbitrary units[AU]/mL) and seropositivity defined as ≥ 7.1 BAU/mL. nAbs were measured using SARS-CoV-2 Surrogate Virus Neutralization Test (cPASS, GenScript, USA) against SARS-CoV-2 Spike protein RBDHRP wild-type (L00847-A), Delta (Z03614), Omicron B.1.1.529 (Z03730) and Omicron BA.2 (Z03741) according to the manufacturers’ instruction; sera were diluted 1:20 with diluent assay and pre-incubated with SARS CoV-2 HRP-RBD in a 1:1 volume ratio at 37 °C for 30 min. The mixture was then added to a capture plate pre-coated with hACE2 protein. Absorbance at 450 nm was measured, which is inversely proportional to the titer of the anti-SARS-CoV-2 neutralizing antibodies. nAb detection was expressed as the signal inhibition (SI, %), with a cut-off of ≥ 30%.
Statistical analysis
Descriptive statistics included proportions for categorical variables and medians and interquartile ranges (IQRs) for continuous variables. Correlations of log10-transformed titers were expressed using the Spearman’s rank correlation coefficient tests (rs). We used Kruskal–Wallis H with Dunn’s post-hoc tests to perform comparisons of anti-S IgG and nAb titers before and after the third mRNA vaccine dose, and pairwise comparisons between groups. P-values were adjusted for multiple comparisons by Benjamini-Hochberg’s method.
To examine antibody waning, we fit a linear mixed model comprising a fixed effect linear decline from 28 days (peak) after vaccination onward and 28-day peak anti-S IgG titers as a random effect, modelled as a constant. Separate models were run for second and third vaccine doses, and for models with and without VBI, where we added a knot at day 90 and 180 to reflect the inflection point of the anti-S IgG titer trends.
To examine the association between most recent anti-S IgG and nAb levels prior VBI and the occurrence of VBI, we fit a complete-case analysis logistic regression model, adjusted for age and PI. VBI participants were matched with noVBI participants based on time since third vaccine dose and calendar month.
All analyses were done using Stata/IC 15.1 (StataCorp, College Station, TX, USA) and visualized using R version 4.2.2 (R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism10. A two-sided P < 0.05 was considered significant.
Results
Participant characteristics
Of 1117 cohort participants who received two-dose CV primary vaccination, we included a randomly selected subset of 138 (12.4%) individuals for whom at least a pre- and a post-vaccination sample was available, comprising 60 CV-CV and 78 CV-CV-mRNA vaccinees (Figure S1, Table 1). 52 (37.7%) had PI and 78 (56.5%) received a third (mRNA-1273) vaccine dose. Median age was 34 years (IQR26-49) and 78 (56.5%) were women. Table S2 summarizes the key characteristics of the included participants compared to the whole cohort, and Table S1 the number of included participants per timepoint. Figure S2 shows the cohort timeline from December 2020 through January 2023, in the context of the successive epidemic waves in Indonesia.
Dynamics of anti-spike IgG titers and neutralizing antibodies after two or three vaccine doses
Whereas pre-vaccine (Day 0) anti-S IgG titers showed strong correlations with all tested nAbs (i.e. Wild-type, Delta, Omicron B.1.1.529 and BA.2) among both CV-CV and CV-CV-mRNA vaccinees (each rs > 0.7), anti-S IgG titers on Day 28 after the second vaccine dose showed strong correlations with wild-type, Delta and BA.2 nAbs (each rs > 0.8) and moderate correlations with B.1.1.529 nAbs (rs = 0.541) in CV-CV vaccinees; and moderate correlation with B.1.1.529 nAbs (rs = 0.440) and weak correlation with BA.2 nAbs (rs = 0.303) in CV-CV-mRNA vaccinees (wild-type and Delta not estimable) (Figure S3).
After the second (CV) vaccine dose, the day-28 anti-S IgG peak titer was 1.6-fold lower than after the third (mRNA-1273) vaccine dose (median 2.3 [IQR2.1–2.9] vs 3.6 [3.4–3.7] log10 BAU/mL vs; p < 0.001). On day 28 after the second (CV) vaccine dose, wild-type nAbs were detected in all participants, Delta nAbs were detected in all but one individual, whereas B.1.1.529 nAbs were not detected in 23 individuals (43.9% [18/41] seropositivity) and BA.2 nAbs in 12 individuals (70.7% [29/41] seropositivity). Between day 28 and 90 after the second (CV) vaccine dose, B.1.1.529 (78.0% [32/41] seropositivity) and BA.2 (87.8% [36/41] seropositivity) nAbs remained reduced, and subsequently increased between day 90 and 180 (100% [9/9] seropositivity each) (Fig. 1, Table S3). This latter finding possibly reflects unrecorded virus exposure during the BA.1/BA.2 wave in February–March 2022 (Figure S2).
Antibody responses against SARS-CoV-2 variants in CV-CV and CV-CV-mRNA vaccinees
The figure shows data separately for the CV-CV vaccinees (blues dots) and the CV-CV-mRNA vaccinees (red dots). Serum anti-spike IgG titers (A) and neutralizing antibodies (nAbs) against SARS CoV-2 wild-type (B), Delta (C), Omicron B.1.1.529 (D) and BA.2 (E) were measured: i) Pre-vaccination (Day 0), i.e. before the first dose in CV-CV vaccinees (n=60) and before third dose in CV-CV-mRNA vaccinees (n=42) ii) Post-vaccination day 28 (CV-CV n=41 and CV-CV-mRNA n=40), day 90 (CV-CV n=41 and CV-CV-mRNA n=40), day 180 (CV-CV n=9 and CV-CV-mRNA n=40), and day 360 (CV-CV-mRNA n=43). Dashed line represents the seropositivity of IgG titer [≥7.1 BAU/mL = 0.85 log10 BAU/mL] and cut-off value for presence of neutralizing antibodies [signal inhibition >30%]. P-values were derived from Kruskal-Wallis H followed by Dunn’s post-hoc tests, adjusted by Benjamini-Hochberg method for multiple comparisons. *, p<0.05; **, p<0.01; ***, p<0.001
Abbreviations: BAU, binding antibody units; CV, CoronaVac; mRNA, mRNA-1273 (Moderna)
After the third (mRNA-1273) vaccine dose, we observed a significant day-28 peak in anti-S IgG and nAbs against all variants, compared to before the third dose (Fig. 1, Table S3). On day 28 wild-type and Delta nAbs and BA.2 nAbs (100% [40/40] seropositivity and median SI 99.0% [IQR95.0–100.0]) were detected in all participants (100% [40/40] seropositivity), whereas B.1.1.529 nAbs were not detected in two individuals (95.0% [38/40] seropositivity). Between day 28 and 90 after the third (mRNA-1273) dose, anti-S IgG titers had decreased significantly (from median 3.6 [IQR3.4–3.7] to 2.8 [2.4–3.0] log10 BAU/mL; p < 0.001), and then reached a plateau between day 90 and 360 days (p = 0.161). Whereas Wild-type and Delta nAbs persisted up to day 360 (100% [40/40] seropositivity), Omicron nAbs had declined significantly between day 28 and 90 for B.1.1.529 and BA.2 (Fig. 1, Table S3).
Effect of prior SARS-CoV-2 infection on antibody responses
Compared with individuals without PI at the time of the first CV dose, individuals with PI had 2.1-fold higher anti-S IgG titers (median 1.9 [IQR1.5–2.6] vs 0.9 [0.8–1.25] log10 BAU/mL; p < 0.001) and higher nAbs against all variants, i.e. Wild-type (80.0% [28/35] vs 28.0% [7/25] seropositivity; p < 0.001); Delta (74.3% [26/35] vs 16.0% [4/25] seropositivity; p < 0.001); B.1.1.529 (20.0% [7/35] vs 0.0% [0/25] seropositivity; p < 0.001); and BA.2 (77.1% [27/35] vs 28.0% [7/25] seropositivity; p < 0.001) (Figure S4, Table S4).
Similarly, compared with individuals without PI at the time of the third (mRNA-1273) vaccine dose, individuals with PI had 2.1-fold higher anti-S IgG titers (median 2.7 [IQR2.0–2.3] vs 1.3 [0.9–2.2] log10 BAU/mL; p = 0.002) and nAbs against wild-type (100% [16/16] seropositivity vs 61.5% [16/26] seropositivity; p = 0.002); Delta (93.8% [15/16] vs 57.7% [15/26] seropositivity; p = 0.002); and BA.2 (68.8% [11/16] vs 23.1% [6/26]; p = 0.006); but there was no statistical difference for B.1.1.529 nAbs (50.0% [8/16] vs 23.1% [7/26] seropositivity; p = 0.329) (Figure S4, Table S4).
By contrast, on day 360 of follow-up after the third (mRNA-1273) vaccine dose, there were no longer statistically significant differences between participants with or without PI, both in regard to anti-S IgG titers (p = 0.056) and nAbs against wild-type (p = 0.077), Delta (p = 0.333), B.1.1.529 (p = 0.488), and BA.2 (p = 0.377) (Figure S4, Table S4).
Omicron breakthrough infections after the third mRNA vaccine dose
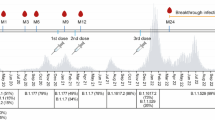
After the third (mRNA-1273) vaccine dose, 27 (34.6%) of 78 individuals experienced a VBI, mostly during the BA.1/BA.2 wave in February–March 2022 (Table S5), at a median of 181 (170–202) days after the mRNA-1273 boost (Fig. 2). None of the VBI cases were severe, four cases were asymptomatic, 22 cases were mild, and one case was moderate. Whole genome sequencing demonstrated BA.1 (n = 10), BA.2 (n = 12) and BE (n = 1), and from 4 samples the virus could not be sequenced.
Antibody responses against SARS-CoV-2 in CV-CV-mRNA vaccinees before, during and after Omicron breakthrough infection, compared to those who never experienced breakthrough infection
Figure shows anti-S IgG titers (A) and neutralizing antibodies (nAbs) against wild-type (B), Delta (C), Omicron B.1.1.529 (D) and Omicron BA.2 (E) in participants who experienced an Omicron BA.1/BA.2 vaccine breakthrough infection (VBI; red dots, n=27) and those who did not (noVBI;blue dots, n=56). VBI (red dots) were those who had a PCR-confirmed infection at least 14 days after the third dose (CV-CV-mRNA vaccinees). The median time of VBI occurrence was 181 days (IQR170-202) since the third mRNA vaccine dose. NoVBI participants (blue dots) were those who had never experienced a VBI during or prior the Omicron wave (February-March 2022). Pre-VBI antibody levels represent the most recent measurement prior to VBI occurrence (median 55 days [IQR33-88] before VBI). Post-VBI antibody level represents the most recent measurement after VBI occurrence (median 101 days [IQR39-175] after VBI). P-values were derived from Kruskal-Wallis H followed by Dunn’s post-hoc tests, adjusted by Benjamini-Hochberg method for multiple comparisons. *, p<0.05; **, p<0.01; ***, p<0.001
Abbreviations: BAU, binding antibody units; CV, CoronaVac; mRNA, mRNA-1273 (Moderna)
After adjusting for age and prior COVID-19 history, VBI occurrence was statistically significantly associated with lower pre-VBI antibody titers (median 55 days [IQR33-88] before VBI), specifically anti-S IgG (aOR5.42 for each log10 titer decrease [95%CI1.08–27.21], p = 0.040), B.1.1.529 nAbs (1.93 increased odds for each 25% SI decrease [1.18–3.14], p = 0.009) and BA.2 nAbs (2.13 increased odds for each 25% SI decrease [0.95–4.79], p = 0.068), but not associated with Wild-type or Delta nAbs (Table 2).
Subsequently, participants who had experienced a VBI developed significantly higher post-VBI nAbs levels against Omicron variants (median 101 days [IQR39-175] after VBI), compared to those who had never had a VBI, i.e. B.1.1.529 (median SI 90.0% [IQR79.5–97.0] vs 59.0% [38.0–92.5]; p = 0.013) and BA.2 (97.5% [91.5–99.3] vs 90.2% [74.7–96.9]; p = 0.021) (Fig. 2, Table S5).
Changes of anti-spike IgG titers after second and third vaccine dose
In a linear mixed model that adjusted for age and monthly new COVID-19 cases in Jakarta, PI was associated with higher anti-S IgG titers on day-28 (peak) after primary CV vaccination compared to not having a history of PI (least square mean [LSM] 2.49 [95%CI2.32, 2.68] vs 2.11 [1.85, 2.38] log10 BAU/mL). After the second (CV) vaccine dose, anti-S IgG titers increased between day 28 and 90 (0.74% increase per day [0.39% to 1.09%], p < 0.001) (due to possible unrecorded Omicron exposure), and declined thereafter between day 90 and 180 (0.44% decline per day [0.11% to 0.76%], p = 0.008). PI had no significant effect on anti-S IgG decline between day 90 and 180 after two-dose CV primary vaccination (0.64% decline per day [0.24% to 1.04%; p = 0.002 for PI participants vs 0.25% decline per day [-0.70% to 0.20%, p = 0.272) for noPI participants (Table S6).
The third (mRNA-1273) vaccine dose was associated with a higher anti-S IgG day-28 (peak) titer compared to primary CV vaccination (LSM3.64 log10 BAU/mL [95%CI3.51, 3.78] vs 2.33 log10 BAU/mL [2.16, 2.49]). (Fig. 3, Table S5). Anti-S IgG titers declined significantly between day 28 and 90 after the third (mRNA-1273) vaccine dose (1.47% decline per day [1.23% to 1.71%]; p < 0.001) and reached a plateau between day 90 and 360 (p > 0.05). VBI occurrence was associated with a subsequent anti-S IgG titer increase of 0.13% per day (-0.01% to 0.27%; p = 0.076), although this association was of borderline significance, compared to stable anti-S IgG titers in participants without VBI (0.03% increase per day during day 181–360 [-0.19% to 0.26%]; p = 0.766).
Dynamic changes in anti-spike IgG titers after second (CV) and third (mRNA-1273) vaccine dose
Figure shows dynamic changes in anti-spike IgG titers between: (A) CV-CV and CV-CV-mRNA vaccinees after the latest vaccine dose on day 28 (n=41 and n=40), day 90 (n=41 and n=40), day 180 (n=9 and n=40), and day 360 (n=0 and n=41); (B) CV-CV vaccinees with (PI) and without (noPI) prior SARS-CoV-2 infection after the latest vaccine dose on day 28 (n=27 and n=14), day 90 (n=24 and n=17), and day 180 (n=5 and n=4); (C) CV-CV-mRNA vaccinees with (PI) and without (noPI) prior SARS-CoV-2 infection after the latest vaccine dose on day 28 (n=0 and n=40), day 90 (n=0 and n=40), day 180 (n=2 and n=38), and day 360 (n=9 and n=34); (D) CV-CV-mRNA vaccinees with VBI (n=27) and without VBI (noVBI) (n=56). The slopes were modelled with piecewise linear mixed models with each individual’s peak antibody level as a random effect, introducing knots at day 28, 90, and 180 after last vaccination. The black dots in panels (A, C and D) indicate the time of VBI occurrence. The estimated antibody values for each time point and the slopes between timepoints can be found in Table S6
Abbreviations: PI, prior infection; VBI, Omicron vaccine breakthrough infection
Discussion
This real-world study in Indonesian healthcare workers who received a heterologous CoronaVac-mRNA-1273 prime-boost vaccine regimen investigated the magnitude and longevity of humoral immune responses, induced by sequential viral antigen exposure. These exposures comprised prior SARS-CoV-2 infection before vaccination, a heterologous ancestral-strain mRNA vaccine booster, and/or subsequent Omicron BA.1/BA.2 breakthrough infection.
There is evidence indicating that previous SARS-CoV-2 infection enhances the potency, breadth and durability of the humoral response to primary vaccination [14]. However, our study findings among individuals vaccinated with two doses of CoronaVac, and other studies among recipients of mRNA vaccines, suggest that this effect may be short-lived, with decreases in spike-specific IgG and nAbs observed regardless of prior infection [15]. Moreover, evidence suggests that vaccination elicits more robust SARS-CoV-2 antibody responses, in terms of specificity, breadth and maturation against viral variants, as well as cellular immune responses, compared to natural infection [15, 16].
In contrast to several previous studies indicating rapid decline in antibody responses within six months after two-dose CoronaVac priming [8,9,10], our study observed sustained presence of B.1.1.529 and BA.2 nAbs up to 180 days after the second CV dose. This persistence may be attributed to potential unrecorded virus exposure during the BA.1/BA.2 wave in February–March 2022 among our study cohort.
The heterologous mRNA-1273 vaccine booster elicited robust peak titers of anti-S IgG and nAbs against variants of concern, consistent with earlier findings on heterologous mRNA booster strategies [17]. However, between day 28 and 90 post-booster, we observed a rapid decline in B.1.1.529 nAbs, resulting in loss of seropositivity in seven out of 40 (17%) participants, and anti-S IgG titer waning at a rate of 1.47% per day. These findings align with a study among HCWs in Israel who received a homologous mRNA (BNT162b2, BioNTech/Pfizer) prime-boost regimen, also demonstrating that the third vaccine dose conferred greater durability than the second dose, with anti-S IgG waning at 1.32% per day, and that Omicron exhibited greater resistance to neutralization compared to wild-type and Delta variants [18].
During the Omicron BA.1/BA.2 epidemic wave in Jakarta in February and March 2022, one in three HCWs who received an mRNA-1273 booster experienced a breakthrough infection. Those who had breakthrough infections exhibited significantly lower titers of anti-Spike IgG, B.1.1.529 or BA.2 nAbs measured before the breakthrough compared to those who did not have a breakthrough. These findings in our population, who were primed with inactivated vaccines, corroborate and expand upon similar observations in populations receiving primary mRNA vaccine regimens in high-income settings [9, 19]. Our data reinforce the understanding that boosting with an ancestral-strain vaccine (such as mRNA-1273) after inactivated vaccine priming, does not confer adequate immune protection against emerging Omicron variants [20]. Nevertheless, none of the 27 participants who experienced breakthrough infections developed severe COVID-19, suggesting that a heterologous ancestral-strain vaccine booster dose after inactivated vaccine priming may still offer protection against severe disease from Omicron BA.1/BA.2. This finding is consistent with findings from observational studies in Singapore [21] Hong Kong [22] and Indonesia [23], which demonstrated that three doses of either homologous vaccination with inactivated or mRNA vaccines [21, 22], or heterologous vaccination with viral vector or mRNA booster after inactivated vaccine priming [23], provided protection against severe or fatal outcomes, despite breakthrough infections with B.1.1.529, BA.2 or BA.5 variants.
There are limited data concerning the longevity and breadth of the immune response elicited by CoronaVac and subsequent Omicron breakthrough infection. A previous prospective study in China among individuals who received three doses of CoronaVac found that spike-specific antibodies and cellular responses were present in the majority of vaccinated individuals twelve months after the third dose; furthermore, the study highlighted that infection with Omicron BA.5 significantly enhanced the magnitude, cross-reactivity, and persistence of serum neutralization, Fc-mediated phagocytosis, nasal spike-specific IgA responses, memory B cells, activated cTfh cells, memory CD4 + T cells, and memory CD8 + T cells, for both the ancestral strain and Omicron subvariants, compared to unvaccinated individuals [24].
Bivalent booster vaccines incorporating ancestral and Omicron strains have been deployed as a strategy to enhanced antibody magnitude and cross-reactivity against emerging variants [25, 26]. Data on the immune responses to bi- or multivalent vaccine boosters following priming with inactivated vaccines are warranted.
Several study limitations should be noted. First, despite regular screening, some breakthrough infections may have gone unnoticedor unreported, potentially leading to misclassification of participants [27]. Second, while accumulating evidence suggests that the anti-spike IgG and nAbs responses correlate with protection against infection, a definited correlate of long-term vaccine protection has yet to be established. Protection is likely influenced by additional factors such as cellular immunity, which is increasingly recognized for its role in preventing infection and severe disease [28, 29] Third, cross-reactivity against recently emerged Omicron subvariants, which exhibit increasing degrees of immune evasion [30], was not evaluated in this study. Lastly, the demographic characteristics of the study volunteers may not fully represent the broader Indonesian population, with a lack of representation of elderly individuals.
In conclusion, this longitudinal study offers valuable real-world insights into the longevity of anti-spike IgG and neutralising antibody responses to CoronaVac primary vaccination and subsequent ancestral-strain mRNA booster and/or Omicron breakthrough infection in Indonesia. Our findings suggest that, after two-dose inactivated vaccine priming, an mRNA-1273 booster dose did not offer immunity against Omicron BA.1/BA.2 subvariants, particularly among individuals with lower anti-Spike IgG and B.1.1.529/BA.2 nAbs. Current evidence indicates that ancestral-strain vaccine boosters, still widely used in many LMIC, may offer partial protection against severe disease. There is an urgent need to expand access to and coverage of vaccine boosters among vulnerable populations in Indonesia and other LMIC, to enhance protection against emerging, immune-evasiding SARS-CoV-2 variants.
Availability of data and materials
Data is provided within the manuscript or supplementary information files.
References
Ekawati LL, Arif A, Hidayana I, Nurhasim A, Munziri MZ, Lestari KD, et al. Mortality among healthcare workers in Indonesia during 18 months of COVID-19. PLOS Glob Public Heal. 2022;2:e0000893. https://doi.org/10.1371/journal.pgph.0000893.
Levin EG, Lustig Y, Cohen C, Fluss R, Indenbaum V, Amit S, et al. Waning immune humoral response to BNT162b2 Covid-19 Vaccine over 6 Months. N Engl J Med. 2021;385:e84. https://doi.org/10.1056/NEJMoa2114583.
Chemaitelly H, Tang P, Hasan MR, AlMukdad S, Yassine HM, Benslimane FM, et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N Engl J Med. 2021. https://doi.org/10.1056/NEJMoa2114114.
Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N Engl J Med. 2022;386:1532–46. https://doi.org/10.1056/NEJMoa2119451.
Costa Clemens SA, Weckx L, Clemens R, Almeida Mendes AV, Ramos Souza A, Silveira MBV, et al. Heterologous versus homologous COVID-19 booster vaccination in previous recipients of two doses of CoronaVac COVID-19 vaccine in Brazil (RHH-001): a phase 4, non-inferiority, single blind, randomised study. Lancet. 2022;399:521–9. https://doi.org/10.1016/S0140-6736(22)00094-0.
Jara A, Undurraga EA, González C, Paredes F, Fontecilla T, Jara G, et al. Effectiveness of an Inactivated SARS-CoV-2 vaccine in Chile. N Engl J Med. 2021;385:875–84. https://doi.org/10.1056/NEJMoa2107715.
Lim WW, Mak L, Leung GM, Cowling BJ, Peiris M. Comparative immunogenicity of mRNA and inactivated vaccines against COVID-19. Lancet Microbe. 2021;2:e423. https://doi.org/10.1016/S2666-5247(21)00177-4.
Sauré D, O’Ryan M, Torres JP, Zuniga M, Santelices E, Basso LJ. Dynamic IgG seropositivity after rollout of CoronaVac and BNT162b2 COVID-19 vaccines in Chile: a sentinel surveillance study. Lancet Infect Dis. 2022;22:56–63. https://doi.org/10.1016/S1473-3099(21)00479-5.
Sembera J, Baine C, Ankunda V, Katende JS, Oluka GK, Akoli CH, et al. Sustained spike-specific IgG antibodies following CoronaVac (Sinovac) vaccination in sub-Saharan Africa, but increased breakthrough infections in baseline spike-naive individuals. Front Immunol. 2023;14:1–12. https://doi.org/10.3389/fimmu.2023.1255676.
Sauré D, O’Ryan M, Torres JP, Zuñiga M, Soto-Rifo R, Valiente-Echeverría F, et al. COVID-19 lateral flow IgG seropositivity and serum neutralising antibody responses after primary and booster vaccinations in Chile: a cross-sectional study. Lancet Microbe. 2023;4:e149–58. https://doi.org/10.1016/S2666-5247(22)00290-7.
Reynolds CJ, Pade C, Gibbons JM, Otter AD, Lin KM, Sandoval DM, et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science (80- ). 2022;377. https://doi.org/10.1126/science.abq1841.
Sinto R, Utomo D, Suwarti, Nelwan EJ, Surendra H, Natasha C, et al. Antibody responses and reactogenicity of a heterologous, full-dose messenger RNA-1273 booster in heavily SARS-CoV-2–exposed Coronavac-vaccinated health-care workers in Indonesia: A real-world observational study. Am J Trop Med Hyg. 2023;108:115–23. https://doi.org/10.4269/ajtmh.22-0256.
Mantus G, Nyhoff LE, Edara V-V, Zarnitsyna VI, Ciric CR, Flowers MW, et al. Pre-existing SARS-CoV-2 immunity influences potency, breadth, and durability of the humoral response to SARS-CoV-2 vaccination. Cell Reports Med. 2022;3:100603. https://doi.org/10.1016/j.xcrm.2022.100603.
Wang J, Huang L, Guo N, Yao YP, Zhang C, Xu R, et al. Dynamics of SARS-CoV-2 antibody responses up to 9 months post-vaccination in individuals with previous SARS-CoV-2 infection receiving inactivated vaccines. Viruses. 2023;15:1–16. https://doi.org/10.3390/v15040917.
Richardson JR, Götz R, Mayr V, Lohse MJ, Holthoff H-P, Ungerer M. SARS-CoV2 wild type and mutant specific humoral and T cell immunity is superior after vaccination than after natural infection. PLoS ONE. 2022;17:e0266701. https://doi.org/10.1371/journal.pone.0266701.
Röltgen K, Nielsen SCA, Silva O, Younes SF, Zaslavsky M, Costales C, et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025-1040.e14. https://doi.org/10.1016/j.cell.2022.01.018.
Kaku CI, Champney ER, Normark J, Garcia M, Johnson CE, Ahlm C, et al. Broad anti–SARS-CoV-2 antibody immunity induced by heterologous ChAdOx1/mRNA-1273 vaccination. Science (80- ). 2022;375:1041–7. https://doi.org/10.1126/science.abn2688.
Gilboa M, Regev-Yochay G, Mandelboim M, Indenbaum V, Asraf K, Fluss R, et al. Durability of immune response after COVID-19 booster vaccination and association with COVID-19 omicron infection. JAMA Netw open. 2022;5:e2231778. https://doi.org/10.1001/jamanetworkopen.2022.31778.
Gilboa M, Gonen T, Barda N, Cohn S, Indenbaum V, Weiss-Ottolenghi Y, et al. Factors associated with protection from SARS-CoV-2 omicron variant infection and disease among vaccinated health care workers in Israel. JAMA Netw open. 2023;6:e2314757. https://doi.org/10.1001/jamanetworkopen.2023.14757.
Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med. 2022. https://doi.org/10.1038/s41591-022-01705-6.
Ng OT, Marimuthu K, Lim N, Lim ZQ, Thevasagayam NM, Koh V, et al. Analysis of COVID-19 Incidence and Severity among Adults Vaccinated with 2-Dose mRNA COVID-19 or Inactivated SARS-CoV-2 Vaccines with and Without Boosters in Singapore. JAMA Netw Open. 2022;5:E2228900. https://doi.org/10.1001/jamanetworkopen.2022.28900.
McMenamin ME, Nealon J, Lin Y, Wong JY, Cheung JK, Lau EHY, et al. Vaccine effectiveness of one, two, and three doses of BNT162b2 and CoronaVac against COVID-19 in Hong Kong: a population-based observational study. Lancet Infect Dis. 2022;22:1435–43. https://doi.org/10.1016/S1473-3099(22)00345-0.
Karuniawati A, Pasaribu AP, Lazarus G, Irawany V, Nusantara DU, Sinto R, et al. Characteristics and clinical outcomes of patients with pre-delta, delta and omicron SARS-CoV-2 infection in Indonesia (2020–2023): a multicentre prospective cohort study. Lancet Reg Heal - Southeast Asia. 2024;22:100348. https://doi.org/10.1016/j.lansea.2023.100348.
Chen Y, Zhao T, Chen L, Jiang G, Geng Y, Li W, et al. SARS-CoV-2 Omicron infection augments the magnitude and durability of systemic and mucosal immunity in triple-dose CoronaVac recipients. MBio. 2024:e0240723. https://doi.org/10.1128/mbio.02407-23b.
Branche AR, Rouphael NG, Diemert DJ, Falsey AR, Losada C, Baden LR, et al. Comparison of bivalent and monovalent SARS-CoV-2 variant vaccines: the phase 2 randomized open-label COVAIL trial. Nat Med. 2023;29:2334–46. https://doi.org/10.1038/s41591-023-02503-4.
Chalkias S, Harper C, Vrbicky K, Walsh SR, Essink B, Brosz A, et al. Three-month antibody persistence of a bivalent Omicron-containing booster vaccine against COVID-19. Nat Commun. 2023;14:1–7. https://doi.org/10.1038/s41467-023-38892-w.
Ho-Yan Fong C, Zhang X, Chen LL, Wing-Shan Poon R, Pui-Chun Chan B, Zhao Y, et al. Effect of vaccine booster, vaccine type, and hybrid immunity on humoral and cellular immunity against SARS-CoV-2 ancestral strain and Omicron variant sublineages BA.2 and BA.5 among older adults with comorbidities: a cross sectional study. eBioMedicine. 2023;88:104446. https://doi.org/10.1016/j.ebiom.2023.104446.
Brasu N, Elia I, Russo V, Montacchiesi G, Stabile SA, De Intinis C, et al. Memory CD8+ T cell diversity and B cell responses correlate with protection against SARS-CoV-2 following mRNA vaccination. Nat Immunol. 2022;23:1445–56. https://doi.org/10.1038/s41590-022-01313-z.
Méndez C, Peñaloza HF, Schultz BM, Piña-Iturbe A, Ríos M, Moreno-Tapia D, et al. Humoral and cellular response induced by a second booster of an inactivated SARS-CoV-2 vaccine in adults. eBioMedicine. 2023;91:1–14. https://doi.org/10.1016/j.ebiom.2023.104563.
Lasrado N, Collier A ris Y, Hachmann NP, Miller J, Rowe M, Schonberg ED, et al. Neutralization escape by SARS-CoV-2 Omicron subvariant BA.2.86. Vaccine. 2023;41:6904–9. https://doi.org/10.1016/j.vaccine.2023.10.051.
Acknowledgements
The authors thank the study participants, the clinical staff at the collaborating clinical sites, Aang Khumaidi, Rahmi Dwi Winarsih and Afrida Yanthi (Cakung primary healthcare centre), Syara Hilwiyah and the late Irma Rodearni (Ciracas primary healthcare centre); the study support staff, Nicolas Tarino, Ichsan Kalbuadji, Ana Yuliana, Yanie Tayipto, and Mutia Rahardjani (OUCRU Indonesia); and Jem Chalk (Oxford) for creating the electronic data capture. We acknowledge Genomik Solidaritas Laboratory (GSI) and Prodia Clinical Laboratory, both in Jakarta, for support in laboratory testing.
Members of the Southeast Asia initiative to combat SARS-CoV-2 variants (SEACOVARIANTS): Nguyen To Anh, Nguyen Thi Thu Hong, Truong Hoang Chau Truc, Nguyen Thi Han Ny, Do Duong Kim Han, Le Kim Thanh, Lam Anh Nguyet, Cao Thu Thuy, Le Nguyen Truc Nhu, Tran Tan Thanh, Lam Minh Yen, Vu Thi Ty Hang, Pham Tieu Kieu, Vo Tan Hoang, Nguyen Thi Thao, Mary Chambers, Vu Duy Thanh, Tran Chieu Hoang, C. Louise Thwaites, Guy Thwaites, Le Van Tan, H. Rogier van Doorn, Trinh Son Tung, Juthathip Mongkolsapaya, Gavin Screaton, Aiete Dijokaite-Guraliuc, Raksha Das, Chang Liu, Piyada Supasa, Muneeswaran Selvaraj, Susanna J Dunachie, Paul Klenerman, E. Yvonne Jones, David I Stuart, Barbara Kronsteiner-Dobramysl, Martha Zewdie, Priyanka Abraham, Jennifer Hill, Yanie Tayipto, Isana Paramita, Wang Lin-Fa, Tan Chee Wah, Yap Wee Chee, Lim Beng Lee, Raph L Hamers, Anuraj H Shankar, Suwarti, Eva Simarmata, Ragil Dien, Wanwisa Dejnirattisai, Warangkana Chantima, Narisara Chantratita, Prapassorn Poolchanuan, Vichapon Tiacharoen, Adul Dulsuk, Sophon Iamsirithaworn, Nick Day, Phaik Yeong Cheah, Tassawan Poomchaichote, Kanpong Boonthaworn, Nghiem My Ngoc, Alba Grifoni, Alessandro Sette.
Funding
This work was funded by the Wellcome Trust, UK (106680/Z/14/Z, 222574/Z/21/Z and 2/226120/Z/22/Z). SD is funded by an NIHR Global Research Professorship (NIHR300791). The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to the study data. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Consortia
Contributions
EJN and RLH are the INVITE principal investigators. LVT is the SEACOVARIANTS principal investigator. TLV, AHS and RLH obtained the funding. S, GL, EJN, AHS and RLH conceptualized the study. RS, F, TD, DO, J, S, JY, HP, AY, NDS, SS, and DS established the cohort, and collected the clinical samples and data. S, SZ and DS performed and supervised the antibody assays, with advice from SD. AP supervised the SARS-CoV-2 whole genome sequencing. NN, NT and WH managed the clinical database and contributed to data verification. S, SZ, and GL performed the statistical analysis and data visualizations, with input from HS, AHS, EJN and RLH. S, GL and RLH wrote the first draft of the manuscript. S, GL and RLH had full access to all of the study data and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors provided valuable input to interpretation of the data and critically reviewed the paper and figures for important intellectual content. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The INVITE study was approved by the research ethics committee of the Faculty of Medicine Universitas Indonesia (841/UN2.F1.ETIK/PPM.00.02/2021) and the Oxford Tropical Research Ethics Committee (22–21). All participants provided written informed consent.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Suwarti, S., Lazarus, G., Zanjabila, S. et al. Anti-SARS-CoV-2 antibody dynamics after primary vaccination with two-dose inactivated whole-virus vaccine, heterologous mRNA-1273 vaccine booster, and Omicron breakthrough infection in Indonesian health care workers. BMC Infect Dis 24, 768 (2024). https://doi.org/10.1186/s12879-024-09644-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-024-09644-y